Abstract
The MuDR element responsible for Mutator activities in maize encodes two genes, mudrA and mudrB. Each encodes multiple transcripts hypothesized to regulate, directly or indirectly, the unique late timing and switch in transposition mechanism during maize development. mudrA, which encodes the MURA transposase, is unstable in bacterial plasmids, a technical problem solved by using phage M13 as a vector to prepare DNA for biolistic transformation. In transgenic maize, a single 2.7-kb mudrA cDNA predicted to encode an 823–amino acid protein is sufficient to catalyze late somatic excisions, despite removal of the native promoter, alternative transcription start sites, known introns, polymorphic 5′ and 3′ untranslated sequences, and the mudrB gene. These results suggest that post-translational regulation confers Mu excision timing. The transgene is active in lines containing silencing MuDR elements. This suggests that endogenous MuDR transposons do not measurably immunize the host against expression of a homologous transgene.
INTRODUCTION
Mutator lines of maize contain a high-copy-number DNA transposon family (Robertson, 1978). Nine Mu element subfamilies exist. All share homologous flanking ∼215-bp terminal inverted repeat (TIR) sequences. Subfamilies Mu1 to Mu8 are nonautonomous and require a source of transposase to catalyze transposition (reviewed in Bennetzen et al., 1993). The Mu transposase is encoded by the 4.9-kb MuDR element (Chomet et al., 1991; Hershberger et al., 1991; Qin et al., 1991; Hsia and Schnable, 1996), which is present in multiple copies in highly mutagenic Mutator lines.
Mu elements are an efficient transposon-tagging tool, because multicopy MuDR lines have a forward mutation frequency 20- to 50-fold higher than either Ac or Spm (Robertson and Mascia, 1981). Moreover, Mu elements transpose equally to linked and unlinked sites (Lisch et al., 1995). They exhibit an extremely high insertion bias (>90%) for low-copy-number transcribed regions of the genome (Cresse et al., 1995). Finally, Mu germinal insertion events occur late, resulting in independent insertions in sibling progeny (Robertson, 1981, 1985).
A fascinating component of Mutator biology is that MuDR catalyzes distinct transposition behaviors of Mu elements in somatic and germinal cells. The full somatic program involves activation, activity, and epigenetic silencing. In a line with methylated Mu elements, introduction of a transcriptionally active MuDR results in Mu element TIR demethylation in leaves (Chandler and Walbot, 1986; Bennetzen, 1987). Demethylated Mu elements can then excise at high frequencies, but only during the terminal cell divisions of somatic tissues, as observed in anthers, aleurone, and leaves (Levy and Walbot, 1990). In the cells that give rise to gametes, Mu follows a different program, because germinal revertants are exceedingly rare (Schnable et al., 1989; reviewed in Walbot, 1991). Instead, Mu elements duplicate and insert in late pregerminal, meiotic, and gametic cells but rarely in the vegetative precursor cells that give rise to the inflorescences (Robertson, 1981; Alleman and Freeling, 1986; Lisch et al., 1995). After amplification, multiple unlinked MuDR elements in some progeny or leaf sectors within progeny plants undergo coordinate epigenetic transcriptional silencing, which results in the remethylation of Mu element TIRs and loss of Mutator activity (Walbot, 1991; reviewed in Fedoroff and Chandler, 1994; Martienssen and Baron, 1994).
As shown in Figure 1A, MuDR consists of two convergently oriented genes, mudrA and mudrB, flanked by promoter-containing TIRs (Hershberger et al., 1991; Benito and Walbot, 1994). By homology and analysis, the function of mudrB remains unknown. In contrast, mudrA is the candidate transposase gene, because it is related to bacterial transposons (Eisen et al., 1994). Furthermore, analysis of lines carrying deletions in MuDR demonstrated that mudrA, but not mudrB, is required to catalyze somatic excisions (Lisch et al., 1999).
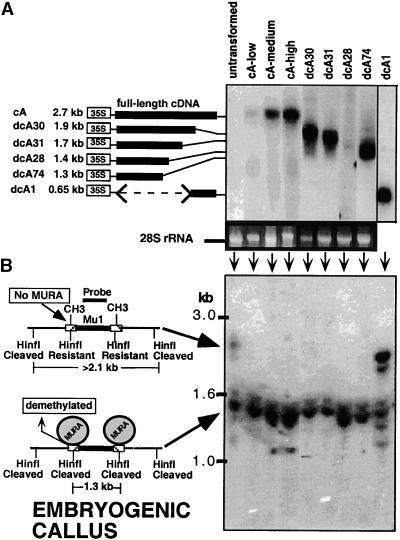
Figure 1.
Structure of MuDR, Endogenous mudrA and mudrB Transcripts, the CaMV 35S–mudrA Construct in cA+ Transgenic Maize Lines, and the Probes Used for RNA Gel Blots.
(A) Structure of an endogenous MuDR element. The element has two open reading frames, termed mudrA and mudrB, encoded in antiparallel orientation. The intergenic region between the two genes is composed of diverse short repetitive elements. The promoters are located within the ∼215-bp TIRs. The mudrA region with high similarity to bacterial transposases is shown in white. The DNA probes for RNA analysis in this study are located above the element. Numbering is according to Hershberger et al. (1991).
(B) The diversity of endogenous mudrA and mudrB transcripts in active Mutator seedlings (Hershberger et al., 1995). Intron sequences shown in solid black are in-frame with exons. Alternative mudrA transcription initiation sites (+169 and +252) produce transcripts with a short or long 5′ leader sequence. aa, amino acids.
(C) The structure of the CaMV 35S–mudrA cDNA in M13 transformed into maize to make cA lines. In construct phMR53, the native promoter, alternative start sites, 5′ UTR, and introns were removed. The CaMV 35S promoter and 130-bp leader sequences were substituted. The mudrA 3′ UTR (polymorphic region) was truncated and fused to the nopaline synthase (nos) terminator.
mudrA encodes diverse transcripts resulting from alternative transcription initiation, intron splice failure, and alternative polyadenylation sites (Figure 1B; Hershberger et al., 1995). Thus, mudrA produces transcripts with polymorphic 5′ and 3′ untranslated regions (UTRs) and a coding region predicted to produce at least two large polypeptides of 736 and 823 amino acids. Although MuDR was identified in 1991 (Chomet et al., 1991; Hershberger et al., 1991; Qin et al., 1991) and fully sequenced (Hershberger et al., 1991; James et al., 1993; Hsia and Schnable, 1996), there has been no progress in using a transgenic approach to determine which transcripts are sufficient to catalyze or regulate specific Mu activities. The major limitation is that all mudrA plasmids grown in Escherichia coli develop frameshift or deletion mutations (reviewed in Bennetzen, 1996). For this reason, it has also not been possible to transfer Mutator activity to heterologous hosts for transposon-tagging experiments.
In this article, we demonstrate that bacteriophage M13 is a suitable vector to manipulate mudrA and to make transgenic plants. We then use transgenic maize to test the function of the fully spliced transcript, capable of encoding the 823– but not the 736–amino acid protein. When expressed in yeast, this cDNA encodes a 120-kD polypeptide that has been shown to specifically bind a Mu TIR sequence in vitro (Benito and Walbot, 1997). To determine whether the polymorphic noncoding sequences of mudrA are required for developmental regulation, we excluded the alternative 5′ and 3′ UTRs from the transgene and replaced the native promoter with a heterologous cauliflower mosaic virus (CaMV) 35S promoter (Figure 1C). In this report, we analyze transgenic plants expressing full-length and truncated versions of this cDNA to determine whether these transgenes are sufficient to program the four molecular and developmental activities catalyzed by MuDR: demethylation, somatic excision, germinal insertion, and epigenetic reprogramming.
RESULTS
mudrA Clones Are Unstable as Plasmids in E. coli but Stable in M13
Since the identification of MuDR, several groups independently reported the frustrating inability to maintain any mudrA plasmid cDNA or genomic clone in E. coli (reviewed in Bennetzen, 1996). The mudrA genomic clone with introns is toxic, perhaps as a result of internal initiation in the second exon at two Shine-Dalgarno-like sequences at +938 and +1437 (numbering according to Hershberger et al., 1991); the resulting translational product of up to 619 amino acids includes the bacterial transposase–related region (Figure 1A). Attempts to stabilize mudrA in low-copy or transcriptionally repressed pET vectors (R.J. Hershberger, R. Taylor, and V. Walbot, unpublished results) or in Agrobacterium tumefaciens (A. Lloyd, C.D. Goodman, and V. Walbot, unpublished data) have failed; to date, all sequenced plasmids have contained internal deletion or frameshift mutations that disrupt the second exon long open reading frame. The mudrA cDNA corresponding to fully spliced transcript is stable in Saccharomyces cerevisiae in a low-copy vector (Benito and Walbot, 1997); however, the low yield of the yeast plasmid made it difficult to manipulate or use for the biolistic transformation of maize callus. By restriction enzyme analysis, we first discovered that this cDNA clone was stable in an M13 vector through two rounds of cloning. Four randomly chosen colonies produced full-length translation products in vitro and therefore did not contain frameshift mutations; one of these was sequenced and contained no mutations. We conclude that M13 is a useful vector for the cloning of this otherwise toxic gene.
Analysis of Transformed Callus Lines
We used biolistic transformation of HiII (hybrid of two inbred lines; A188 × B73) type II embryogenic callus (Armstrong and Green, 1985) to make transgenic maize (Fromm et al., 1990; Gordon-Kamm et al., 1990). This genotype lacks intact copies of MuDR by DNA gel blotting or by sequence analysis of polymerase chain reaction (PCR) products from degenerate MuDR elements (G. Rudenko and V. Walbot, manuscript in preparation). As shown in Figure 2A, hybridization analysis failed to detect mudrA or mudrB RNA transcripts. The M13-based mudrA expression construct was cotransformed with Bar gene plasmid pAHC20 (Christensen and Quail, 1996), which confers Basta (AgrEvo USA, Apple Valley, MN) herbicide resistance (De Block et al., 1987; Thompson et al., 1987). RNA from herbicide-resistant calli was analyzed for expression of mudrA by RNA gel blot hybridization. Of 64 herbicide-resistant lines, 18 expressed reasonable levels of the expected 2.7-kb mudrA transcript, and these were termed cA (for cDNA mudrA) lines.
Figure 2.
Test for Interaction of CaMV 35S–mudrA—Encoded Full-Length and Truncated Proteins with the MURA DNA Binding Sites of Mu1 and Mu2 TIRs in Embryogenic Callus.
(A) RNA gel blot hybridization analysis of mudrA transgene transcripts in maize leaves from T0 plants. Lines tested correspond to plants regenerated from each callus line shown in (B). Line dcA1 was probed with the mudrA+B (BX1.0) probe (Figure 1A), which hybridizes with the 3′ end of mudrA. All other lanes were probed with the 1.3-kb 5′ mudrA probe (Figure 1A). The estimated sizes of the truncated transgenes, based on RNA gel blot analysis, are shown at left. At bottom is the ethidium bromide–stained formaldehyde agarose gel indicating the 28S rRNA.
(B) At left is a model to explain the effect of MURA binding on the methylation status of HinfI sites in the TIRs of Mu1 and Mu2. MURA binds to an ∼30-bp region overlapping the HinfI site in each TIR (Benito and Walbot, 1997). At right is a DNA gel blot of callus DNA digested with methylation-sensitive HinfI and hybridized with probe pA/B5, which recognizes both Mu1 and Mu2. DNA was prepared from calli 10 to 12 weeks after stable transformation with the CaMV 35S–mudrA vector.
We also performed RNA gel blot screening to identify lines with 5′ or 3′ deletions in mudrA, because a deleted transgene with partial function could help define essential protein domains. We isolated eight dcA (for deletion of cA) lines that hybridized to the 1.3-kb 5′ mudrA probe (Figure 2A) but not to the probe corresponding to the terminal 350 bp of the gene. An additional line, dcA1, expressed an abundant ∼650-bp transcript, which hybridized to a probe spanning the last 350 bp of mudrA but not to a probe corresponding to the first 1.3 kb of the gene (Figure 2A). These results are consistent with a >2-kb 5′ deletion in the dcA1 line. Thus, after biolistic transformation, the random breakage of vector DNA during chromosome integration can generate a useful deletion series of the transgene.
Expression of the 823–Amino Acid MURA-Encoding cDNA Results in Demethylation of Its Binding Site in Embryogenic Callus
A hallmark phenotype of inheriting an active MuDR is DNA demethylation at the HinfI site of Mu1 TIRs (Chandler and Walbot, 1986; Bennetzen, 1987). This restriction site overlaps the MURA binding site defined in vitro (Benito and Walbot, 1997). It is hypothesized that binding of MURA in vivo blocks maintenance methylation. Because the HiII stock contains several endogenous, methylated copies of Mu1 and Mu2 (Chandler et al., 1986; Masterson et al., 1988), we tested the HinfI methylation status of these elements in stably transformed embryogenic calli before we proceeded to regenerate whole plants. DNA gel blot analysis was performed on callus DNA isolated 10 to 12 weeks after bombardment. As shown in Figure 2B, callus lines expressing low, medium, or high levels of mudrA were fully demethylated at the HinfI sites, whereas Mu elements in untransformed callus DNA remained more methylated. Therefore, the mudrA transgene contributed a known MuDR phenotype.
Surprisingly, four 3′ truncated dcA callus lines also underwent demethylation at the Mu1 and Mu2 TIRs. The 5′ deleted dcA1 line remained methylated, suggesting that the initial introduction of M13 full-length mudrA cDNA or lingering extrachromosomal transgene DNA was not responsible for the HinfI site demethylation in these lines. To rule out the possibility that the dcA lines also had a poorly expressed intact mudrA transgene, we further analyzed line dcA74 containing the shortest dcA transgene. By reverse transcription (RT)–PCR, a 3′ primer at +1090 amplified mudrA transcript from this line, whereas a downstream primer at +1412 failed to amplify a band; in the same experiment, full-length mudrA-expressing plants produced product from both primers (data not shown). Therefore, line dcA74 does not possess an intact, expressed mudrA transgene. The simplest explanation of the demethylation by lines with mudrA 3′ deletions is that the DNA binding domain of MURA is present at its N terminus; this hypothesis must be confirmed in vitro by gel shift experiments.
The demethylation data imply that MURA can bind to its target motif in the absence of other MuDR-encoded proteins, as expected from gel shift binding assays in vitro using the 823–amino acid protein (Benito and Walbot, 1997). Rapid TIR demethylation in embryogenic callus further suggests that the MURA transposase is both properly translated and competent to bind its target in undifferentiated cells. This is interesting, because Mu element excisions are restricted to terminally differentiated somatic cells undergoing their last few cell divisions (Levy and Walbot, 1990), even though MuDR transcripts and MURB protein are abundant in meristematic tissues (Donlin et al., 1995; Hershberger et al., 1995; Joanin et al., 1997). Among hypotheses to explain the absence of excisions early in development are the following: that MURA transposase is not translated, or that it does not interact with its TIR binding site in undifferentiated dividing cells. Our callus demethylation data indirectly rules out both of these theories.
Characterization of Regenerated Transgenic Lines
Because full-length and 3′ truncated mudrA transgenes resulted in Mu element TIR demethylation, a subset of cA and dcA lines (seven and seven, respectively) was regenerated and carried forward into the T3 and T4 generations; two cA lines and two dcA lines with low plant fertility or immediate transgene silencing were discarded. As shown in Table 1, only lines in which the presence of mudrA transcripts perfectly cosegregated with herbicide resistance were chosen. Herbicide resistance is indicated by a (+). We anticipated large-scale field experiments, and a tightly linked, scorable marker facilitated tracking the transgene locus. In the lines selected, the herbicide resistance plasmid appears to have inserted at the same single locus as the mudrA transgene. For example, in line cA36, a total of 36 outcross progeny at generation T1 were analyzed for herbicide resistance and expression of mudrA by RT-PCR. Of these, 16 were herbicide resistant, and the same 16 plants but no others expressed mudrA. In our experience, at least 90% of cobombarded plasmids integrate at tightly linked sites, although one member of the pair may not be expressed (data not shown).
Table 1.
Characterization of Genetic Lines Used in This Study
| Linkage Test
|
Generation T1 to T4 Behavior
|
DNA Gel Blot Analysis
|
|||
|---|---|---|---|---|---|
| Line | Cosegregationa of CaMV 35S–mudrAbwith Herbicide Reisistancec | Transgene/Herbicide Stability |
Segregation of Herbicide Resistance |
No. of Copies of Bar Gened |
No. of Copies of CaMV 35S–mudrA Transgenee |
| cA36 | 16+/16+ (36) | Poor | <1:1 | >11 | 4 |
| cA75A | 17+/17+ (31) | Good | 1:1 | >4 | 4 |
| cA75B | 17+/17+ (27) | Excellent | 1:1 | >4 | 4 |
| dcA28 | 3+/3+ (5) | Good | 1:1 | >8 | >7 |
| dcA30 | 4+/4+ (7) | Good | 1:1 | 2 | >2 |
| dcA31 | 5+/6+ (9)f | Moderate | 1:1 | >5 | >4 |
| dcA74 | 4+/4+ (10) | Poor | <1:1 | 1 | >2 |
The first plus (+) indicates the number of plants that contain the CaMV 35S–mudrA transgene, and the second plus (+) indicates the subset of CaMV 35S–mudrA plants that are also Basta resistant. The number in parentheses indicates the total number of outcross plants tested in the T1 generation.
CaMV 35S–mudrA transgene inheritance tested by RT-PCR or genomic PCR.
Resistance to Basta applied to the leaf surface.
Blot probed with pUC18 DNA to detect the Bar transgene.
Blot probed with CaMV 35S DNA to detect CaMV 35S–mudrA construct.
Indicates possible transgene silencing. The presence of CaMV 35S–mudrA was tested by RT-PCR.
Three transgenic lines with full-length mudrA but distinctive transcript abundance and susceptibility to silencing were used for Mu transposition assays. An RNA gel blot of these lines at the T0 generation is shown in Figure 3. Line cA36 is highly prone to silencing but can produce significant levels of mudrA transcript. Sister lines cA75A and cA75B were independently regenerated and maintained, but they express mudrA transcript at different levels. In addition, line cA75B undergoes epigenetic silencing infrequently compared with line cA75A. In our experience, two plants regenerated from the same maize callus line can express the transgene of interest at very different levels; hence, each T0 plant and its progeny must be analyzed and tracked individually.
Figure 3.
RNA Gel Blot Hybridization Analysis of MuDR and CaMV 35S–mudrA Full-Length cDNA Transgenic Lines.
Control total RNA is from a leaf of a standard inbred tester. MuDR RNA is from an immature ear of an active, high-copy MuDR line. The transgene samples are from mature leaves of T0 plants. Both mudrA and mudrB are from the same blot, probed with the cross-hybridizing mudrA+B (BX1.0) probe. No mudrB transcript was detected in any cA lines. The 28S rRNA panel is from the ethidium bromide–stained agarose gel.
CaMV 35S–mudrA cDNA Programs Somatic Excision of Mu1 Elements
The a1-mum2 allele contains a Mu1 element at the a1 locus (Chomet et al., 1991). In the absence of transposase, a1-mum2 kernels are colorless; as shown in Figure 4A, when crossed with a MuDR source, Mu1 somatic excisions occur and are visualized as reddish purple spots on the kernel aleurone. To test for transgene mudrA-mediated somatic excision, we crossed lines expressing mudrA to a1-mum2 stocks in which the single copy of MuDR originally present had been segregated out of the stock (Chomet et al., 1991; Qin et al., 1991). We asked if CaMV 35S–mudrA could activate Mu1 excisions at the a1-mum2 reporter gene.
Figure 4.
Genetic Test of the Ability of CaMV 35S–mudrA to Catalyze Excisions of Mu1 Elements at the a1 Locus in the Absence of Intact MuDR Elements.
(A) At left is a diagram demonstrating the expected aleurone phenotypes of different A1 genotypes. At right is the genetic experiment in which sibling plants carrying the a1-mum2 excision reporter were crossed to or by transgenic T2 generation plants segregating 1:1 for the transgene. cA+ indicates herbicide resistance and cA− indicates herbicide sensitivity.
(B) At left are the leaf phenotypes of transgenic line cA75B parents 5 to 7 days after application of Basta herbicide. At right are the phenotypes of progeny of crosses between a1-mum2 and +/− transgene lines. The ears are the T4 generation.
(C) Percentage of spotted kernels per ear of cA+ CaMV 35S–mudrA lines. All kernels have one copy of a1-mum2 and one copy of the CaMV 35S–mudrA locus; both genes were transmitted through pollen in crosses onto a1 tester ears. Twenty-five percent of kernels were expected to be spotted. Each bar represents an individual ear.
For each transgenic line, we planted a family (kernels derived from the same ear) segregating 1:1 for the transgene (Figure 4A) and crossed all individuals reciprocally with a line homozygous for a1-mum2 but lacking MuDR. As shown in Figure 4B and summarized in Table 2, all three herbicide-resistant full-length cDNA transgenic lines did catalyze excisions, with the best line, cA75B+, having activity in 22 of 22 ears. Negative control crosses using herbicide-sensitive parents resulted in no spotted kernels in the 50 progeny ears examined (∼13,000 kernels). None of the transgenic lines containing deletions in CaMV 35S–mudrA yielded spotted kernels (0 out of ∼7000 kernels).
Table 2.
Summary of Mu1 Excision Activity at a1-mum2a
| Progeny
|
|||
|---|---|---|---|
| Transgenic Parent | Spotted Ears | Unspotted Ears | Percentage of Ears Spotted |
| cA36− | 0 | 13 | 0 |
| cA36+ | 6 | 15 | 29 |
| cA75A− | 0 | 13 | 0 |
| cA75A+ | 34 | 4 | 89 |
| cA75B− | 0 | 10 | 0 |
| cA75B+ | 22 | 0 | 100 |
| dcA28+ | 0 | 7 | 0 |
| dcA30+ | 0 | 8 | 0 |
| dcA31− | 0 | 8 | 0 |
| dcA31+ | 0 | 8 | 0 |
| dcA74+ | 0 | 2 | 0 |
The experiment is described in Figure 4A.
When a single-copy MuDR a1 line is crossed to an a1-mum2 tester (MuDR a1/a1 × a1-mum2/a1-mum2), 50% spotted kernels are expected and are typically observed (Lisch et al., 1995; Hsia and Schnable, 1996). The expectation for a subset of the transgene crosses (hemizygous CaMV 35S–mudrA, a1-mum2/A1 × a1/a1) was that herbicide-resistant plants should produce 25% spotted kernels, 50% purple, and 25% colorless. As shown in Figure 4C, progeny of cA36+-derived ears exhibited a low excision frequency (26 of 1018 kernels, or 2.6%), whereas 11% of cA75A+ kernels (493 of 4458) and 18% of cA75B+ kernels (406 of 2222) were spotted. Two ears exhibited the expected 25% excision frequency.
We hypothesized that transgene silencing was responsible for the low excision frequency catalyzed by line cA36. In fact, all spotted and unspotted kernel progeny of a cA36+ ear were herbicide sensitive (nine out of nine). The two most spotted of these progeny expressed only low levels of mudrA transcript (data not shown). In contrast, for both cA75A+ and cA75B+, all seven spotted kernels tested gave rise to herbicide-resistant seedlings, each of which was shown to also carry the CaMV 35S–mudrA transgene by PCR and have abundant mudrA expression on RNA gel blots (data not shown). This suggests that in the cA36-derived family shown in Table 2, the bar and mudrA transgenes were in the process of silencing.
To demonstrate conclusively that CaMV 35S–mudrA was directly responsible for the somatic excisions seen, we grew seedlings from spotted a1-mum2/a1 cA75 kernels to maturity and crossed them with an a1 non-Mutator stock. If a MuDR element separate from the CaMV 35S–mudrA transgene element were responsible for the excision phenotype, then independent assortment of an unlinked element or meiotic recombination would separate the two. As shown in Table 3, among 357 spotted progeny kernels tested, nearly all were herbicide resistant, indicating that they inherited the transgene locus. The nine partially herbicide-sensitive plants all inherited the CaMV 35S–mudrA transgene, based on PCR analysis; we conclude that the bar gene was simply silencing in these individuals. Consequently, any contaminating MuDR element would have to be located within 1 centimorgan (P < 0.05) of the transgene in cA75, which is highly unlikely. We consider that these results directly prove that the fully spliced mudrA cDNA is sufficient to program somatic excisions of Mu elements in maize.
Table 3.
Cosegregation of a1-mum2–Conferred Excision Phenotype with Inheritance of the CaMV 35S–mudrA Transgene in Line cA75+a
| Kernel Phenotype of Progeny |
Expected Transgene Inheritance (%) |
Total Tested |
Herbicide Resistant |
Herbicide Sensitive |
Observed Transgene Inheritance |
Phenotype Linkage to Transgene |
|---|---|---|---|---|---|---|
| Random | 50 | 13 | 7 | 6 | 7+/13 | unlinked |
| Spotted only | 100 | 357 | 348 | 9(0)b | 357+/357 | <1 centimorgan (P < 0.05) |
The cross performed was (spotted, a1-mum2/a1, hemizygous CaMV 35S–mudrA) × (unspotted, a1/a1).
Partial herbicide sensitivity. Genomic PCR was performed using CaMV 35S–mudrA—specific primers; the resulting gel was DNA gel blotted and probed with the mudrA probe. All nine plants carried the CaMV 35S–mudrA transgene.
Developmental Timing of Excisions
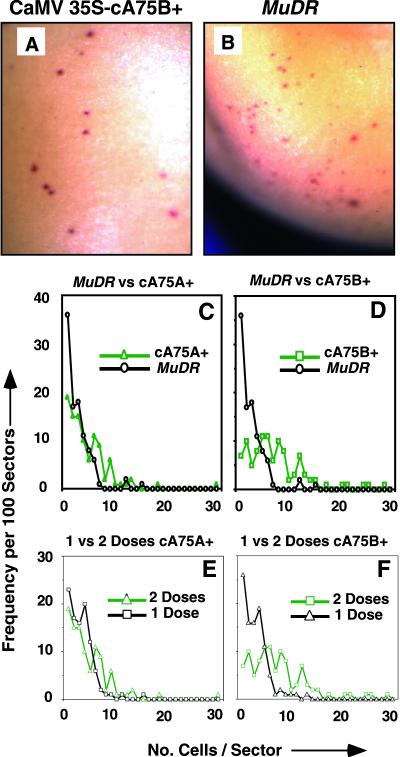
On quick visual inspection, the mudrA cDNA programmed somatic excisions with the same characteristically late developmental timing as active Mutator lines (Levy et al., 1989; Levy and Walbot, 1990). Microscopic scoring of Mu1 excisions at a1-mum2 indicated that >80% of the sectors were composed of ⩽10 cells programmed by either CaMV 35S–mudrA or a single copy of MuDR. As shown in Figure 5, the sector size distribution seen with line cA75A+ was nearly identical to the single-copy MuDR line, whereas a slightly broader distribution was observed in line cA75B+. Even if the latter pattern is a true representation of this line, it reflects a shift in timing by only one or two cell divisions. Slight differences in anthocyanin pigment diffusion could easily account for the small variation seen.
Figure 5.
Comparative Analysis of the Developmental Timing of Excision from Aleurone Cells of Mu1 from a1-mum2 in MuDR and CaMV 35S–mudrA Lines.
(A) cA75B+ aleurone. Most of the excisions shown consist of single cells.
(B) One-copy MuDR aleurone. Most sectors consist of one or two cells.
(C) and (D) Sector size distribution comparison. One hundred randomly chosen sectors were scored per line. The MuDR source was selfed, and kernels contain one to three copies of MuDR plus one to three copies of a1-mum2. The cA75+ line contains two doses of CaMV 35S–mudrA and two to three copies of a1-mum2. The MuDR data in both (C) and (D) are the same. The MuDR kernel had one additional 48-cell sector (not shown on graph).
(E) and (F) Sector size distribution as a function of CaMV 35S–mudrA transgene locus dose. The two-dose data are from (C) and (D), respectively. The one-dose kernels contain one dose of CaMV 35S–mudrA and two to three copies of a1-mum2. In the two-dose kernels, the transgene had been transmitted through the female. Single-dose kernel transgenes had been transmitted through male germinal cells.
In Mutator lines, excision timing is not affected by varying MuDR or excision reporter copy number (Levy and Walbot, 1990), in contrast to the dosage responses of Ac (Brink and Nilan, 1952). It seemed possible that the more variably competent CaMV 35S–mudrA transgenes might exhibit dosage dependence; however, a change from two doses (maternal transmission) to one dose (paternal transmission) had no measurable impact on either excision timing or its frequency (Figures 5E and 5F).
In Mutator lines, excisions are largely limited to somatic cells; germinal revertants have either not been observed in large populations or occur with a frequency of <10−4 (reviewed in Bennetzen, 1996). A germinal excision at a1-mum2 that restores gene expression (a revertant) would be a solid purple kernel or sector of purple kernels on an ear. An initial experiment in which both the a1-mum2 allele and the transgene were transmitted through the female lineage yielded only one out of ∼6800 purple kernels, which we attribute to contamination, because no further instances of putative germinal excisions were observed in the next generation. These results suggest that at best, the CaMV 35S–mudrA transgene catalyzes germinal excisions at a frequency equal to or less than native MuDR stocks.
Excision Frequency Is Not Correlated to mudrA Transcript Abundance
In terms of average excision frequency per spotted kernel, it is visually clear that some ears with a cA75B+ transgene match the most spotted examples of single-copy MuDR lines, whereas the frequency in lines cA36+ and cA75A+ is much lower (data not shown). Nevertheless, it appears that the CaMV 35S–mudrA transgene can fully complement the MuDR-catalyzed excision intensity per kernel.
One difference, however, is that excision frequency is visually more variable in transgenic lines than in a single-copy MuDR line. We hypothesized that the variable levels of excision catalyzed by CaMV 35S–mudrA in some progeny could reflect differences in transgene expression. We compared excision frequency to mudrA transcript levels. Sector number was determined for eight sibling a1-mum2 kernels for both cA75A+ and cA75B+. In line cA75B+, there is a 47-fold range in spot number, from nine spots per kernel to 439, but as shown in Figure 6, there is less than a twofold increase in seedling transcript abundance between them. Eight ranked kernels of line cA75A+ showed similar results (data not shown). Therefore, above the minimum threshold required for somatic excision, sector frequency is not positively correlated with transcript levels. These results suggest that CaMV 35S–mudrA excision activity is post-transcriptionally determined. The caveat in this experiment is that we compared endosperm excision frequency to transcript abundance using RNA prepared from the tips of leaf 4.
Figure 6.
Aleurone Mu1 Excision Frequency Compared with mudrA Transcript Abundance.
All kernels were from a single cA75B+ ear of the T3 generation, and they are ranked by the number of excision sectors in the aleurone, from zero at left to 439 at right. Seedlings were scored for herbicide resistance, and RNA was isolated from the tips of leaf 4. The same RNA gel blot was probed with the 1.3-kb mudrA probe and subsequently with a maize actin probe used as an RNA loading control.
Mu Insertions Are Not Detected in the Germline of CaMV 35S–mudrA Transgenic Plants
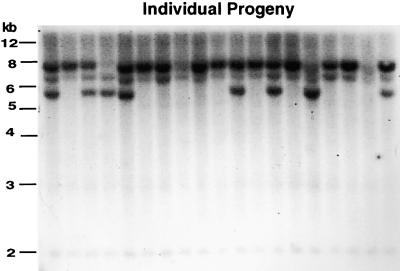
To test the ability of the CaMV 35S–mudrA cDNA to catalyze germinal insertions, we grew spotted kernels of genotype CaMV 35S–mudrA/-;a1-mum2/a1 and crossed them to the a1 tester; these plants had three unlinked copies of Mu1 and the molecularly similar element Mu2. Figure 7 exemplifies the DNA gel blot hybridization screen for new Mu1 or Mu2 insertion fragments in the progeny against the background of segregating parental fragments. No new fragments were detected in 38 progeny, drawn equally from maternal and paternal transmission of the transgene, or in a second test of 34 progeny grown from the most highly spotted kernels. If the CaMV 35S–mudrA transgene can catalyze germinal insertions of Mu elements, then the frequency must be <5% (P < 0.05) per plant or <2.5% per donor Mu1 element per generation. This is less than the weakest active Mu line in which a single copy of MuDR in a poor chromosomal position catalyzes new insertions at a frequency of 6 to 14% per Mu1 element per generation (Lisch et al., 1995).
Figure 7.
DNA Gel Blot Screen for Mu1 and Mu2 Germinal Insertions in the Progeny of CaMV 35S–mudrA Plants.
Line cA75B+ progeny were used, and all kernels were spotted. DNA was digested with HindIII and probed with the Mu1/Mu2–specific probe pA/B5. Only parental fragments, visualized as segregating bands in the progeny, are seen. DNA gel migration size standards are indicated at left.
CaMV 35S–mudrA Transgene Reactivates Mu1 Excisions in an Epigenetically Silenced MuDR Background
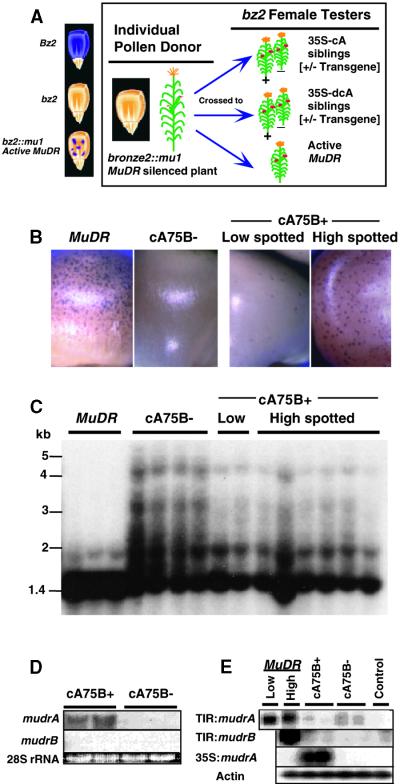
Because the fully spliced mudrA cDNA and its deletion derivatives were sufficient to demethylate Mu1 elements in callus (Figure 2), we asked if they were also sufficient to reactivate epigenetically silenced Mutator lines or whether the transgenes were susceptible to homology-dependent silencing (reviewed in Vaucheret et al., 1998) by silenced copies of MuDR. First, do CaMV 35S–mudrA transgenes restore somatic excision activity in silenced lines? We crossed the transgenes into 10 families containing multiple copies of silenced MuDR elements and visually scored reactivation of excision of a Mu1 element at a pigment locus (bronze2) that had been somatically stable (zero spots per ear) or silencing (zero to 20 spots per kernel) in the previous generation. As illustrated in Figure 8A, the silenced MuDR sources were homozygous bz2::mu1 and were crossed as pollen onto three types of bz2 females: those hemizygous for the CaMV 35S–mudrA transgene (cA+ or dcA+), their herbicide-sensitive siblings (cA− or dcA−) (negative controls), and a multicopy active MuDR source (positive control). As shown in Table 4, 17 out of 20 ears hemizygous for the cA75A+ and cA75B+ transgenes showed reactivation of somatic excisions at bz2::mu1 (1328 of 7071 kernels, or 19%), whereas none of the 18 herbicide-sensitive siblings produced any spotted kernels (zero of 5125 kernels).
Figure 8.
Stability of the CaMV 35S–mudrA Transgene in the Presence of Multiple Silencing MuDR Elements.
(A) At left is a cartoon of aleurone genotypes and phenotypes at the Bz2 locus. At right is a schematic of the genetic experiment.
(B) Kernel progeny showing reactivation of excisions at bz::mu1 from a previously silenced MuDR line. Pollen from a silenced bz2::mu1 individual was crossed onto bz2 testers containing either high-copy active MuDR or the cA75B+ transgene. The cA75B+/− kernels are siblings from the same ear.
(C) DNA gel blot analysis of the methylation status at HinfI sites within the MURA binding sites in the TIRs of Mu1 and Mu2 elements (see Figure 2 for assay description). Kernels pictured in (B) were planted; DNA from leaf 4 was digested with HinfI, and DNA gel blots were probed with the Mu1/Mu2–specific probe pA/B5. DNA gel migration size standards are indicated at left.
(D) RNA gel blot analysis of immature ear tissue from plants derived from cA75B+/− kernels shown in (B) and analyzed in (C). The mudrA- or mudrB-specific probes were hybridized against 15 μg of total RNA. Each lane represents ear RNA pooled from two plants.
(E) RT-PCR analysis of cA75B+/− immature ears. Samples are from the plants analyzed in (B) through (D). This analysis was designed to detect the presence of reactivated mudrB (TIR:mudrB) transcripts from the silenced MuDR elements and to distinguish between mudrA transcripts originating from the CaMV 35S–mudrA transgene or from the silenced MuDR (TIR:mudrA) elements. For RT-PCR, we used promoter-specific 5′ PCR primers. PCR products were DNA gel blotted and probed with the mudrA-, mudrB-, or actin-specfic probes. The MuDR sample is from a high-copy MuDR line. The control is a non-MuDR tester. Actin primers were included during RT-PCR as an internal control.
Table 4.
Reactivation of Somatic Excisions at bz2::mu1a
| Parent
|
Progeny
|
||
|---|---|---|---|
| Female Tester | Spotted Ears | Unspotted Ears | Percentage of Ears Reactivated |
| cA75A− | 0 | 9 | 0 |
| cA75A+ | 3 | 3 | 50 |
| cA75B− | 0 | 9 | 0 |
| cA75B+ | 14 | 0 | 100 |
| cA36− | 0 | 10 | 0 |
| cA36+ | 0 | 13 | 0 |
| dcA31− | 0 | 1 | 0 |
| dcA31+ | 0 | 2 | 0 |
| dcA74− | 0 | 3 | 0 |
| dcA74+ | 0 | 1 | 0 |
| MuDR | 13 | 1 | 90 |
The experiment is described in Figure 8A.
On a per ear basis, the reactivation by cA75+ (85%) was comparable to reactivation by a source of multicopy, active MuDR (90%). On a per kernel basis, 21% of the maximum 50% cA75B+ kernels were spotted (934 of 4461), whereas 54% of the maximum 100% multicopy MuDR kernels were spotted (937 of 1740). In contrast, none of 13 ears carrying the epigenetically unstable cA36+ transgene reactivated (zero of 3812 kernels). In a small test, neither of two previously described truncated transgene lines reactivated somatic excision: dcA31+ (zero of 862 kernels) and dcA74+ (zero of 127 kernels). An additional mudrA transgenic line carrying a 1.8-kb 3′ truncation, dcA76+, also failed to reactivate excision at bz2::mu1 (zero of 2811 kernels). Therefore, in this experiment, a stable, full-length mudrA transgene, but not deleted transgenes, was able to reactivate Mu1 element excisions in epigenetically silenced MuDR lines.
Methylation of Mu1 TIRs is a molecular correlate of MuDR epigenetic silencing. Therefore, by DNA gel blot hybridization analysis, we asked if expression of the full-length mudrA transgene could demethylate Mu1 element TIR HinfI sites, which had become methylated after MuDR silencing. From one cA75B+ ear (segregating 1:1 for the transgene) that had inherited a pool of methylated Mu1 and silenced MuDR elements, unspotted, herbicide-sensitive progeny showed a high degree of TIR methylation in leaf 4 DNA. In contrast, spotted, herbicide-resistant siblings showed partial to nearly complete DNA demethylation at the Mu1/Mu2 TIRs in leaf 4 DNA samples (Figures 8B and 8C). Although demethylation by the transgene was weaker than by the active MuDR source, expression of the transgene clearly resulted in the demethylation of multiple Mu elements. Thus, based on excision and demethylation, CaMV 35S–mudrA does reactivate epigenetically silenced Mutator lines.
Does the transgene first reactivate MuDR and then become silenced by homology-dependent transgene silencing? We asked if the CaMV 35S–mudrA transgene had reactivated transcription of epigenetically silenced MuDR elements in the same spotted or control kernel siblings used in the demethylation test. As shown in Figure 8D, we performed RNA gel blot analysis on these plants using total RNA from pooled immature ears, a tissue previously shown to express very high levels of MuDR transcript in active Mutator lines (Hershberger et al., 1995). As expected, immature ears from cA75B+ but not cA75B− plants expressed high levels of mudrA transcript. RT-PCR demonstrated, however, that the source of all transcript was the CaMV 35S–mudrA transgene and not reactivated MuDR elements (Figure 8E). In addition, RNA gel blot analysis and RT-PCR experiments using total RNA from immature ear tissue failed to detect mudrB transcript, which would be expected from restoration of MuDR transcription (Figures 8D and 8E). Therefore, the CaMV 35S–mudrA transgene does not appear to transcriptionally reactivate the resident epigenetically silenced MuDR elements during the somatic growth of the plant, although the transgene itself remains transcriptionally active.
The continued transcriptional silencing of MuDR elements implies that much of the excision and demethylation activity seen in these progeny was programmed directly by the CaMV 35S–mudrA transgene and not by reactivating MuDR elements. Further support for this comes from the result that the deleted transgenes were unable to cause excisions in inactive Mutator lines (Table 4), even though they could demethylate Mu1 elements (Figure 2B). Because the full-length mudrA transgene did not reactivate transcription of silenced MuDR elements but did cause the demethylation and excision of Mu1/Mu2 elements, we conclude that the transgene remains functional in nuclei containing homologous silenced copies of MuDR. Therefore, CaMV 35S–mudrA is not measurably silenced, transcriptionally or post-transcriptionally, by these homologs.
DISCUSSION
We have successfully generated transgenic maize expressing the Mutator transposase MURA. Experiments were made possible by the discovery that whereas the mudrA gene is toxic to bacteria in plasmid form, it is stable in M13. Direct DNA transfer of M13 clones may finally permit Mu transposon tagging in other species and determination of whether maize-specific host factors are required for the regulation of Mutator phenomena.
MuDR encodes two genes, mudrA and mudrB, both of which produce multiple transcripts (Figure 1B). We have demonstrated that the fully spliced mudrA cDNA coding region (Figure 1C), predicted to encode 823 amino acids, is sufficient to catalyze a high frequency of developmentally late somatic excisions in transgenic maize (Figures 4 and 5). Consequently, transcriptional and post-transcriptional regulation acting through the 5′ or 3′ UTRs and intron retention are not required to set excision timing. The 2.7-kb cDNA clone complements two other known functions of an active 4.9-kb MuDR element: demethylation of Mu TIRs (Figure 2B) and partial reactivation of epigenetically silenced Mutator lines (Figure 8 and Table 4). We find no evidence that the cDNA clone under the control of a CaMV 35S promoter catalyzes germinal insertions (Figure 7).
CaMV 35S–mudrA Is Sufficient to Catalyze Somatic Excisions
We have demonstrated that the CaMV 35S–mudrA transgene programs a high frequency of Mu1 somatic excisions from the anthocyanin pigment locus a1 (Figure 4 and Table 2) and, more recently, from two maize Lc::mu1 transgenic loci (M.N. Raizada and V. Walbot, unpublished results). By a genetic test, excisions at a1-mum2 appear to be directly caused by the CaMV 35S–mudrA cDNA transgene and not by a contaminating or cryptic MuDR element (Table 3). The cDNA corresponds to the fully spliced mRNA, which represents ∼61% of mudrA transcript in multicopy MuDR seedlings (Hershberger et al., 1995). The transgene is predicted to encode an 823–amino acid protein, but we cannot rule out the involvement of smaller polypeptides resulting from internal initiation. By design, the alternative 736–amino acid protein predicted to be translated when mudrA intron 3 is retained could not be produced in our transgenic lines. Intron 3 has an intriguingly rare 5′ splice site consensus sequence (Jackson, 1991) and fails to splice in ∼20% of seedling transcripts (Hershberger et al., 1995).
Somatic excision frequency from a1-mum2 is similar when the transposase is supplied by CaMV 35S–mudrA or a single copy of MuDR. Therefore, MuDR-encoded MURB is not required for frequent somatic excision nor for its timing. The caveat in our conclusion is that all maize lines tested contain MURB polypeptides expressed from mudrB homologs (hmudrB); because some homologs contain only a single amino acid substitution, they could conceivably substitute for wild-type MURB (G. Rudenko and V. Walbot, unpublished results).
Plants Expressing the mudrA cDNA Retain Late Excision Timing
A striking feature of Mutator activity is that excisions are restricted to the final cell divisions of somatic tissues. This developmental control was hypothesized to result from programmed retention of intron 3, use of alternative transcription or polyadenylation sites, or translational control exerted through the 5′ leader sequence (Hershberger et al., 1995). These possibilities are ruled out by our results, because these features are missing from the expression vector. In maize, the CaMV 35S promoter has strong activity in both mature tissues and meristematic cells, such as at the root apex (Omirulleh et al., 1993), and yet it programs the late excision timing characteristic of Mutator.
Other hypotheses to explain late excision timing are that mudrA is not translated in meristematic cells or that MURA does not interact with its target chromatin site in these cells. For example, during human V(D)J recombination to generate immunoglobin genes, recombinase activity is controlled by changes in chromatin structure (Stanhope-Baker et al., 1996). Benito and Walbot (1997) previously demonstrated that the 823–amino acid MURA polypeptide is a DNA binding protein in vitro, specific to the TIRs. In this study, we have demonstrated in vivo that mudrA expression results in the demethylation of the MURA binding site in both embryogenic callus cells and differentiated leaf cells, implying that MURA is in fact translated and interacts with its DNA target in both cell types (Figure 2). It appears, therefore, that Mu excision timing is regulated post-translationally.
Our callus result is consistent with two previous studies (James and Stadler, 1989; Planckaert and Walbot, 1989) that found that Mu1 elements in calli derived from active Mutator plants can remain stably unmethylated at the HinfI TIR sites for many months in tissue culture. Furthermore, plants regenerated from these calli have unmethylated TIRs. Cultures derived from an inactive Mutator line retain largely methylated TIRs in culture and during plant regeneration. Collectively, all results suggest that the methylation status at the TIR HinfI sites reflects the presence or absence of MuDR proteins, even in meristematic cells.
The control of transposition timing in plants is poorly understood (reviewed in Fedoroff, 1989). An increase in copy number leads to later excisions in the case of Ac (Brink and Nilan, 1952; Scofield et al., 1993) but earlier excisions by Dotted (Coe and Neuffer, 1977). We speculated that our transgenic MURA source might be present at subthreshold levels. When we altered the parental transmission and hence gene dosage of the CaMV 35S–mudrA transgene, we found no change in excision timing (Figures 5E and 5F). This result is in accordance with all previous observations in which excision timing is independent of MuDR copy number. With a reliable and active MURA transgene (line cA75B+) in maize, which is incapable of self-transposition, it may now be possible to select trans-acting excision timing mutants. To date, the only alteration in Mu timing is a single line with the bz1::mu1 reporter gene in which excision is much earlier than normal in the aleurone (Walbot, 1992).
Excision Frequency Is Not Correlated to mudrA Transcript Levels
Although MuDR transcript levels vary widely, most Mutator lines catalyze the excision of Mu elements in the aleurone at comparable frequencies. In this report, we have directly demonstrated that there is not a correlation between mudrA transcript levels and excision frequency. We propose that an additional post-transcriptional, rate-limiting step, conferred by a host-encoded factor or a feature of the mudrA coding region, controls excision frequency. One possibility is the third base codon usage of MuDR exons, which is very biased toward A or U, as found in rarely expressed maize genes (Fennoy and Bailey-Serres, 1993); it is possible that this rare codon usage contributes to the lack of correlation observed between MuDR transcript and activity levels. Ac and Spm also exhibit biased codon usage (Fennoy and Bailey-Serres, 1993). The poor translation of transposase transcripts may be an adaptive feature to buffer the maize host from quantitative changes in transcript levels resulting from transposon duplications.
No Evidence for Germinal Insertions
We detected no Mu germinal insertions in 72 progeny tested (Figure 7). This result could be interpreted as a requirement for MURB or another form of MURA. It is interesting that the d201 MuDR deletion line reported by Lisch et al. (1999), which has a complete mudrA gene but lacks mudrB, also failed to cause germinal insertions of Mu elements in 193 plants tested. However, Mu1 elements in line d201 retain HinfI site methylation, and somatic excision is less robust than in our CaMV 35S–mudrA lines. Therefore, we propose that silencing rather than the lack of mudrB per se might be responsible for the lack of germinal insertions observed by Lisch et al. (1999).
In our transgenic lines, the lack of germinal insertions may also be due to poor expression of the transgene in germinal cells. Whereas the CaMV 35S promoter has strong expression in maize leaves, there is almost no expression in maize microspores in transient assays (Fennell and Hauptmann, 1992; Jardinaud et al., 1995). Furthermore, there is no expression in mature pollen of stably transformed Arabidopsis and only very weak expression in transgenic tobacco pollen (Wilkinson et al., 1997). By contrast, the MuDR TIR promoters have weak activity in suspension cells (Benito and Walbot, 1994) but produce abundant transcripts in pollen (M.N. Raizada and V. Walbot, manuscript submitted for publication).
The CaMV 35S–mudrA transgene may also be translated at levels less than those required for germinal insertion but not for somatic excision. The construct has an altered translation start sequence and possesses no introns, which are known to boost expression of transgenes in maize (Callis et al., 1987). In addition, the translation start sequence of mudrA in the transgene has been changed from its native CCAUGG to UCAUGG; in an analysis of 85 maize genes, this first-position cytosine was found in 58% of AUG start codons, whereas uracil, the rarest base used, was found only 8% of the time (Luehrsen and Walbot, 1994).
Aside from germinal insertions, we recently demonstrated that MuDR catalyzes a high frequency of somatic insertions by using a transgenic Mu1 element that permits us to plasmid rescue new Mu insertions (RescueMu::Lc; M.N. Raizada and V. Walbot, manuscript in preparation). After crossing CaMV 35S–RescueMu::Lc to CaMV 35S–mudrA, however, we were unable to detect somatic insertions in leaves with this assay. This leaves open the possibility that wild-type mudrB may be required for any Mu insertion activity.
mudrA Transgene Is Active in the Presence of Methylated, Silencing Copies of MuDR
Host-induced silencing of viruses and transgenes is seen as part of an endogenous defense mechanism against pathogens and transposons (reviewed in Martienssen, 1996; Vaucheret et al., 1998). A viral transgene can silence its homolog in a subsequently introduced virus (Ratcliff et al., 1997). In this report, we expressed a plant transposase as a transgene in its native host, in a background containing endogenous copies of the transposon. We predicted dramatic transgene silencing for several reasons. First, although the mechanisms of silencing are not understood, it is certain that sequence homology recognition is a key component (Napoli et al., 1990; Meyer et al., 1993; Park et al., 1996). It is also hypothesized that transgene repeats and high RNA expression can trigger silencing (reviewed in Vaucheret et al., 1998). Finally, multicopy MuDR lines also produce abundant transcript (Hershberger et al., 1995), and unlinked MuDR elements undergo simultaneous methylation and cosilencing even in the absence of a homologous transgene (reviewed in Walbot, 1991; Fedoroff and Chandler, 1994).
We therefore speculated that a multicopy mudrA cDNA, with nearly 100% identity to the long coding regions of endogenous, presumably methylated copies of MuDR, would additively result in “super-silencing” of the transgene. However, when we introduced the mudrA cDNA into a background containing multiple copies of actively silencing and recently silenced copies of MuDR (Figure 8A), the transgene retained excision activity (Figure 8B and Table 4), TIR demethylation activity (Figure 8C), and expression on RNA gel blots (Figures 8D and 8E). Therefore, we found no evidence of homology-induced transgene silencing transcriptionally or, by inference, post-transcriptionally.
One hypothesis to explain the apparent lack of epigenetic interaction of the CaMV 35S–mudrA with endogenous MuDR elements is that MuDR silencing may require the TIRs. The mudrA and mudrB promoters are nearly identical (166 of 168 bp) and are located within the TIRs. When silencing occurs, both mudrA and mudrB transcripts are undetectable (Hershberger et al., 1991), and methylation of the TIRs is associated with this silencing. The TIRs also encode the 5′ UTRs of all mudrB transcripts and half of the mudrA transcripts (those initiating at position +169). These 5′ UTRs share 40 of 47 bases of identity (Hershberger et al., 1995). Consequently, simultaneous silencing of mudrA and mudrB is likely accomplished through transcriptional and/or post-transcriptional mechanisms acting through the TIRs. If this is true, our transgene, devoid of its native TIR, would not be affected by the silencing process. As a practical implication, such an epigenetically stable source of MURA might prevent silencing of Mu elements during future transposon-tagging experiments by preventing maintenance methylation of the TIRs.
Prospects for Gene Tagging and Understanding MuDR in Transgenic Plants
Although the CaMV 35S–mudrA construct used in this experiment does not program a high frequency of germinal insertions, we are very optimistic that it will be possible to transfer Mutator activities to heterologous hosts. In maize, we know that a single, full-length MuDR element programs germinal insertions (Lisch et al., 1995). With the use of M13, it should now be possible to clone and deliver genomic mudrA and even entire MuDR elements to other organisms by biolistic transformation. Versions of maize transposons Ac and Spm are used in efficient transposon-tagging lines in Arabidopsis and tomato (Osborne and Baker, 1995; Wisman et al., 1998). It will be intriguing to assay the developmental timing of Mu excisions and insertions in these or other hosts. In fact, both Arabidopsis (GenBank) and rice (Eisen et al., 1994; Yoshida et al.,1998) contain mudrA-like sequences, suggesting that Mu elements might have been active previously in diverse dicot and monocot species.
METHODS
Plasmid Vectors
phMR53 (cA transgene) is the mudrA cDNA clone that was reconstructed in vitro and stabilized in a low-copy yeast vector, as previously described (Benito and Walbot, 1997). The yeast plasmid was then used as the template for polymerase chain reaction (PCR) amplification. The 5′ primer sequence was GCGAATTCATGGACTTGACGCCCAG and the 3′ primer sequence was CGGAATTCCTAC-ATAACAGTCTTACAAC. Amplification conditions were 95°C for 45 sec, 51°C for 1 min, and 74°C for 3 min for 28 cycles using Pfu polymerase (Stratagene, La Jolla, CA). The cDNA clone starts at position +449 of the published MuDR sequence (Hershberger et al., 1991) at the putative ATG translational start site, changing the sequence context from CCATGG to TCATGG. The clone ends at position +3209 at the end of cDNA clone C4 and includes 63 bp of the native 3′ untranslated region (UTR) but none of the native polyadenylation signals (Hershberger et al., 1995). The amplified product was subcloned into the EcoRI site of Phagescript SK (Stratagene) to give clone phMR49. All cDNA ligation products checked inserted in the same orientation in the Phagescript vector, as did a separate cloning of a nearly full-length genomic mudrA clone. Attempts made to ligate the cDNA or genomic clones in the opposite orientation were unsuccessful. The double-stranded replicative form of M13 was first in vitro transcribed using T3 RNA polymerase and in vitro translated using rabbit reticulocyte extract to detect the presence of frameshifted clones. All clones gave full-length protein products. The cDNA was then subcloned as an EcoRI fragment from phMR49 into the same sites in phMR52 (M13) to give phage MR53. The cDNA thus contains a new 130-bp cauliflower mosaic virus (CaMV) 35S leader sequence. Large amounts of double-stranded DNA were isolated 4 hr after transfection of XL-1 Blue cells ( ; Stratagene). Both the transfection time and the number of times the clone was transfected were minimized to prevent the selection of internal deletions in M13. The final clone was fully sequenced as double-stranded DNA and found to be intact. The same sample of DNA used for sequencing served as the source for biolistic transformation.
; Stratagene). Both the transfection time and the number of times the clone was transfected were minimized to prevent the selection of internal deletions in M13. The final clone was fully sequenced as double-stranded DNA and found to be intact. The same sample of DNA used for sequencing served as the source for biolistic transformation.
phMR52 is a potentially useful M13 cassette vector for the cloning and expression of toxic genes in plants. The nopaline synthase (nos) terminator was subcloned from plasmid pR as an XbaI-EcoRI fragment, filled in with the Klenow fragment of DNA polymerase I, and then blunt-ligated to the HincII site of pBluescript KS+ (Stratagene) to give clone pMR50. The CaMV 35S promoter from +7072 to +7565 (Franck et al., 1980) from pJD255 was then added as a SstI-PstI subclone into the same sites of pMR50 to give clone pMR51, a new CaMV 35S–nos cassette in pBluescript KS+. This clone includes 130 bases of the native CaMV 35S leader sequence. The CaMV 35S–nos sequence was then subcloned as a XhoI-SacI fragment into the same sites in Phagescript SK to give phage MR52.
pAHC20 is the ubiquitin promoter–Bar herbicide resistance plasmid that was kindly provided by P. Quail (Plant Gene Expression Center, Albany, CA) (Christensen and Quail, 1996).
Stable Maize Transformation
A detailed protocol can be found at http://www.stanford.edu/~walbot/StableMaizeTransf.html. Briefly, embryogenic A188 × B73 (HiTypeII) calli (Armstrong and Green, 1985; Armstrong, 1994) were osmotically treated (Vain et al., 1993) and then transformed using the PDS 1000HE biolistic device (BioRad, Hercules, CA) at 650 psi; this was repeated at 1100 psi in a vacuum of 27 psi (Gordon- Kamm et al., 1990; Sanford et al., 1993). The distance from the rupture disc to the macrocarrier was 1 cm, and that from the mesh screen to the target was 5.9 cm. For three bombardments, 25 μg total of phMR53 and pAHC20 was coprecipitated in equimolar quantities onto 2 mg of 1 μM spherical gold particles (Alameda Scientific Instruments, Richmond, CA) using the procedure of Wan et al. (1994). Transformed calli were selected on 3 mg/mL bialaphos (Meiji Seika Kaisha Ltd., Yokohama, Japan) (Spencer et al., 1990). Individual resistant callus lines (designated cA1, cA2, cA3, etc.) were checked by RNA gel blot hybridization for the presence of transgene expression and selectively regenerated. The initial regenerated plants were called T0, whereas the first seed belonged to the T1 generation.
Leaf Herbicide Test
To test for bialaphos resistance, a 5-cm-diameter marked leaf surface was painted with 0.75% glufosinate ammonium (Ignite 600, 50% solution; Hoescht, Montreal, Canada) with 0.1% Tween 20 using a Q-tip. The area was visually scored for the presence or absence of necrosis 5 to 7 days later.
Herbicide and MuDR Transgene Cosegregation and Stability Tests
Transgenic lines were chosen that exhibited a 1:1 cosegregation pattern of leaf herbicide resistance to stable mudrA transgene expression in 8 to 20 T1 progeny of an outcross. Thereafter, herbicide resistance was used to identify plants carrying the mudrA transgene. To examine transgene expression, we performed reverse transcription (RT)–PCR analysis on total leaf RNA isolated using Trizol (Gibco BRL, Rockville, MD). Two micrograms of total RNA was added to 2 μL of 5 × First Strand Buffer (Gibco BRL), 0.5 μL of 100 mM DTT, and 50 ng of mudrA RT-primer GATATGCATGGACCAAAGGCAC at MuDR position +1530 (MuDR numbering according to Hershberger et al., 1991) in a volume of 8 μL. The mix was heated for 5 min at 70°C and chilled on ice for 30 sec. Then a 2-μL cocktail was added consisting of 0.5 μL of 10 mM deoxynucleotide triphosphates (dNTPs), 0.5 μL RNasin (40 units per μL; Promega, Madison, WI), and 1 μL of Superscript II RT (200 units per μL; Gibco BRL), and the mixture was incubated at 42 to 50°C for 1 hr. For internal verification or to detect dcA line transcripts, two PCR products were generated. Two and a half microliters of the first-strand cDNA was directly added to a 25-μL reaction consisting of AmpliTaqII buffer (Perkin-Elmer, Foster City, CA), 0.2 mM dNTPs, 2 mM MgCl2, 100 ng of 5′ primer CaMV 35S +99 −122 (CGCTCATGTGTTGAGCATATAAG), 100 ng of 3′ primer mudrA +1412 (GCTCGAGTACAAGAGCTGGAAGCT), and 100 ng of a second 3′ primer mudrA +1090 (CTTACAGTATCCAACGTATC) with 1 unit of AmpliTaq. Cycle conditions were 3 cycles of 95°C for 1 min, 42°C for 1 min, and 72°C for 2 min, and 32 cycles of 95°C for 1 min, 54°C for 1 min, and 72°C 2 min.
Verification of Kernel Genotype
Ears exhibiting somatic excisions at a1-mum2 and bz2::mu1 nearly always cosegregated with herbicide resistance. With a few exceptions, susceptible plants gave rise to spotted kernels. To check if these kernels still inherited the CaMV 35S–mudrA transgene but had experienced Bar transgene silencing, we performed PCR analysis on seedlings derived from these spotted kernels. PCR conditions were identical to those listed above, with 5 ng of genomic DNA used per 25-μL reaction. As an internal PCR control, 100 ng of the following maize actin primers were added to the reaction: 5′ primer pMAcI 5′ +464 (GCCTACGTAGGTGATGAGGCTCAGGC) and 3′ primer pMAcI 3′ +867 (CTCACACCATCACCTGAATCCATCAC) (Shah et al., 1983). The PCR products were electrophoresed, and a DNA gel blot was made and hybridized with either the mudrA-specific probe or maize actin probe.
Verification That Cryptic MuDRs Did Not Cause Excisions
To ensure that the presence of excisions on kernels was caused by the mudrA-823 transgene and not by the reactivation of a tightly linked cryptic or contaminating MuDR, a total of 357 heavily spotted a1-mum2 kernels derived from outcrosses to a1 tester were pooled from ∼20 ears of line cA75B, germinated, and tested for herbicide resistance (Table 3). All but nine seedlings displayed full herbicide resistance. The nine partially sensitive seedlings were tested for the presence of the mudrA transgene by PCR analysis, using the 5′ primer CaMV 35S +99 −122 and 3′ primer mudrA +1412, as described above; the presence of CaMV 35S–mudrA was verified by DNA gel blotting.
Nucleic Acid Probes
BX1.0 cross-hybridizes to both mudrA and mudrB transcripts and extends from the BamHI (+2865) to the XbaI (+3945) site in MuDR (Hershberger et al., 1995). The 1.3-kb mudrA-specific probe extends from MuDR sites +450 to +1790 (SphI site) and was isolated as a 1.3-kb PstI-SphI fragment from phage phMR49. The mudrB-specific probe extends from the StuI (+3630) to StuI (+4310) sites of MuDR and was isolated from plasmid pMR29. The Mu1-specific probe is the 650-bp AvaI-BstN1 internal fragment of Mu1 and was isolated as a SmaI fragment from plasmid pA/B5 (Chandler and Walbot, 1986). It cross-hybridizes to Mu1 (1.4 kb), Mu2 (1.75 kb), and Mu1.0 (∼1 kb). The pBluescript KS+ probe is the entire 2.9-kb plasmid (Stratagene) that hybridizes to the pUC18 backbone of the Bar plasmid pAHC20 (Christensen and Quail, 1996). The CaMV 35S probe extends from +7072 to +7565 (Franck et al., 1980) and was isolated as a XbaI-PstI fragment from plasmid pR (Ludwig et al., 1990). The maize actin probe extends from +362 to +1338 (Shah et al., 1983) and was generated by PCR from plasmid pMAc1 obtained from R. Meagher (University of Georgia), using the PCR conditions listed above. Actin primer sequences were 5′-GCCGGTTTCGCTGGTGATGATGCGCC-3′ (5′ primer) and 5′-GTGATCTCCTTGCTCATACGATCGGC-3′ (3′ primer). Ten to 50 ng of probe DNA was prepared using a DecaPrimeII random primer kit (Promega) and 32P-dCTP (Amersham, Little Chalfont, UK), incubated at 37°C for >3 hr, and then purified on a NucTrap push column (Stratagene).
Identification of CaMV 35S–mudrA—Catalyzed MuDR Reactivated Transcripts
To identify endogenous mudrA transcripts, we performed RT-PCR as described above. The mudrA RT primer was GATATGCATGGACCA-AAGGCAC at +1530, and PCR primers were 5′ mudrA +210 CTCCTCTAAATGCTCTCTGG and 3′ mudrA +1412 GCTCGAGTACAAGAGCTGGAAGCT (numbering according to Hershberger et al., 1991). To identify reactivated mudrB transcripts, we used oligo-dT as the RT primer, and PCR primers were 5′ mudrB +4730 CTTGTACAGATCTTGTGACCAGTCGCA and 3′ mudrB +3780 GTCCAC-AAATCGATGTTACGGTCGTT. CaMV 35S–mudrA transcripts were identified as described above. As an internal control, actin was amplified using RT primer oligo-dT and PCR primers pMAcI 5′ +464 and pMAcI 3′ +867 (above).
Sector Size Scoring
To score the a1-mum2 excision sector size distribution, we made close-up slides of randomly chosen kernels and projected them onto a wall surface. The number of cells in each sector were either scored visually if cell walls were clearly visible or scored by comparing the area of a single cell sector with the area of larger sectors in the transect by using a ruler on the projected surface. All sectors in focus were selected for scoring. On the curved aleurone surface, peripheral cells containing diffused anthocyanin pigment were subjectively excluded. For each distribution series, 20 to 25 sectors from each of four to six randomly chosen kernels were scored. All scoring was performed in blind test by undergraduate assistants.
Plant Materials
Kernel aleurone anthocyanin pigmentation requires the presence of the structural genes of the anthyocyanin pigment pathway, including A1 and Bz2, in addition to its transcription factors R and C1. The T0 transgenic plants were in an A188/B73 background (non-MuDR; Bz2 A1 r−/r− C1/c1) and were outcrossed to a W23 inbred bz2 tester line (non-MuDR; bz2/bz2 A1 R− C1) (T1 progeny).
a1-mum2 Somatic Excision Material
T1 herbicide-resistant plants were then outcrossed either to an inbred a1 tester line (non-MuDR; a1/a1 Bz2 R− C1) or to a line containing a Mu1 element at the a1 locus in the a1-mum2 tester line (zero copy MuDR; a1-mum2/a1-mum2 Bz2 R− C1) to generate a non-MuDR, transgene-containing line in an A1/a1 or A1/a1-mum2 background (T2 progeny). To look for somatic excisions in the T3 generation (Figure 4 and Table 2), we crossed both herbicide-resistant and herbicide-sensitive sibling plants of the A1/a1 genotype (T2 generation) reciprocally to the a1-mum2/a1-mum2 tester line to generate the genotype R C1 Bz2/− A1/a1-mum2 or a1/a1-mum2.
For A1/a1-mum2 T2 plants, a subset (herbicide-resistant and herbicide-sensitive siblings) was crossed reciprocally to the a1/a1 tester to generate T3 progeny with the genotype R C1 Bz2/− A1/a1 and a1/a1-mum2 to look for somatic excisions. These ears were scored to determine w hether the expected 25% of kernels were spotted. To verify that excisions required the transgene source, we selected these a1-mum2 spotted kernels (R C1 Bz2 a1/a1-mum2), and the resulting plants were outcrossed reciprocally to the inbred a1 tester line. Spotted-kernel T4 generation progeny (R C1 Bz2 a1/a1-mum2) were randomly chosen and assayed for herbicide resistance (Table 3). The other subset of A1/a1-mum2 T2 generation plants (both herbicide-resistant and herbicide-sensitive siblings) was crossed reciprocally to the a1-mum2/a1-mum2 tester to generate T3 progeny with the genotype R C1 Bz2/− A1/a1-mum2 and a1-mum2/a1-mum2.
bz2 ::mu1 Silenced Mutator Material
T1 herbicide-resistant plants were outcrossed again to a W23 inbred bz2 tester line (non-MuDR; bz2/bz2) to generate a non-MuDR but transgene-containing T2 line in a bz2/bz2 A1 R− C1 background. To study the effects of the transgene on reactivating an epigenetically silenced high-copy MuDR line in the T3 generation, we crossed both herbicide-resistant and herbicide-sensitive sibling T2 females in pairs by pollen from an individual silenced MuDR donor containing a silenced Mu1 element at the bz2 locus in the bz2::mu1 allele (bz2::mu1/bz2::mu1, A1 R− C1; high-copy MuDR “off” line) (Figure 8 and Table 4). Excision from bz2::mu1 could create a wild-type Bz2-expressing pigmented sector.
Acknowledgments
We are especially grateful to Karen Brewer for countless hours counting spots on kernels. We thank the following people for tremendous laboratory and field assistance: Maria-Ines Benito, Ryan Swinney, Margaret Wizenberg, Helen Bailey, Daren Bowlby, Louise Bitting, and Barbara Lilley. We thank Mike Freeling for the gift of a1-mum2 seed, Charles Armstrong for HiII seed, Peter Quail for the gift of the pAHC20 construct, and Chris Somerville for use of the helium gun. We thank Dayakar Pareddy, Peggy Lemaux, Rosalind Williams Carrier, Yuchan Wan, David B. Walden, and Charles Armstrong for advice on maize transformation. We thank Akemi Ono, Soo-Hwan Kim, George Rudenko, and Brieann Linkenhoker for comments on the manuscript. M.N.R. was the recipient of an NSERC 1967 Science and Engineering Predoctoral Fellowship from the Canadian Government and the Joseph R. McMicking Graduate Fellowship from Stanford University. This work was supported by National Institutes of Health Grant No. GM49681 to V.W.
References
- Alleman, M., and Freeling, M. (1986). The Mu transposable elements of maize: Evidence for transposition and copy number regulation during development. Genetics 112 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, C.L. (1994). Regeneration of plants from somatic cell cultures: Applications for in vitro genetic manipulation. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 663–671.
- Armstrong, C.L., and Green, C.E. (1985). Establishment and maintenance of friable, embryogenic maize callus and the involvement of l-proline. Planta 164 207–214 [DOI] [PubMed] [Google Scholar]
- Benito, M.-I., and Walbot, V. (1994). The terminal inverted repeat sequences of MuDR are functionally active promoters in maize cells. Maydica 39 255–264. [Google Scholar]
- Benito, M.-I., and Walbot, V. (1997). Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (1987). Covalent DNA modification and the regulation of Mutator element transposition in maize. Mol. Gen. Genet. 208 45–51. [Google Scholar]
- Bennetzen, J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204 195–229. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., Springer, P.S., Cresse, A.D., and Hendrickx, M. (1993). Specificity and regulation of the Mutator transposable element system in maize. Crit. Rev. Plant Sci. 12 57–95. [Google Scholar]
- Brink, R.A., and Nilan, R.A. (1952). The relation between light variegated and medium variegated pericarp in maize. Genetics 37 519–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis, J., Fromm, M., and Walbot, V. (1987). Introns increase gene expression in cultured maize cells. Genes Dev. 1 1183–1200. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., Rivin, C., and Walbot, V. (1986). Stable nonMutator stocks of maize have sequences homologous to the Mu1 transposable element. Genetics 114 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet, P., Lisch, D., Hardeman, K.J., Chandler, V.L., and Freeling, M. (1991). Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics 129 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgen. Res. 5 213–218. [DOI] [PubMed] [Google Scholar]
- Coe, E.H., and Neuffer, M.G. (1977). The genetics of corn. In Corn and Corn Improvement, 2nd ed, G.F. Sprague, ed (Madison, WI: American Society for Agronomy), pp. 111–223.
- Cresse, A.D., Hulbert, S.H., Brown, W.E., Lucas, J.R., and Bennetzen, J.L. (1995). Mu1-related transposable elements of maize preferentially insert into low copy number DNA. Genetics 140 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block, M., Botterman, J., Vandewiele, M., Dockx, J., Thoen, C., Gossele, V., Rao Movva, N., Thompson, C., Van Montagu, M., and Leemans, J. (1987). Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 6 2513–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin, M.J., Lisch, D., and Freeling, M. (1995). Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell 7 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J.A., Benito, M.-I., and Walbot, V. (1994). Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22 2634–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N.V. (1989). Maize transposable elements. In Mobile DNA, D. Berg and M. Howe, eds (Washington, DC: American Society for Microbiology), pp. 375–411.
- Fedoroff, N.V., and Chandler, V.L. (1994). Inactivation of maize transposable elements. In Homologous Recombination and Gene Silencing in Plants, J. Paszkowksi, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 349–385.
- Fennell, A., and Hauptmann, R. (1992). Electroporation and PEG delivery of DNA into maize microspores. Plant Cell Rep. 11 567–570. [DOI] [PubMed] [Google Scholar]
- Fennoy, S.L., and Bailey-Serres, J. (1993). Synonymous codon usage in Zea mays L. nuclear genes is varied by levels of C- and G-ending codons. Nucleic Acids Res. 21 5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck, A., Guilley, H., Jonard, G., Richards, K., and Hirth, L. (1980). Nucleotide sequence of cauliflower mosaic virus DNA. Cell 21 285–294. [DOI] [PubMed] [Google Scholar]
- Fromm, M.E., Morrish, F., Armstrong, C., Williams, R., Thomas, J., and Klein, T.M. (1990). Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/Technology 8 833–839. [DOI] [PubMed] [Google Scholar]
- Gordon-Kamm, W.J., et al. (1990). Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 2 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger, R.J., Warren, C.A., and Walbot, V. (1991). Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc. Natl. Acad. Sci. USA 88 10198–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger, R.J., Benito, M.-I., Hardeman, K., Warren, C., Chandler, V.L., and Walbot, V. (1995). Characterization of the major transcripts encoded by the regulatory MuDR transposable element of maize. Genetics 140 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia, A.-P., and Schnable, P.S. (1996). DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, I.J. (1991). A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 19 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M.G., and Stadler, J. (1989). Molecular characterization of Mutator systems in maize embryogenic callus cultures indicates Mu element activity in vitro. Theor. Appl. Genet. 77 383–393. [DOI] [PubMed] [Google Scholar]
- James, M.G., Scanlon, M.J., Qin, M., Robertson, D.S., and Myers, A.M. (1993). DNA sequence and transcript analysis of transposon MuA2, a regulator of Mutator transposable element activity in maize. Plant Mol. Biol. 21 1181–1185. [DOI] [PubMed] [Google Scholar]
- Jardinaud, M.-F., Souvre, A., Beckert, M., and Alibert, G. (1995). Optimisation of DNA transfer and transient β-glucuronidase expression in electroporated maize (Zea mays L.) microspores. Plant Cell Rep. 15 55–58. [DOI] [PubMed] [Google Scholar]
- Joanin, P., Hershberger, R.J., Benito, M.-I., and Walbot, V. (1997). Sense and antisense transcripts of the maize MuDR regulatory transposon localized by in situ hybridization. Plant Mol. Biol. 33 23–36. [DOI] [PubMed] [Google Scholar]
- Levy, A.A., and Walbot, V. (1990). Regulation of the timing of transposable element excision during maize development. Science 248 1534–1537. [DOI] [PubMed] [Google Scholar]
- Levy, A.A., Britt, A.B., Luehrsen, K.R., Chandler, V.L., Warren, C., and Walbot, V. (1989). Developmental and genetic aspects of Mutator excision in maize. Dev. Genet. 10 520–531. [DOI] [PubMed] [Google Scholar]
- Lisch, D., Chomet, P., and Freeling, M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., Girard, L., Donlin, M., and Freeling, M. (1999). Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S.E., Bowen, B., Beach, L., and Wessler, S.R. (1990). A regulatory gene as a novel visible marker for maize transformation. Science 247 449–450. [DOI] [PubMed] [Google Scholar]
- Luehrsen, K., and Walbot, V. (1994). The impact of AUG start codon context on maize gene expression in vivo. Plant Cell Rep. 13 454–458. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. (1996). Epigenetic phenomena: Paramutation and gene silencing in plants. Curr. Biol. 6 810–813. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., and Baron, A. (1994). Coordinate suppression of mutations caused by Robertson's Mutator transposons in maize. Genetics 136 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson, R.V., Biagi, K., Wheeler, J.G., Stadler, J., and Morris, D.W. (1988). An embryogenic cell line of maize from A188 (Minnesota) contains Mu1-like elements. Plant Mol. Biol. 10 273–279. [DOI] [PubMed] [Google Scholar]
- Meyer, P., Heidmann, I., and Niedenhof, I. (1993). Differences in DNA methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 4 89–100. [DOI] [PubMed] [Google Scholar]
- Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omirulleh, S., Abraham, M., Golovkin, M., Stefanov, I., Karabaev, M.K., Mustardy, L., Morocz, S., and Dudits, D. (1993). Activity of a chimeric promoter with the doubled CaMV 35S enhancer element in protoplast-derived cells and transgenic plants in maize. Plant Mol. Biol. 21 415–428. [DOI] [PubMed] [Google Scholar]
- Osborne, B.I., and Baker, B. (1995). Movers and shakers: Maize transposons as tools for analyzing other plant genomes. Curr. Opin. Cell. Biol. 7 406–413. [DOI] [PubMed] [Google Scholar]
- Park, Y.-D., Papp, I., Moscone, E.A., Iglesias, V.A., Vaucheret, H., Matzke, A.J.M., and Matzke, M.A. (1996). Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 9 183–194. [DOI] [PubMed] [Google Scholar]
- Planckaert, F., and Walbot, V. (1989). Molecular and genetic characterization of Mu transposable elements in Zea mays: Behavior in callus culture and regenerated plants. Genetics 123 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, M., Robertson, D.S., and Ellingboe, A.H. (1991). Cloning of the Mutator transposable element MuA2, a putative regulator of somatic mutability of the a1-Mum2 allele in maize. Genetics 129 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276 1558–1560. [DOI] [PubMed] [Google Scholar]
- Robertson, D.S. (1978). Characterization of a mutator system in maize. Mutat. Res. 51 21–28. [Google Scholar]
- Robertson, D.S. (1981). Mutator activity in maize: Timing of its activation in ontogeny. Science 213 1515–1517. [DOI] [PubMed] [Google Scholar]
- Robertson, D.S. (1985). Differential activity of the maize mutator Mu at different loci and in different cell lineages. Mol. Gen. Genet. 200 9–13. [Google Scholar]
- Robertson, D.S., and Mascia, P.N. (1981). Tests of four controlling-element systems of maize for mutator activity and their interaction with Mu mutator. Mutat. Res. 84 283–289. [Google Scholar]
- Sanford, J.C., Smith, F.D., and Russell, J.A. (1993). Optimizing the biolistic process for different biological applications. Methods Enzymol. 217 483–509. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., Peterson, P.A., and Saedler, H. (1989). The bz-rcy allele of the Cy transposable element system of Zea mays contains a Mu-like element insertion. Mol. Gen. Genet. 217 459–473. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., English, J.J., and Jones, J.D.G. (1993). High-level expression of the Activator transposase gene inhibits the excision of Dissociation in tobacco cotyledons. Cell 75 507–517. [DOI] [PubMed] [Google Scholar]
- Shah, D.M., Hightower, R.C., and Meagher, R.B. (1983). Genes encoding actin in higher plants: Intron positions are highly conserved but the coding sequences are not. J. Mol. Appl. Genet. 2 111–126. [PubMed] [Google Scholar]
- Spencer, T.M, Gordon-Kamm, W.J., Daines, R.J., Start, W.G., and Lemaux, P.G. (1990). Bialaphos selection of stable transformants from maize cell culture. Theor. Appl. Genet. 79 625–631. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker, P., Hudson, K.M., Shaffer, A.L., Constantinescu, A., and Schlissel, M.S. (1996). Cell type–specific chromatin structure determines the targetting of V(D)J recombinase in vitro. Cell 85 887–897. [DOI] [PubMed] [Google Scholar]
- Thompson, C.J., Rao Movva, N., Tizard, R., Crameri, R., Davies, J.E., Lauwereys, M., and Botterman, J. (1987). Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain, P., McMullen, M.D., and Finer, J.J. (1993). Osmotic treatment enhances particle bombardment–mediated transient and stable transformation of maize. Plant Cell Rep. 12 84–88. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Mourrain, P., Palauqui, J.-C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16 651–659. [DOI] [PubMed] [Google Scholar]
- Walbot, V. (1991). The Mutator transposable element family of maize. Curr. Top. Genet. Eng. 13 1–37. [DOI] [PubMed] [Google Scholar]
- Walbot, V. (1992). Developmental regulation of excision timing of Mutator transposons of maize: Comparison of standard lines and an early excision bz1-Mu1 line. Dev. Genet. 13 376–386. [Google Scholar]
- Wan, Y.C., Widholm, J.M., and Lemaux, P.G. (1994). Type I callus as a bombardment target for generating fertile, transgenic maize (Zea mays L.). Planta 196 7–14. [Google Scholar]
- Wilkinson, J.E., Twell, D., and Lindsey, K. (1997). Activities of CaMV 35S and nos promoters in pollen: Implications for field release of transgenic plants. J. Exp. Bot. 48 265–275. [Google Scholar]
- Wisman, E., Hartmann, U., Sagasser, M., Baumann, E., Palme, K., Hahlbrock, K., Saedler, H., and Weisshaar, B. (1998). Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 95 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Tamaki, K., Watanabe, K., Fujino, M., and Nakamura, C. (1998). A maize MuDR-like element expressed in rice callus subcultured with proline. Hereditas 129 95–99. [DOI] [PubMed] [Google Scholar]