Abstract
Inactivation of the retinoblastoma (Rb) tumor suppressor by Simian virus 40 (SV40) large T antigen is one of the central features of tumorigenesis induced by SV40. Both the N-terminal J domain and the LxCxE motif of large T antigen are required for inactivation of Rb. The crystal structure of the N-terminal region (residues 7–117) of SV40 large T antigen bound to the pocket domain of Rb reveals that large T antigen contains a four-helix bundle, and residues from helices α2 and α4 and from a loop containing the LxCxE motif participate in the interactions with Rb. The two central helices and a connecting loop in large T antigen have structural similarities with the J domains of the molecular chaperones DnaJ and HDJ-1, suggesting that large T antigen may use a chaperone mechanism for its biological function. However, there are significant differences between large T antigen and the molecular chaperones in other regions and these differences are likely to provide the specificity needed for large T antigen to inactivate Rb.
Keywords: chaperone mechanism/Rb tumor suppressor/SV40 large T antigen/viral oncogene
Introduction
Progression through the G1/S phase is a tightly regulated process in the eukaryotic cell cycle that involves a complex network of protein–protein interactions (Pardee, 1989; Sanchez and Dynlacht, 1996). One of the central features of this network is an interaction between the transcription factor E2F and the retinoblastoma (Rb) tumor suppressor (Chellappan et al., 1991; Weinberg, 1995). E2F family members are required for the expression of genes involved in DNA synthesis and transcriptional regulation, and overexpression of E2F can induce quiescent cells to enter S phase (La Thangue, 1994; Dyson, 1998). The underphosphorylated, active form of the Rb tumor suppressor binds to E2F and represses its transcription activity, leading to cell cycle arrest (Mihara et al., 1989). The E2F–Rb complex also binds directly to the promotor of some cell cycle genes and actively represses transcription in part through the action of histone deacetylase (Magnaghi-Jaulin et al., 1998).
In early to mid-G1 phase, Rb is hyperphosphorylated by the CDK–cyclin complexes. This event releases E2F, which is then able to activate cell growth and proliferation (DeCaprio et al., 1992; Mittnacht, 1998). The important role that Rb plays in the control of cell cycle progression is also indicated by the observations that (i) genetic alterations or deletions in Rb or in proteins that regulate the Rb pathway are frequently observed in diverse types of human cancer (Harbour et al., 1988; Lee et al., 1988; Toguchida et al., 1988); and (ii) re-introduction of the gene for Rb into cells lacking Rb reduces or delays the tumor formation (Huang, 1988; Bookstein et al., 1990).
E2F can also be liberated from the Rb tumor suppressor by a number of viral oncoproteins, including the large T antigens of SV40 and of polyoma viruses (Chellappan et al., 1992; Nevins, 1992). SV40 and polyoma viruses both require the cellular DNA replication machinery for replication of their genomes (Brodsky and Pipas, 1998). The large T antigen encoded by SV40 or polyoma viruses inactivates several key proteins involved in cell cycle regulation (DeCaprio, 1999). In this way the viruses keep their hosts in G1/S phase and are able to make use of the host replication system. Some of this control of host replication is through the interaction of the C-terminal half (residues 351–626) of SV40 large T antigen with the p53 tumor suppressor (Kierstead and Tevethia, 1993). The N-terminal region of SV40 large T antigen (residues 1–125) can also induce tumors in transgenic mice and transform established cell lines in culture (Zhu et al., 1992). Transformation by the N-terminal region of a large T antigen is a result, at least in part, of the release of E2F caused by the inactivation of Rb. Two regions in the N-terminal region of large T antigen—the LxCxE motif and the J domain—must act together to inactivate Rb (Srinivasan et al., 1997). The LxCxE motif, which is also present in adenovirus E1A and human papilloma virus (HPV) E7, is the minimal sequence required for Rb binding. These proteins with mutations within the LxCxE motif fail to interact with Rb (Kaelin et al., 1990). Residues 1–82 in large T antigen, which are important for viral replication, transcriptional regulation and virion assembly, share homology with the J domain of molecular chaperone DnaJ from Escherichia coli (Campbell et al., 1997). Mutations in this region alone inactivate the large T antigen, implicating the significance of the J domain in transformation (Stubdal et al., 1996, 1997; Zalvide et al., 1998). Several lines of evidence indicate that the N-terminal region of T antigen could function as a molecular chaperone. For example, a chimeric protein in which the J domain of E.coli DnaJ has been replaced by the J domain of large T antigen can restore the ability of E.coli lacking endogenous DnaJ to form bacteriophage λ plaque (Kelley and Georgopolous, 1997). Furthermore, the J domain of large T antigen binds to hsc70, a member of the hsp70 family, and stimulates its ATP hydrolysis activity (Sawai and Butel, 1989). Thus, it seems possible that large T antigen could inactivate Rb by behaving like a molecular chaperone.
The Rb protein consists of three parts: an N-terminal region, a central pocket region and a C-terminal region. The pocket domain (residues 380–772) is important for Rb’s role as a tumor suppressor (Chow and Dean, 1996). The pocket region is essential for growth suppression and is the minimal region where E2F and/or several viral oncoproteins bind (Hu et al., 1990; Huang et al., 1990; Helin et al., 1992; Kaelin et al., 1992). The pocket domain is highly conserved in other members of the Rb family such as p107 and p130, and many of the mutations in Rb that occur in human cancer are within this region.
Recently, the crystal structure of the Rb pocket region in a complex with an LxCxE peptide from HPV E7 was determined (Lee et al., 1998). However, for SV40 large T antigen, the LxCxE motif alone is not sufficient to inactivate the function of Rb. Thus, structural studies on the complex formed between the Rb pocket domain and the full N-terminal region of large T antigen containing both the J domain and the LxCxE motif should help us to understand how E2F is liberated from Rb by large T antigen. Here, we present the crystal structure of the complex formed by the Rb pocket and the N-terminal region (residues 7–117) of SV40 large T antigen (Table I), and describe the extensive interactions between the tumor suppressor and the oncoprotein, which provide structural insights into how large T antigen inactivates Rb.
Table I. Statistics from crystallographic analysis.
| Data collection | |
| data set | native |
| resolution (Å) | 20.0–3.2 |
| total observations | 50 139 |
| unique observations | 12 666 |
| data coverage (%) | 99.5 |
| Rsym | 0.084 |
| mean I/σ(I) | 15.9 |
| highest resolution bin | 3.29–3.20 |
| completeness | 97.2 |
| Rsym | 0.369 |
| mean I/σ(I) | 2.8 |
| Refinement | |
| resolution range (Å) | 20–3.2 |
| number of reflections (I > 0) | 12 171 |
| Rworking/Rfree (%) | 24.8/31.4 |
| number of atoms | 3627 |
| number of water molecules | – |
| r.m.s.d. bond length (Å) | 0.009 |
| r.m.s.d. bond angles (°) | 1.44 |
| mean protein main-chain temperature factor (Å2) | 41.9 |
Rsym= ΣhΣi |Ih,i – Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. R = Σ|Fobs – Fcalc|/ΣFobs, where Fobs = Fp, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). R.m.s.ds in bond lengths and angles are the deviations from ideal value.
Results
Overall structure of the complex
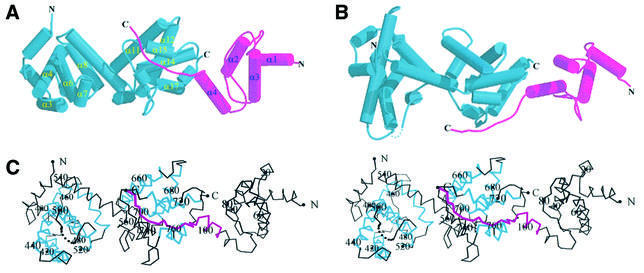
The pocket domain of Rb consists of an A box (residues 379–577) and a B box (residues 645–772), both of which are mostly formed with α-helices. A flexible linker that connects the A and B boxes in the native protein has been deleted in our work to aid crystallization of the complex. Five of the helices in each box (α3, α4, α6, α7 and α8 in the A box, and α11, α12, α14, α15 and α17 in the B box) form the cyclin box fold (Kim and Cho, 1997; Lee et al., 1998), which is also present in transcription factor TFIIB (Nikolov et al., 1995) and in members of the cyclin family (Brown et al., 1995; Jeffrey et al., 1995; Noble et al., 1997; Figure 1). Additional features of the Rb pocket domain include five more helices in the A box, and three helices and a short hairpin in the B box. The N-terminal region of large T antigen has a four-helix domain (residues 7–102) and an extended loop (residues 103–117). All four helices are involved in the formation of a compact core. In addition, residues from two of the helices and an extended loop participate in intermolecular interactions with the Rb pocket domain.
Fig. 1. (A) Schematic drawing of the Rb pocket–SV40 large T antigen complex. Large T antigen is shown in magenta, Rb pocket is shown in blue. The cyclin boxes in Rb pocket and the helices in T antigen are labeled. (B) View of the complex looking down the vertical axis of (A). Both parts were drawn using the programs MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merrit and Murphy, 1994). (C) Stereo view of the Cα trace of the Rb pocket–large T antigen complex in the same orientation as in (A). Every 20th residue is numbered and its Cα atom highlighted as a circle. The regions in large T antigen that contact with Rb are shown in magenta. Cyclin boxes are shown in blue.
Overall, the complex of the Rb pocket domain and large T antigen N-terminal domain forms an elongated shape with the dimensions 40 × 44 × 105 Å. The constituent domains are arranged linearly in the order A box, B box, large T antigen (Figure 1). All the intermolecular interactions in the complex are between the B box and the large T antigen.
Residues from helices α8, α9 and α10 in the A box and helices α11, α12, α13 and α14 in the B box are tightly packed together to form a hydrophobic core at the A–B interface, through which a 13 Å deep groove is formed. Most of the conserved residues of Rb family members are clustered in the A–B interface and it has been proposed that this region may bind to other proteins such as E2F (Lee et al., 1998). Interestingly, we observed that the L3 region in large T antigen with a related crystallographic symmetry binds tightly to this interface. Although it is unclear whether this interaction is correlated with any biological functions or whether it is simply a result of the crystal packing effect, our observation indicates that the interface between the A and B box of Rb could be involved in binding to other molecules.
While most of the conserved residues in the A box form an interface for the A–B box and the core of the A box, a number of conserved residues in the B box are clustered to form a shallow surface groove where large T antigen interacts with Rb. This groove, which mainly accommodates the LxCxE motif of large T antigen, accounts for approximately two-thirds of the overall interaction between large T antigen and the Rb pocket domain.
The structure of the N-terminal region of SV40 large T antigen
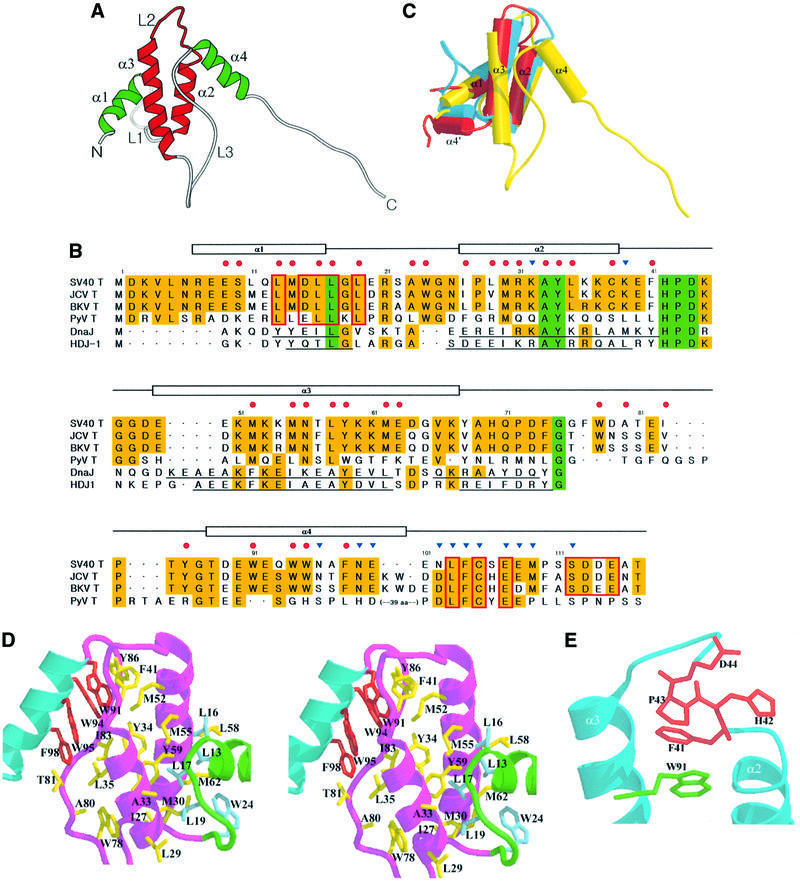
The N-terminal region of large T antigen can be divided into two parts: the J domain-like region (residues 7–102), where four α-helices form a compact core structure, and an extended loop region (residues 103–117) containing the LxCxE motif, which can independently bind to the Rb pocket (Figure 2A). The central feature of the J domain of large T antigen is an anti-parallel bundle. This is formed by two finger-like helices, α2 and α3, and is stabilized by the hydrophobic core of the conserved side chains. The two helices are at an angle of –157° to each other and are connected by a loop (L2) containing the HPD sequence (residues 42–44), which is invariant in all members of the J domain family (Figure 2B; Silver and Way, 1993; Tsai and Douglas, 1996). Helix α1 crosses the middle of these central helices at an angle of 53° to helix α2 and –113° to helix α3. Helix α4 is positioned at the opposite face of the two central helices from α1, and interacts directly with residues from helix α2 but not helix α3. Two loops, L1 and L3, also contribute to the formation of a core.
Fig. 2. (A) Overall structure of the N-terminal portion of SV40 large T antigen (residues 7–117). The two central helices and connecting loop are colored red, surrounding helices are in green and the other loops are colored gray. (B) Sequence alignment of T antigen with other J domain-containing proteins. The conserved residues are boxed with yellow and green. Residues participating in core formation (side chains with <20% of accessible surface area) are indicated by red circles and those interacting with Rb by blue triangles. JCV, large T antigen of JC virus; BKV, large T antigen of BK virus; PyV, large T antigen of murine polyoma virus. The conserved residues in HPV E7 and adenovirus E1 are boxed in red lines. (C) Structural comparison of SV40 large T antigen (yellow) with the J domain of DnaJ (blue) and HDJ-1 (red). Helix α4′ is from DnaJ or HDJ-1, and helix α4 is from large T antigen. (D) Stereo view of the hydrophobic core structure of large T antigen. Helix α1 is colored green, α2 and α3 magenta, and α4 light blue. (E) The local structure of the L2 loop of large T antigen. The side chain of Phe41 is stacked between Trp91 (green) in helix α4 and Pro43.
Sequence comparison and biochemical studies suggest that the J domain of large T antigen is structurally and functionally similar to the J domain from two molecular chaperones: DnaJ from E.coli and HDJ-1, the human Hsp40 (Figure 2B; Srinivasan et al., 1997). Indeed, the structure of the J domain of large T antigen is very similar to other J domain structures in the region of the two central helices and a connecting loop with root mean square differences (r.m.s.ds) of 1.25 and 1.38 Å for 25 residues in α2 and α3 of DnaJ and HDJ-1, respectively (Pellecchia et al., 1996; Qian et al., 1996). However, there are clear differences in other regions (Figure 2C). First, helix α1 in large T antigen is twice as long as the equivalent region in other J domains, where a loop is followed by a short helix formed by five residues. Secondly, helix α3 in T antigen is one turn longer than the equivalent helix in DnaJ or HDJ-1. Thirdly, and most significantly, while the region after helix α3 in DnaJ and HDJ-1 is rich in glycine/phenylalanine and is unstructured, the equivalent region in T antigen, which is not homologous to DnaJ or HDJ-1, is well structured, participating in the formation of the core. Furthermore, residues 60–67 in DnaJ (60–66 in HDJ-1) form a helical structure, and when three J domain structures are superimposed, this helix (α4′) is directed to the N-terminus of the J domain. By contrast, the region (residues 68–90) containing the equivalent residues in T antigen forms a long loop, L3, which occupies the opposite face of helices α2 and α3 from helix α1. Residues 68–78 in this loop make contacts with the interface of the A–B box in the Rb pocket, which is related with a crystallographic symmetry, through several intermolecular hydrogen bonds, and the rest of the loop is inserted between helices α3 and α4, thus stabilizing the J domain of large T antigen (Figure 2A). Residues 91–102 in large T antigen form helix α4, which contributes significantly to both intra- and intermolecular interactions.
The core of the N-terminal region of large T antigen is formed mainly by hydrophobic interactions together with a few polar interactions (Figure 2D). The strongly conserved residue Leu17 fits into a hydrophobic pocket formed by Leu13, Met14, Leu19, Ala23, Trp24, Leu29, Met30 and Ala33. Residues Tyr34, Met52, Tyr59, Ile83 and Trp94, which interact with each other to occupy the core formed by helices α2, α3 and α4, are surrounded by Cys38, Phe41, Trp91, Trp95 and Phe98. In addition, hydrogen bonds (Gln12–Lys54, Glu49–Tyr86, Tyr59–Glu63, Thr81–Asn102) and electrostatic interactions (Glu9–Lys61, Lys36–Glu40) further stabilize the core of the N-terminal region of large T antigen. A number of non-functional SV40 large T antigen mutants have been reported in the N-terminal region (database: bigdaddy. bio.pitt.edu/SV40). The effect of these mutations can be explained by perturbation in local structure. For example, the mutations M30I and K51D both result in a replication defective mutant (Gluzman and Ahrens, 1982). From our results, we would predict that Ile30 could fit loosely into the neighboring environment and that the contact with Trp24 would be decreased. Similarly, Asp51 would fail to make a hydrogen bond to the carbonyl oxygen of Leu16.
The importance of loop L2 is indicated by the strong conservation of the HPD sequence in all J domains (Tsai and Douglas, 1996). Large T antigen mutants in this region are unable to dissociate E2F from Rb and are, as a consequence, replication defective. Also, DnaJ mutants in this region have lost their chaperone function (Srinivasan et al., 1997). The HPD motif (residues 42–44) in large T antigen forms a helical turn in which the carbonyl oxygen of His42 makes a hydrogen bond to the nitrogen of Lys45 (Figure 2E). The imidazole ring of His42 donates a hydrogen bond to the main chain carbonyl oxygen of Glu40 in the C-terminal portion of helix α2. The side chain of Phe45 is located between Pro43 and the side chain of Trp91, which in turn is packed by series of rings from Trp94, Trp95 and Phe98. The side chains of both Asp44 and Lys45 are exposed in an opposite direction to the axis of helix α3 and could interact with other proteins. From our structure, it seems that one of the roles of His–Pro is to stabilize the C-terminal portion of helix α2 and provide some rigidity to the L2 loop, which plays a key role in recognizing hsc70.
The global structural differences observed in the regions after the helix α3 between T antigen and DnaJ/HDJ-1 result in local differences including the orientations and characters of some side chains. In particular, the side chain of Tyr32 of DnaJ (Tyr30 in HDJ-1) is exposed to solvent whereas the ring of the equivalent residue, Phe41 of large T antigen, is rotated >90° from the superimposed side chain of Tyr32 of DnaJ and directed to the core. As a result, Lys37 in T antigen, the residue equivalent to Leu28, which is located next to the ring of Tyr32 and partly buried in DnaJ, is exposed. Also, residues Ile27 and Arg31 from helix α2, and Met52, Asn56, Tyr59 and Glu63 from helix α3, are buried in T antigen whereas the equivalent residues are exposed in DnaJ.
The imidazole ring of His33 in DnaJ is oriented parallel to the ring of Pro34 in DnaJ and HDJ-1. If the equivalent large T antigen residue His42 were in the same orientation, it would collide with residues from helix α4. Thus, the side chain of His42 is rotated by ∼90° in large T antigen from the ring of Pro43 (equivalent to Pro34 in DnaJ and HDJ-1). Overall, these local changes lead the main chain of the L2 loop to shift ∼10 Å toward helix α1 when compared with the equivalent region in DnaJ. As will be discussed later, we presume that these local and global differences in structure between large T antigen and DnaJ/HDJ-1 may provide the distinctive function necessary for large T antigen to disassemble Rb–E2F upon ATP hydrolysis by hsc70, a process that can not be fully accomplished by DnaJ (Sullivan et al., 2000).
Although many conserved polar residues are present in helices α2 and α3 near the L2 loop, only a few of them are exposed and interact with residues from other proteins. These include Lys36, Lys37 and Glu40 from α2, and Glu50, Lys51, Lys53 and Lys54 from helix α3. Several hydrogen bonds link the backbone of the L3 loop with the side chains of residues from helix α3. The carbonyl oxygen of Ile83 and Pro84 accepts hydrogen bonds from the side chains of Lys60 and Asn56, respectively, and the amide of Tyr86 donates hydrogen to the side chain of Asn56. The stacking force from the line of rings of Trp91, Trp94, Trp95 and Phe98 in helix α4 is also a key element in the stability of L3.
Interface between large T antigen and the Rb pocket complex
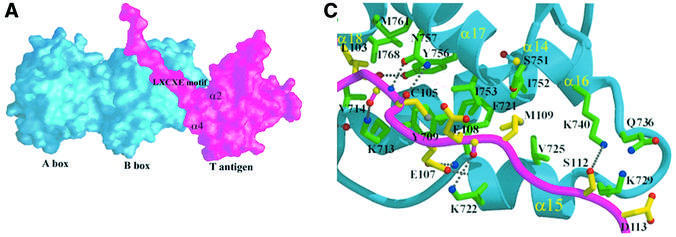
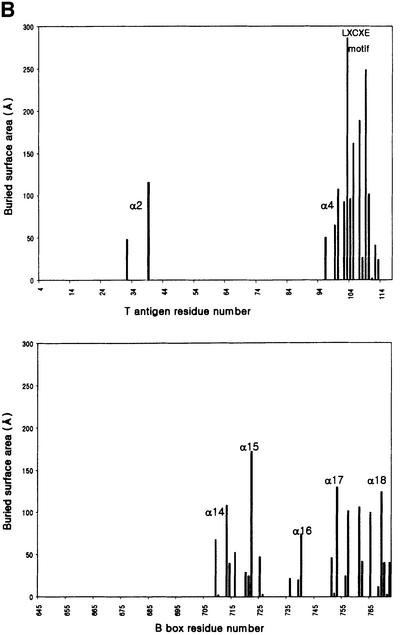
The in vitro binding analyses show that the N-terminal region of large T antigen binds to the Rb pocket. They also show that large T antigen and E2F bind to the Rb pocket simultaneously (Figure 3). Our crystal structure shows that the N-terminal region of large T antigen binds to the B box of the Rb pocket domain. The regions within large T antigen that are involved in the intermolecular interaction can be divided into three parts: helix α2, helix α4 and the extended loop containing the LxCxE motif (Figure 4A and B). In total, an area of 1600 Å2 is buried upon complex formation: 59% of this area is contributed by non-polar residues, 27% by polar groups and 14% by charged groups, indicating the importance of hydrophobic interactions in the large T antigen–Rb complex.
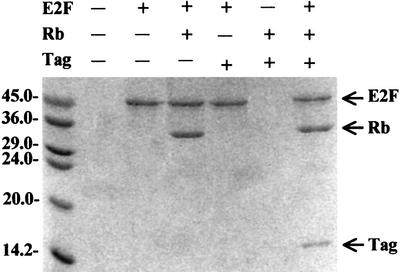
Fig. 3. The in vitro protein interaction assays for the large T antigen N-terminal region and E2F-1 binding to the Rb pocket. Lane 1, molecular weight marker; lane 2, the control resin; lane 3, control resin where E2F-1 was bound; lane 4, the Rb pocket was added to the E2F-1-bound resin; lane 5, large T antigen was added to the E2F-1-bound resin; lane 6, large T antigen and the Rb pocket added to the empty resin; lane 7, large T antigen and the Rb pocket added to the E2F-1-bound resin.
Fig. 4. (A) Surface representation of the Rb pocket (blue) and large T antigen (magenta) complex. Regions (α2, α4 and LxCxE motif) in large T antigen that interact with the Rb pocket are labeled. The figure was drawn using GRASP (Nicholls et al., 1991). (B) Bar graph showing the differences in solvent accessibility in large T antigen and Rb pocket residues in the complex compared with the isolated subunits. The structural elements discussed in the text are indicated. (C) Detailed view of the interface of the complex showing the region of conserved motifs, the LxCxE and the SDDE in large T antigen. The main chain trace of the loop in T antigen is shown in magenta, the main chain of Rb is in blue, carbon atoms in Rb are shown in green, carbon atoms in T antigen are in yellow, oxygen atoms in red, nitrogen atoms in blue and sulfur atoms in gray. (D) The second and third part of the interface (described in the text) between large T antigen and the Rb pocket. Coloring scheme is as in (C).
The interaction between the LxCxE motif and Rb is the most extensive part of the interaction and accounts for approximately two-thirds (1190 Å2) of the total buried surface area. The loop containing the LxCxE motif forms an extended conformation and binds to the shallow surface groove on the B box created by five helices, three of which are from the cyclin box. As seen in Figure 4C, helices α17 and α15 of Rb occupy the top and bottom of the groove, respectively, α16 and α18 are at either end, and helix α14 is located at the back. Although Leu103, Cys105, Glu107 and Met109 in large T antigen interact tightly with neighboring residues, and Ser112 is close to Lys740 of Rb, other residues in this loop are exposed or interact with symmetry-related molecules. Leu103 makes van der Waals contacts with side chains of Val714 and Leu769 in Rb, and Cys105 interacts with the side chains of Ile753 and Tyr709. A number of hydrogen bonds are also involved in the formation of the complex: four carbonyl groups of the main chain of the loop accept hydrogen atoms from the side chains of Rb (Leu103–Tyr756, Phe104–Lys713, Cys105–Asn757 and Glu108–Lys722); an amide of Cys105 donates hydrogen to the side chain of Asn757; and the side chain of Glu107 accepts hydrogens from the amide groups of the Phe721 and Lys722 main chains. Mutation of Glu107 to Lys107 leads to the dissociation of large T antigen from Rb and is frequently used in biochemical experiments (Zalvide et al., 1998). We would predict that in this mutation not only are two hydrogen bonds lost but also the side chain of Lys107 could make unfavorable interactions with the amide of Phe721 or Lys722. Met109 in large T antigen is replaced by Leu in HPV E7 protein and by Phe in adenovirus E1A, respectively. In large T antigen, Met109 is surrounded by the Rb side chains of Phe721, Val725, Phe739, Ser751, Ile752 and Ile753. When compared with E7 Leu28 and its environment in Rb–E7 peptide structure, the positions of the side chain of Met109 and its interacting residues are very similar to those of equivalent residues with the largest r.m.s.d. of 1.2 Å (Lee et al., 1998). The end of the loop in large T antigen contains a highly conserved acidic sequence SDDE. Since the LxCxE region is located around the cluster of lysine residues in the Rb B box, it has been proposed that these acidic residues may interact with this lysine patch (Lee et al., 1998). However, our structure shows that the region containing the SDDE sequence is very flexible, as indicated from high temperature factors, and that only the side chains of Ser112 and Asp113 are close to the side chains of Lys740 (3.7 Å) and Gln736 (4.5 Å), respectively. However, since the two serine residues, Ser111 and Ser112, are targets for casein kinase II, it is possible that phosporylation may induce a conformational change and that this might allow the acidic region to interact tightly with the lysine cluster. Mutational analysis of the lysine patch surrounding the LxCxE binding site has revealed that these lysine residues are critical for binding of the phosphorylated C-terminal portion of Rb (Harbour et al., 1999).
In the second part of the Rb–large T antigen interface, residues from the C-terminal portion of helix α4 of the large T antigen interact with helices α14 and α18 of the Rb pocket (Figure 4D). The side chains of Asn96 and Glu100 in large T antigen accept hydrogen bonds from Lys765 and Gln762 in Rb, respectively. The aliphatic group of Asn99 makes van der Waals contact with Leu769, and a hydrogen bond links the side chain of Asn102 and the main chain carbonyl oxygen of Asn713.
In the third part of the Rb–large T antigen interface, two residues from helix α2 of large T antigen lie close to helices α14 and α18 of Rb (Figure 4D). The side chain of Lys32 is 4.2 Å away from the side chain of Asn716 and the side chain of Lys39 is 3.6 Å away from the carbonyl group of Leu769. These distances are relatively large and thus contributions from these interactions are not as significant as other interactions described above. Nevertheless, these interactions may help to fit the J domain of large T antigen into the pocket region of Rb. Mutation of Gln32 in PyV (Lys32 in SV40) to Glu32 significantly decreases the E2F activation, indicating the important functional role of this residue (Sheng et al., 1997). In addition, if the rest of the Rb structure (residues 380–785; Lee et al., 1998) is superimposed onto our structure, Arg775 forms an ion-pair with Glu39 of large T antigen.
Discussion
In this paper, we have described how the N-terminal region of large T antigen could associate with Rb, and have provided an insight into how the J domain and the LxCxE motif of large T antigen act together to inhibit the function of Rb. One of the most important roles of Rb in the regulation of the cell cycle is its association with E2F and, in this respect, it is of considerable interest to understand how Rb binds to and dissociates from E2F and how large T antigen affects this. The in vitro interaction assay in Figure 3 as well as some previous data demonstrate that both the N-terminal portion of large T antigen (residues 7–117) and a peptide containing the LxCxE sequence bind to Rb or Rb family members at a different site from E2F (Harris et al., 1998; Lee et al., 1998; Sullivan et al., 2000). Therefore, it is unlikely that large T antigen binds competitively to the same site within Rb as E2F. Consistent with this idea, the HPD motif in large T antigen, a critical region for dissociation of E2F from Rb, is >15 Å away from the nearest region of the Rb pocket in our structure.
The structural similarities in the two helical regions of the J domains of large T antigen and the chaperones DnaJ and HDJ-1 suggest that large T antigen and chaperones may use similar mechanisms for their function. DnaJ not only assists protein folding but also disassembles molecular complexes. For example, it binds to inactive, dimeric replication protein RepA and dissociates it into active monomers in the presence of DnaK, GrpE and ATP (Wickner et al., 1991). Likewise, one of the primary roles of large T antigen may be to recruit hsc70, a DnaK homolog that is expressed constitutively in mammalian cells, and to stimulate hsc70 to induce ATP hydrolysis, and this would lead to disassembly of the Rb–E2F complex. Since the dissolved Rb and E2F are both functionally active, it is unlikely that the dissociation of the complex by large T antigen involves any denaturation or degradation process similar to that observed with the Rb homolog p130 (Zalvide et al., 1998). Based on the proposed mechanism for the role of DnaJ–DnaK in the disassembly of complexes (Laufen et al., 1999), the following model can be suggested for large T antigen action in dissociation of E2F from Rb (Figure 5A): (i) upon binding of hsc70 to the large T antigen–Rb–E2F complex, the Rb–E2F complex is transferred from large T antigen to the substrate binding site of hsc70; (ii) Rb–E2F is then moved from the substrate binding site of hsc70 to its ATPase domain; (iii) ATP hydrolysis and conformational changes in hsc70 occur, including the closing of the substrate binding cavity, and dissociation of each molecule from the complex. Recent studies show that Rb and E2F are released as individual molecules from hsc70, in contrast to p130–E2F, in which the two molecules are released as a complex, implying that the inactivation mechanism by large T antigen is not identical for Rb and its homologs (Sullivan et al., 2000).
Fig. 5. (A) Schematic representation of the dissociation mechanism of the Rb–E2F complex by large T antigen and hsc70. The model is based on the DnaJ–DnaK chaperone mechanism proposed by Laufen et al. (1999; see text). (B) Surface representation of a model for the interaction between the Rb pocket domain–large T antigen complex and hsc70 (DnaK). Residues Arg167, Asn170 and Thr173 are critical for the interaction with the J domain, whereas Asn147, Asp148 and Glu152 do not a play significant role in binding to the J domain (Suh et al., 1998). The model is based on information from NMR perturbation and mutational analysis of the DnaJ–DnaK interaction (see text). The solvent-accessible surface was calculated using GRASP (Nicholls et al., 1991) with a water probe radius of 1.4 Å, and is colored by electrostatic potential of large T antigen and hsc70 in the range less than –10 to >10 KbT, where Kb is the Boltzman constant and T is the temperature. The positive potential is shown in blue and negative in red.
How does the large T antigen recognize hsc70? NMR perturbation and mutational analysis of the interactions between DnaJ and DnaK indicate that residues in helix α2 and the loop containing the HPD motif in DnaJ bind to the cleft of the ATPase domain in DnaK by electrostatic interactions (Greene et al., 1998; Suh et al., 1998). In particular, Asp35 in DnaJ and Arg167, Asn170 and Thr173 in DnaK are crucial for the interaction (Suh et al., 1998). These residues are conserved in large T antigen (Asp44) and hsc70 (Arg167, Asn170, Thr173), respectively, and thus it seems likely that large T antigen and hsc70 interact in a similar mode to DnaJ–DnaK (Figure 5B).
The side chain of Asp44 in T antigen is exposed and points in the opposite direction to the axis of helix α3. The position of this residue may be critical for the interaction with hsc70 given that the side chain of Asp35 in DnaJ lies in different directions in inactive and active DnaJ (Huang et al., 1999). Furthermore, a large T antigen mutant in which Asp44 is replaced by Asn44 is unable to bind to hsc70 and stimulate ATP hydrolysis (Sheng et al., 1997). The local stability provided by the hydrogen bonds between His42 and Glu40, and His42 and Lys45, and the stacking force between Pro43 and Phe41 should provide rigidity to the structure of the L2 loop of large T antigen including the positions of Asp44 and Lys45, and this rigidity would presumably help to guide these residues to interact with hsc70. NMR perturbation studies on DnaJ indicate conformational changes at His33 and Asp35 upon binding of DnaK. In our structure for large T antigen, the main chain of the L2 loop is shifted ∼10 Å toward helix α1 when compared with the L2 loop in DnaJ and HDJ-1. This is the most significant structural difference within the central region, α2–L2–α3, of the large T antigen. The high temperature factors in this region indicate that the L2 loop is flexible. A large part of this flexibility is probably contributed from Gly46 and Gly47 immediately after Lys45. Mutations of these glycine residues reduce the activity of large T antigen (Peden and Pipas, 1992). We presume that although His42 and Pro43 provide some structural rigidity to the L2 loop and help in the recognition of hsc70, the flexibility of the loop is necessary to allow the local conformational changes required for the stimulation of ATP hydrolysis of hsc70.
Chemical shift studies have identified several additional residues in DnaJ whose positions are perturbed upon binding of DnaK (Greene et al., 1998), and that may be involved in binding to DnaK. These include Val12, Ser13, Ala16, Arg19, Glu20, Ile21, Arg22, Ala24, Tyr25, Lys26, Arg27, Leu28, Met30, Tyr32, Tyr54 and Thr58. The equivalent residues in large T antigen are Val19, Glu20, Ala23, Pro28, Leu29, Met30, Arg31, Ala33, Tyr34, Leu35, Lys36, Lys37, Lys39, Phe41, Tyr59 and Glu63, where the majority of them are involved in core formation (Figure 2B). In DnaJ, Arg22, Tyr25, Tyr32, Tyr54 and Thr58 are exposed whereas the equivalent residues in large T antigen are buried by loop L3 and helix α4. Thus, it is unlikely that these buried residues interact directly with hsc70 unless significant conformational changes occur in this region. Two positively charged residues in this region, Lys32 and Lys39, make contact with Rb, one of which, Lys39, is implicated to shift upon binding to hsc70. Therefore, we suspect that Lys39 may be important for transferring Rb–E2F to hsc70. To date, the NMR perturbation studies have only been carried out on two chaperones, DnaJ and DnaK, in the absence of any substrate. It will be interesting to see whether the addition of a substrate affects the chemical shifts on any of the residues implicated in DnaJ–DnaK complex formation. Two more lysine residues, Lys36 and Lys37, are exposed and their equivalent residues in DnaJ are shifted upon binding to DnaK. Thus, it seems possible that these residues in large T antigen may participate in electrostatic interactions with residues in hsc70. However, these positively charged residues are both replaced by glutamine in the PyV large T antigen, which can form a complex with hsp70, and it is unclear whether positively charged residues in these positions are required for the viral large T antigen–hsc70 interaction as they are for the DnaJ–DnaK interaction. Evidence from functional studies suggests that there may be differences between the binding mode of two complexes, hsc70–large T antigen and DnaJ–DnaK. Functional studies of a chimeric T antigen, where the J domain of DnaJ was substituted for the equivalent region of SV40 large T antigen show that although the J domain of DnaJ can bind to hsc70, it does not stimulate the ATP hydrolysis of hsc70 as efficiently as does the J domain from large T antigen (Sullivan et al., 2000). Thus, there is specificity in the recognition of hsc70 and in the stimulation of ATP hydrolysis of hsc70 by the J domain of large T antigen. How is this specificity achieved? In DnaJ, the presence of a Gly/Phe region after helix α3 affects the local conformation around the loop region connecting two central helices (Huang et al., 1999). DnaJ without a Gly/Phe region can bind to DnaK but it can not stimulate the ATPase activity of DnaK. In large T antigen, the characteristics of the L3 loop and α4 helix would contribute to the local and global structural differences between the J domains of T antigen and DnaJ as described above, and such differences may provide the basis of specificity for ATP hydrolysis of hsc70 by large T antigen, an essential step for disassembly of Rb–E2F.
From our structural studies, we propose that large T antigen uses a molecular chaperone mechanism to disassemble the E2F–Rb complex. However, other viral oncoproteins that inactivate Rb, such as HPV E7 and adenovirus E1A, do not possess an internal J domain, and it is unclear whether the molecular chaperone mechanism used by SV40 large T antigen to disassemble E2F and Rb can be applied to these other viral oncoproteins. Interestingly, the recent finding that the C-terminal portion of HPV E7 can form a complex with a human DnaJ family member, hTid-1, suggests that E7 could also use a DnaJ-mediated chaperone mechanism to deregulate the tumor suppression function of Rb (Schilling et al., 1998). This result, therefore, indicates the possibility that the chaperone-mediated mechanism of Rb–E2F dissociation may extend to other viral oncoproteins.
Materials and methods
Expression and purification
The pocket domain (residues 378–772) of the human Rb tumor suppressor gene was inserted into PET15b (NcoI–BamHI digested) and expressed in E.coli BL21. To aid the crystallization, residues 578–644 were removed by deletion mutagenesis. The pocket domain was purified using cation exchange chromatography (mono S column) and gel filtration chromatography (Superdex 75 column). The N-terminal region of SV40 large T antigen (residues 7–117) was inserted into PET15b (NdeI and BamHI digested) and expressed in E.coli BL21 as a His-tagged form. The protein was initially isolated using a nickel column. After thrombin digestion, T antigen was further purified using anion exchange chromatography (mono Q column), followed by gel filtration (Superdex 75 column).
Crystallization and data collection
The purified Rb pocket (20 mg/ml) and T antigen (30 mg/ml) were mixed at a 1:1.5 molar ratio in a solution of 25 mM Tris–HCl, 150 mM NaCl, 5 mM dithiothreitol (DTT) pH 7.5 and applied to a gel filtration column. The fraction containing the complex was concentrated by ultrafiltration. Crystals of the complex were grown at 4°C using the hanging drop vapor diffusion method. The crystals were obtained by mixing the protein complex solution with an equal volume of the buffer used in the crystallization well (4–6% PEG 6K, 70 mM potassium phosphate, 10 mM DTT pH 7.2). Crystals of 0.1 × 0.1 × 0.05 mm were formed over 2 weeks. To improve the size and quality of crystals, both micro- and macro-seeding were necessary. The crystals formed in space group I4 with a = b = 127.13 Å, c = 96.19 Å, and contained one complex in the asymmetric unit. The native data set was collected with the Mar30 imaging plate area detector mounted on a Rigaku RU200 rotating anode X-ray generator. A crystal was flash frozen in reservoir buffer supplemented with 40% sucrose and 5% glycerol at –170°C. Data collection at synchrotron sources (DESY/photon factory) was also tried. However, because of the properties of crystal, which are very fragile and sensitive to cryo-cooling, this did not improve the qualities of data. Reflection data were indexed, integrated and scaled using DENZO and SCALEPACK (Otwinowski and Minor, 1997).
Structure determination and refinement
The structure of the complex was determined by a molecular replacement method. The Rb pocket domain was located with the program AMoRe (CCP4, 1994) using the structure of the isolated pocket model as a search model (Lee et al., 1998). The molecular replacement model was refined by rigid body protocol followed by simulated annealing and the R factor was dropped to 42%. The structure was built with the program CHAIN (Sack, 1988) using electron density maps calculated with phases derived from the initially refined model of the pocket domain of Rb (Lee et al., 1998). Successive rounds of rebuilding and simulated annealing refinement with the program CNS (Brünger et al., 1998) allowed complete interpretation of the N-terminal portion of T antigen as well as of the pocket domain of Rb. In each round of refinement, the Rfree value was monitored, and the omit map was used to check systematically every part of the complex molecule at the end of the refinement. The refined model contains residues 7–117 of SV40 large T antigen and residues 379–577 and 645–772 of the Rb pocket (Table I). Residues 503–509 are disordered and are not modeled.
In vitro binding assay
All procedures except SDS–PAGE analysis were carried out at 4°C. The E2F-1 gene (residues 88–428) was inserted into PET15b and expressed in E.coli BL21. The His-tagged E2F-1 was purified using a Ni-NTA– agarose column followed by mono Q and Superdex 75 columns. The purified His-tagged E2F-1 (1 mg) was loaded onto the Ni-NTA–agarose column (1 ml) equilibrated with buffer A (50 mM sodium phosphate, 300 mM NaCl, 2 mM β-mercaptoethanol pH 8.0). The purified Rb pocket (1 mg) and/or large T antigen (0.5 mg, residues 7–117) was then applied to the E2F-1-bound Ni-column, and washed five times with buffer A. The proteins were eluted with buffer B (50 mM sodium phosphate, 300 mM NaCl, 10% glycerol, 7 mM β-mercaptoethanol and 250 mM imidazole pH 8.0). Eluted proteins were analyzed by 15% SDS–PAGE using standard procedure.
Coordinates have been deposited with the Protein Data Bank (PDB). The accession code is 1GH6.
Acknowledgments
Acknowledgements
We are grateful to M.S.Woo for her technical help for the purification of a large T antigen and to J.M.Bradbury for help with preparation of the manuscript. This work was supported by funds from the Korean Ministry of Science and Technology (Biotech2000 program), the Ministry of Education (BK21 program), POSTECH, Korean Science and Engineering Foundation (PNRC of Yonsei University and Basic research program) and the Korean Academy of Science and Technology (a Young Scientist Award to Y.C.).
REFERENCES
- Bookstein R., Shew,J.Y., Chen,P.L., Scully,P. and Lee,W.-H. (1990) Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science, 247, 712–715. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L. and Pipas,J.M. (1998) Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol., 72, 5329–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.R., Noble,M.E., Endicott,J.A., Garman,E.F., Wakatsuki,S., Mitchell,E., Rasmussen,B., Hunt,T. and Johnson,L.N. (1995) The crystal structure of cyclin A. Structure, 3, 1235–1247. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Campbell K.S. et al. (1997) DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev., 11, 1098–1110. [DOI] [PubMed] [Google Scholar]
- Chellappan S.P., Hiebert,S., Mudryj,M., Horowitz,J.M. and Nevins,J.R. (1991) The E2F transcription factor is a cellular target for the RB protein. Cell, 65, 1053–1061. [DOI] [PubMed] [Google Scholar]
- Chellappan S.F., Kraus,V.B., Kroger,B., Munger,K., Howley,P.M. and Nevins,J.R. (1992) Adenovirus E1A, simian virus 40 tumor antigen and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl Acad. Sci. USA, 89, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.N. and Dean,D.C. (1996) Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol. Cell. Biol., 16, 4862–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- DeCaprio J.A. (1999) The role of the J domain of SV40 large T in cellular transformation. Biologicals, 27, 23–28. [DOI] [PubMed] [Google Scholar]
- DeCaprio J.A., Furukawa,Y., Ajchenbaum,F., Griffin,J.D. and Livingston,D.M. (1992) The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc. Natl Acad. Sci. USA, 89, 1795–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. and Ahrens,B. (1982) SV40 early mutants that are defective for viral DNA synthesis but competent for transformation of cultured rat and simian cells. Virology, 22, 78–92. [DOI] [PubMed] [Google Scholar]
- Greene M.K., Maskos,K. and Landry,S.J. (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA, 95, 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour J.W., Lai,S.L., Whang-Peng,J., Gazdar,A.F., Minna,J.D. and Kaye,F.J. (1988) Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science, 241, 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour J.W., Luo,R.X., Dei Santi,A., Postigo,A.A. and Dean,D.C.C. (1999) Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell, 98, 859–869. [DOI] [PubMed] [Google Scholar]
- Harris K.F., Christensen,J.B., Radany.E.H. and Imperiale,M.J. (1998) Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol. Cell. Biol., 18, 1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K., Lees,J.A., Vidal,M., Dyson,N., Harlow,E. and Fattaey,A. (1992) A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell, 70, 337–350. [DOI] [PubMed] [Google Scholar]
- Hu Q.J., Dyson,N. and Harlow,E. (1990) The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J., 9, 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.J. (1988) Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science, 242, 1563–1566. [DOI] [PubMed] [Google Scholar]
- Huang K., Flangan,J.M. and Prestegard,J.H. (1999) The influence of C-terminal extension on the structure of the ‘J-domain’ in E.coli DnaJ. Protein Sci., 8, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Wang,N.P., Tseng,B.Y., Lee,W.H. and Lee,E.H. (1990) Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J., 9, 1815–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey P.D., Russo,A.A., Polyak,K., Gibbs,E., Hurwitz,J., Massague,J. and Pavletich,N.P. (1995) Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature, 376, 313–320. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G., Hu,Q.J., Dyson,N. and Harlow,E. (1990) The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J., 9, 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G. Jr et al. (1992) Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell, 70, 351–364. [DOI] [PubMed] [Google Scholar]
- Kelley W.L. and Georgopolous,C. (1997) The T/t common exon of SV40, JCV and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl Acad. Sci. USA, 94, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierstead T.D. and Tevethia,M.J. (1993) Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol., 67, 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-Y. and Cho,Y. (1997) Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box. Nature Struct. Biol., 4, 390–395. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- La Thangue N.B. (1994) DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem. Sci., 19, 108–114. [DOI] [PubMed] [Google Scholar]
- Laufen T., Mayer,M.P., Beisel,C., Klostermeier,D., Mogk,A., Reinstein,J. and Bukau,B. (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl Acad. Sci. USA, 96, 5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.Y., To,H., Shew,J.Y., Bookstein,R., Scully,P. and Lee,W.H. (1988) Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science, 241, 218–221. [DOI] [PubMed] [Google Scholar]
- Lee J.O., Russo,A.A. and Pavletich,N.P. (1998) Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature, 391, 859–865. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0 A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Mihara K., Cao,X.R., Yen,A., Chandler,S., Driscoll,B., Murphree,A.L., T’Ang,A. and Fung,Y.K. (1989) Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science, 246, 1300–1303. [DOI] [PubMed] [Google Scholar]
- Mittnacht S. (1998) Control of pRB phosphorylation. Curr. Opin. Genet. Dev., 8, 21–27. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F: A link between the Rb tumour suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamics properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nikolov D.B., Chen,H., Halay,E.D., Usheva,A.A., Hisataka,K., Lee,D.K., Roeder,R.G. and Burley,S.K. (1995) Crystal structure of a TFIIB–TBP–TATA–element ternary complex. Nature, 377, 119–128. [DOI] [PubMed] [Google Scholar]
- Noble M.E., Endicott,J.A., Brown,N.R. and Johnson,L.N. (1997) The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem. Sci., 22, 482–487. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pardee A.B. (1989) G1 events and regulation of cell proliferation. Science, 246, 603–608. [DOI] [PubMed] [Google Scholar]
- Peden K.W. and Pipas,J.M. (1992) Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes, 6, 107–118. [DOI] [PubMed] [Google Scholar]
- Pellecchia M., Szyperski,T., Wall,D., Georgopoulos,C. and Wuthrich,K. (1996) NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol., 260, 236–250. [DOI] [PubMed] [Google Scholar]
- Qian Y.Q., Patel,D., Hartl,F.U. and McColl,D.J. (1996) Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J. Mol. Biol., 260, 224–235. [DOI] [PubMed] [Google Scholar]
- Sack J.S. (1988) CHAIN: a crystallographic modeling program. J. Mol. Graph., 6, 224–225. [Google Scholar]
- Sanchez I. and Dynlacht,B.D. (1996) Transcriptional control of the cell cycle. Curr. Opin. Cell Biol., 8, 318–324. [DOI] [PubMed] [Google Scholar]
- Sawai E.T. and Butel,J.S. (1989) Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J. Virol., 63, 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B., De-Medina,T., Syken,J., Vidal,M. and Munger,K. (1998) A novel human DnaJ protein, hTid-1, a homolog of the Drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology, 247, 74–85. [DOI] [PubMed] [Google Scholar]
- Sheng Q., Denis,D., Ratnofsky,M., Roberts,T.M., DeCaprio,J.A. and Schaffhauen,B. (1997) The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J. Virol., 71, 9410–9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver P.A. and Way,J.C. (1993) Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell, 74, 5–6. [DOI] [PubMed] [Google Scholar]
- Srinivasan A. et al. (1997) The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol., 17, 4761–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubdal H., Zalvide,J. and DeCaprio,J.A. (1996) Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J. Virol., 70, 2781–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubdal H., Zalvide,J., Campbell,K.S., Schweitzer,C., Roberts,T.M. and DeCaprio,J.A. (1997) Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol., 17, 4979–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh W.C., Burkholder,W.F., Lu,C.Z., Zhao,X., Gottesman,M.E. and Gross,C.A. (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl Acad. Sci. USA, 95, 15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C.S., Tremblay,J.D., Fewell,S.W., Lewis,J.A., Brodsky,J.L. and Pipas,J.M. (2000) Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol. Cell. Biol., 20, 5749–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguchida J., Ishizaki,K., Sasaki,M.S., Ikenaga,M., Sugimoto,M., Kotoura,Y. and Yamamuro,T. (1988) Chromosomal reorganization for the expression of recessive mutation of retinoblastoma susceptibility gene in the development of osteosarcoma. Cancer Res., 48, 3939–3943. [PubMed] [Google Scholar]
- Tsai J. and Douglas,M.G. (1996) A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem., 271, 9347–9354. [DOI] [PubMed] [Google Scholar]
- Weinberg R.A. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- Wickner S., Hoskins,J. and McKenney,K. (1991) Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature, 350, 165–167. [DOI] [PubMed] [Google Scholar]
- Zalvide J., Stubdal,H. and DeCaprio,J.A. (1998) The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol., 18, 1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Rice,P.W., Gorsch,L., Abate,M. and Cole,C.N. (1992) Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J. Virol., 66, 2780–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]