Abstract
Transit sequences of chloroplast-destined precursor proteins are phosphorylated on a serine or threonine residue. The amino acid motif around the phosphorylation site is related to the phosphopeptide binding motif for 14-3-3 proteins. Plant 14-3-3 proteins interact specifically with wheat germ lysate–synthesized chloroplast precursor proteins and require an intact phosphorylation motif within the transit sequence. Chloroplast precursor proteins do not interact with 14-3-3 when synthesized in the heterologous reticulocyte lysate. In contrast, a precursor protein destined for plant mitochondria was found to be associated with 14-3-3 proteins present in the reticulocyte lysate but not with 14-3-3 from wheat germ lysate. This indicates an unrecognized selectivity of 14-3-3 proteins for precursors from mitochondria and plastids in plants in comparison to fungi and animals. The heterooligomeric complex has an apparent size of 200 kD. In addition to the precursor protein, it contains 14-3-3 (probably as a dimer) and a heat shock protein Hsp70 isoform. Dissociation of the precursor complex requires ATP. Protein import experiments of precursor from the oligomeric complex into intact pea chloroplasts reveal three- to fourfold higher translocation rates compared with the free precursor, which is not complexed. We conclude that the 14-3-3–Hsp70–precursor protein complex is a bona fide intermediate in the in vivo protein import pathway in plants.
INTRODUCTION
In plant cells, the plastids—chloroplasts in green leaves—represent the most prominent family of organelles. Plastids must import >90% of their protein constituents from the cytosol in a post-translational event. Faithful post-translational targeting must be achieved for a number of subcellular compartments, for example, the nucleus, vacuole, peroxisome, endoplasmic reticulum, mitochondria, and the plastids. In the case of mitochondria and plastids, N-terminal, cleavable targeting signals are both necessary and sufficient to support import into the organelles in vitro (Keegstra et al., 1989). Cells in which post-translational protein translocation occurs must overcome a number of problems: one of these is to prevent missorting between organelles, which seems to occur very rarely in a homologous system (de Castro Silva-Filho et al., 1997). In addition, the cells must retard folding of the organellar proteins before these polypeptides reach their final destination to avoid the biological function of the proteins in the wrong compartment, namely, the cytosol. Concomitantly, the precursor proteins must retain a suitable secondary and tertiary structure that is competent for recognition by and translocation into the organelle. How these goals are achieved is poorly understood, but it seems likely that the targeting signals of the different preproteins play a prominent role in these events.
The protein translocon at the outer envelope of chloroplasts (the Toc complex) mediates specific recognition, binding, and translocation of preproteins in a nucleotide-dependent manner (Chen and Schnell, 1999; Keegstra and Cline, 1999). Subunits of the Toc complex are Toc160, a putative precursor receptor (Perry and Keegstra, 1994; Bölter et al., 1998); Toc75, the preprotein translocation channel (Perry and Keegstra, 1994; Hinnah et al., 1997); and Toc34, a GTP binding protein that might influence precursor binding and translocation (Seedorf et al., 1995; Kouranov and Schnell, 1997).
Heat shock proteins of the Hsp70 class are involved in the transport of proteins into mitochondria, endoplasmic reticulum, and chloroplasts (Deshaies et al., 1988; Zimmermann et al., 1988; Waegemann et al., 1990), and precursor proteins are known to interact with Hsp70 during translocation (Beckmann et al., 1990). Whereas this interaction seems to be a general and not an organelle-specific phenomenon (Ellis and van der Vies, 1991), three cytosolic factors that act on mitochondrial preproteins have been described. First, the cytosolic mitochondrial fusion protein targeting factor (Mft52) from yeast (Cartwright et al., 1997) in cooperation with the nascent chain–associated complex seems to initiate mitochondrial protein targeting in vivo, as has been shown for hybrid proteins containing mitochondrial presequences from ATPase subunit β (F1β) fused to LacZ and subunit IV of cytochrome oxidase fused to mouse dihydrofolate reductase, respectively (George et al., 1998). Furthermore, the presequence binding factor and the mitochondrial import stimulating factor have been purified from rabbit reticulocyte lysate and rat liver cytosol, respectively (Murakami et al., 1992; Hachiya et al., 1993). Both factors recognize mitochondrial targeting sequences and stimulate the import of preproteins into yeast mitochondria. Mitochondrial import stimulating factor, a member of the 14-3-3 protein family, delivers a bound precursor to the yeast TOM70–TOM37 (for translocon at the outer membrane of mitochondria) receptor subcomplex in a reaction requiring ATP hydrolysis by mitochondrial import stimulating factor. In contrast, Hsp70-bound preproteins seem to interact preferentially with the TOM20–TOM22 receptor subcomplex (Hachiya et al., 1995; Haucke and Schatz, 1997; Komiya et al., 1997).
The import of several nuclear proteins, for example, lamin B2 (Hennekes et al., 1993), simian virus 40 T-antigen (Jans et al., 1991; Rihs et al., 1991), the yeast SW15 transcription factor (Moll et al., 1991), and Cdc25 (cell division cycle protein; Yang et al., 1999), are inhibited by phosphorylation of a serine or threonine residue near the nuclear localization signal. Binding of 14-3-3 protein to a phosphorylated Cdc25 is responsible for a largely cytosolic localization by preventing nuclear import, whereas in comparison, the nonphosphorylated form favors the nuclear compartment. 14-3-3 proteins are ubiquitous in all eukaryotic cells, and 10 14-3-3 isoforms have been described for Arabidopsis (Wu et al., 1997). 14-3-3 proteins act as a novel type of molecular chaperone, first identified in modulating interactions between components of signal transduction pathways (reviewed in Aitken, 1996; Ferl, 1996). An amino acid consensus motif for this site has been deduced from phosphopeptide binding studies for 14-3-3 proteins (Muslin et al., 1996; Yaffe et al., 1997); this motif shows intriguing similarities to a phosphopeptide motif present in a variety of chloroplast precursor proteins (Waegemann and Soll, 1996).
A protein kinase localized in the cytosol of pea leaf mesophyll cells or wheat germ embryo cells phosphorylates the transit peptides of major chloroplast precursor proteins (Waegemann and Soll, 1996) but not those of plant mitochondrial or peroxisomal preproteins. However, phosphorylated precursor proteins were incapable of chloroplast translocation, although the recognition process at the organellar surface was not disturbed. Therefore, import of a phosphorylated precursor is stopped at an early stage of translocation but after receptor recognition, and it can proceed only after dephosphorylation of the transit peptide. A consensus motif was deduced for the phosphorylation site from the different chloroplast precursor proteins studied, that is, those of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (preSSU), the light-harvesting chlorophyll a/b binding protein, and the oxygen-evolving complex 23- and 33-kD subunits (preOE23 and preOE33; Waegemann and Soll, 1996). The significant similarities between the 14-3-3 phosphopeptide binding motif and the determined phosphorylation sites in plastid-destined precursor proteins pointed to a possible interaction between these partners.
In this study, we show that chloroplast precursor proteins, when synthesized in the homologous wheat germ system, interact with 14-3-3 proteins, whereas plant mitochondrial preproteins do not. Interaction is completely dependent on the phosphorylation motif in the targeting signal. The precursor proteins form a high molecular mass oligomeric complex with 14-3-3 proteins and a cytosolic Hsp70. The complex-bound precursor is much more competent for import into isolated organelles than is the free precursor protein. Our findings indicate that in plants, “nuclear-encoded” plastid proteins form a specific oligomeric complex, which keeps the precursor proteins in a highly import-competent state.
RESULTS
We observed a strong similarity between the 14-3-3 phosphopeptide binding site and the chloroplast precursor protein kinase phosphorylation motif in the transit peptide of SSU, OE23, and OE33 (Figure 1A). Therefore the preSSU, mature SSU, and the plant mitochondrial ATPase subunit β precursor preF1β from tobacco were heterologously expressed and attached to Ni2+ metal–chelate matrix via a C-terminal hexahistidine tag. A translationally active wheat germ lysate was incubated with the different affinity matrices. After extensive washing, bound proteins were eluted with 1 mM ATP followed by elution in a salt gradient from 50 to 500 mM NaCl in the absence of ATP. Eluted proteins were analyzed by SDS-PAGE and immunoblotting by using antisera raised against Hsp70 and 14-3-3 (Hirsch et al., 1992). Hsp70 was present in all fractions tested, indicating its presence in multiple functional forms. Upon elution with ATP, a slight increase of Hsp70 in this fraction was detectable irrespective of the subcellular origin of the protein or the presence of a targeting signal. However, 14-3-3 proteins were detected only in the salt eluate of the chloroplast precursor preSSU but not from the mature form of the SSU or the mitochondrial preF1β, indicating a preferential interaction with the plastid-targeting peptide (Figure 1B). For further studies, cDNA clones encoding for 14-3-3 proteins from pea were isolated and sequenced (GenBank accession numbers AJ238681 and AJ238682). The deduced amino acid sequences of both cDNAs show 74% sequence identity to other 14-3-3 isoforms from either plants or mammals. A new antiserum was raised against the heterologously expressed 14-3-3 protein encoded by the cDNA Pisum sativum mRNA for 14-3-3–like protein with accession number AJ238681, which specifically recognized 14-3-3 isoforms in wheat germ lysate on immunoblots, as demonstrated by a competition experiment performed in the presence of excess pea 14-3-3 protein (Figure 2).
Figure 1.
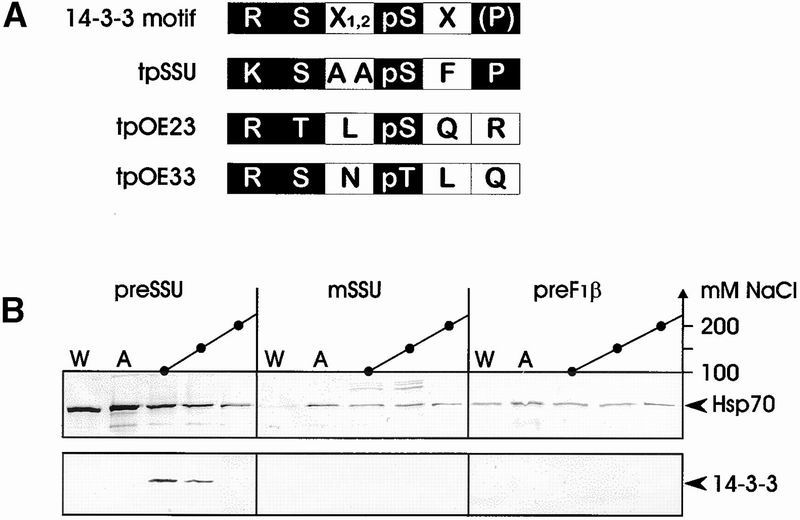
14-3-3 Proteins Interact with a Chloroplast Precursor–Containing Affinity Matrix.
(A) Amino acid comparison between the phosphopeptide binding motif of 14-3-3 proteins and the phosphorylation site in the transit peptide (tp) of plastid precursor proteins (Waegemann and Soll, 1996; Yaffe et al., 1997). Letters in white indicate amino acids that are important for selectivity, letters in parentheses indicate those that are not essential, and letters in black indicate those that are variable.
(B) PreSSU, mature SSU (mSSU), and ATPase subunit β precursor (preF1β) containing a C-terminal hexahistidine tag were bound to a Ni2+-chelating matrix. Wheat germ lysate was passed over the matrix, washed extensively, and eluted with 1 mM ATP followed by elution with a linear gradient of 50 to 500 mM NaCl. The last wash fraction (W), the ATP eluates (A), and the salt eluates (100 to 200 mM NaCl; black dots) were precipitated with trichloroacetic acid. Proteins were separated by SDS-PAGE followed by immunoblot analysis with antisera raised against Hsp70 and 14-3-3 proteins, as indicated by arrowheads. The immunoblot is shown.
Figure 2.
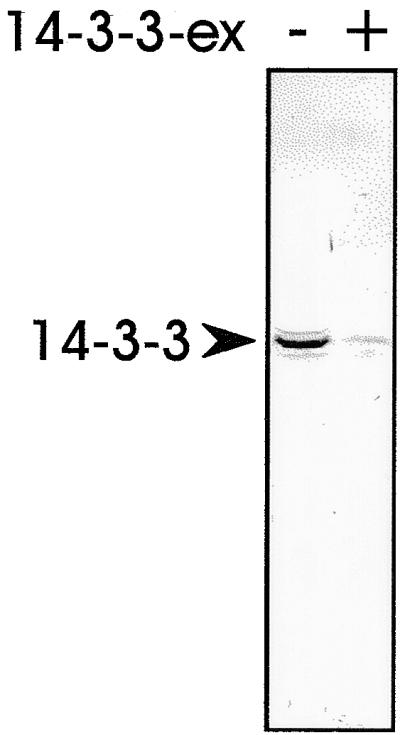
Anti–14-3-3 Antiserum Specifically Recognizes Wheat Germ 14-3-3 Proteins.
Soluble proteins from a wheat germ lysate were separated by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with an antiserum against a pea 14-3-3 protein either in the absence (−) or presence (+) of 2 μg of recombinant protein (14-3-3-ex).
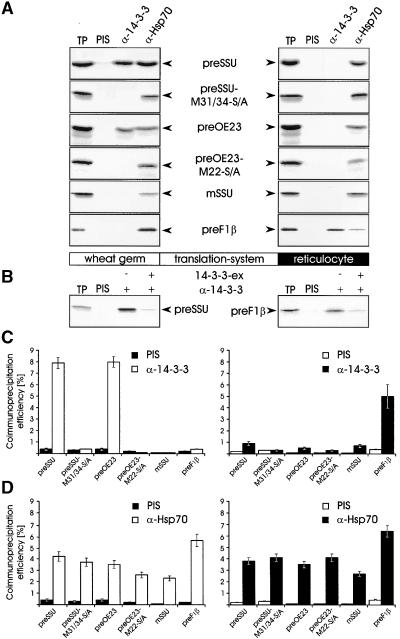
To obtain evidence for selective binding of 14-3-3 proteins to chloroplast precursor proteins, we translated mRNAs encoding preSSU, preOE23, and the nonphosphorylatable mutants M31/34-S/A and preOE23–M22-S/A, which contain a serine to alanine mutation (M) at the positions indicated (Waegemann and Soll, 1996), in either a wheat germ or a reticulocyte lysate system. Postribosomal supernatants of the various translation products were subjected to coimmunoprecipitation experiments using antisera raised against 14-3-3, Hsp70, or the preimmune serum, respectively. The appearance of labeled translation products was taken as a degree of interaction between the partner proteins (Figure 3A). The results demonstrate that the preSSU and preOE23 when translated in a wheat germ system are stably associated with 14-3-3 proteins, whereas they do not interact with 14-3-3 proteins to a significant extent when translated in a reticulocyte lysate system. These results are consistent with our earlier findings (Waegemann and Soll, 1996) that a reticulocyte lysate is not capable of phosphorylating chloroplast-targeting signals. It could indicate further that in a plant system, 14-3-3 proteins interact preferentially with plastid transit sequences but not with mitochondrial targeting signals.
Figure 3.
Chloroplast but Not Mitochondrial Precursor Proteins Interact with Plant 14-3-3 Proteins.
The different proteins were translated in vitro in either the wheat germ or the reticulocyte lysate system. 35S-labeled translation products of the preSSU, preSSU–M31/34-S/A, mature SSU (mSSU), and preF1β as well as 3H-labeled translation products of preOE23 and preOE23–M22-S/A (as indicated by arrowheads) were coimmunoprecipitated by using antisera against 14-3-3 (α-14-3-3), Hsp70 (α-Hsp70), or preimmune serum (PIS). Ten percent of the labeled translation product (TP) was used as internal standard to calculate the immunoprecipitation efficiencies. Radioactive bands were cut out, dissolved in dimethyl sulfoxide, and quantified by liquid scintillation counting.
(A) Typical autoradiograms.
(B) Immunoprecipitations of the preSSU and preF1β translation product (as indicated by arrowheads) with antisera against 14-3-3 (α-14-3-3) were performed either in the absence (−) or presence (+) of 1 μg of recombinant pea 14-3-3 protein (14-3-3-ex). An autoradiogram is shown.
(C) and (D) Quantification of immunoprecipitation efficiency for α-14-3-3 (C) and α-Hsp70 (D) antibodies, respectively. The calculations were conducted from a minimum of five or three independent experiments, respectively.
Support for this idea comes from the result obtained with preF1β. This plant mitochondrial precursor protein interacts with 14-3-3 proteins when translated in the heterologous reticulocyte lysate but not when it is synthesized in the wheat germ lysate (Figure 3A). Neither the nonphosphorylatable proteins preSSU–M31/34-S/A and preOE23–M22-S/A nor the mature form of SSU interacts with 14-3-3 proteins to a detectable level when translated in either system (Figure 3A). Recombinant 14-3-3 proteins from pea could effectively compete with endogenous 14-3-3 in coimmunoprecipitation reactions (Figure 3B), indicating the specificity of the antiserum. The immunoprecipitation efficiency using the anti–14-3-3 antiserum was between 5 and 8% for the preSSU, preOE23, and preF1β but did not exceed 0.5% for the preSSU–M31/34-S/A, preOE23–M22-S/A, and mSSU (Figure 3C; mean of a minimum of five independent experiments).
Coimmunoprecipitation of the different translation products by an antiserum to Hsp70 demonstrates that all proteins independent of the translation system and independent of the presence and nature of a transit sequence associate with the chaperone protein (Figure 3A). Quantification of the immunoprecipitation experiments using the anti-Hsp70 antiserum showed that the extent of Hsp70 translation product association in both translation systems was similar (Figure 3D; mean of a minimum of three independent experiments). From these data, we suggest that in vitro–translated proteins exist in at least two different heterooligomeric forms. The first contains 14-3-3 and perhaps some additional subunits. Formation of this complex is favored by a phosphorylatable wild-type-like chloroplast targeting sequence. A second form contains the general chaperone Hsp70, which is independent of the nature of the translated polypeptide and might be similar to that reported earlier (Deshaies et al., 1988; Zimmermann et al., 1988). Hsp70 is most likely not involved in an organelle-selective transport pathway, but it seems to be a general intermediate during or after translation and could lead to the 14-3-3–containing complex.
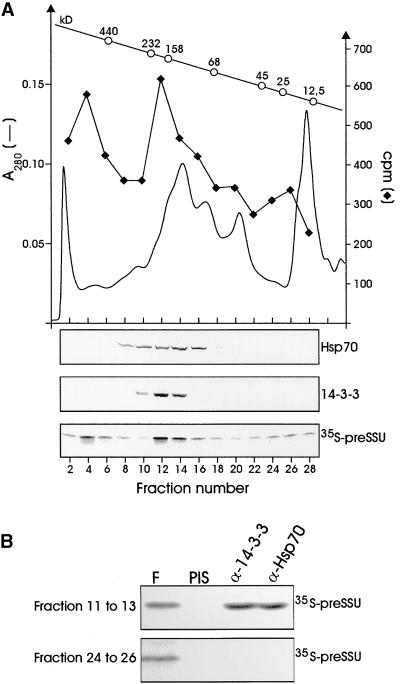
To determine whether the preSSU translation product exists in different oligomeric forms and complexes, we subjected a postribosomal supernatant of preSSU freshly synthesized in wheat germ lysate to size exclusion chromatography (Figure 4). Three major peaks of different apparent molecular size containing labeled preSSU were detected. The nature and composition of the high molecular mass fraction complex (⩾600 kD) were not analyzed because we could detect neither significant proportions of 14-3-3 proteins nor Hsp70 in these fractions by immunoblotting (Figure 4B; α-Hsp70 and α-14-3-3). A major proportion of the preSSU migrated in a complex with an apparent size between 170 and 200 kD. These fractions also contain significant amounts of 14-3-3 and Hsp70. PreSSU interacts closely with 14-3-3 and Hsp70 in these complex fractions, as shown by the ability of the respective antiserum to coimmunoprecipitate the precursor protein (Figure 4B). Uncomplexed, free preSSU eluted from the size exclusion column at ∼20 kD, which is its calculated molecular mass. These low molecular mass fractions did not contain Hsp70 or 14-3-3 proteins, as determined by immunoblot analysis (Figure 4A). Accordingly, antisera raised against 14-3-3 and Hsp70 could not coimmunoprecipitate preSSU (Figure 4B). The ratio of the preSSU distribution between these three most prominent peaks varied to a great extent, depending on the translation conditions. Prolonged translation times (>60 min) and elevated incubation temperatures (>25°C) resulted in a large increase in the presence of preSSU in the high molecular mass fraction as well as in the noncomplexed fraction (data not shown). Under standard conditions (<60 min at 25°C), >50% of the synthesized preSSU was found in the 170- to 200-kD complex fraction.
Figure 4.
PreSSU Forms a High Molecular Weight Complex in Wheat Germ Lysate.
(A) A translation assay (50 μL volume) was fractionated by size exclusion chromatography calibrated with protein standards of different molecular mass (open circles). Thirty fractions of 500 μL were collected and precipitated with trichloroacetic acid, separated by SDS-PAGE, blotted, and analyzed by immunostaining with antisera raised to Hsp70 and to the 14-3-3 protein as well as by autoradiography (35S-preSSU). Radioactivity (filled diamonds) was quantified by liquid scintillation counting after dissolving the nitrocellulose in dimethyl sulfoxide.
(B) Fractions 11 to 13 and 24 to 26 were combined, and the two pools were split into equal aliquots. Combined fractions were subjected to coimmunoprecipitation by antisera to 14-3-3 (α-14-3-3) and Hsp70 (α-Hsp70) as indicated. Ten percent of each combined fraction (F) was used as internal standard. An autoradiogram is shown. PIS, control experiment using preimmune serum.
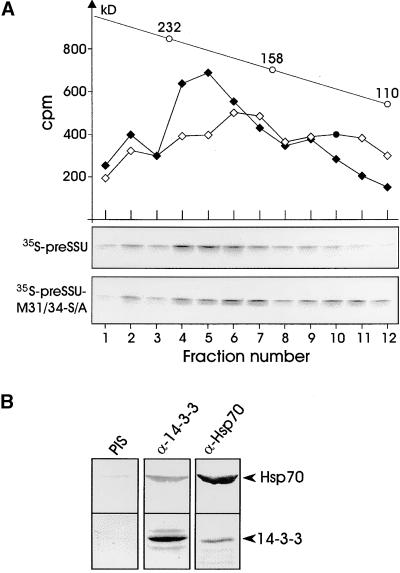
The data in Figures 4A and 4B suggested that the peak fractions of 14-3-3 and Hsp70 did not comigrate exactly (cf. immunoblots in Figure 4A, fractions 10 to 16), indicating that two complexes of similar size might overlap in these fractions. Therefore, the preSSU and the nonphosphorylatable mutant preSSU–M31/34-S/A were translated in wheat germ lysate and subjected to size exclusion chromatography (Figure 5A). We expected to find two complexes of similar but distinguishable size, because the wild-type precursor can interact with 14-3-3 protein as well as Hsp70 (cf. interaction shown in Figure 3A), whereas the nonphosphorylatable mutant associates only with Hsp70 (see Figure 3A) but not with 14-3-3. This was found to be the case (Figure 5A). The oligomeric complex containing the preSSU migrated at a slightly higher apparent molecular mass (∼200 kD, fractions 4 to 6), with a shoulder overlapping with the apparent molecular size range of the most prominent preSSU–M31/34-S/A complex (∼170 kD, fractions 6 and 7). We conclude that two soluble oligomeric protein complexes with the wild-type precursor can coexist during in vitro translation, with the 14-3-3 protein–containing precursor complex being the most prominent (ratio of ∼3:1).
Figure 5.
14-3-3 Forms a Heterooligomeric Complex with preSSU and Hsp70.
(A) Wheat germ lysate–synthesized 35S-preSSU (filled diamonds) and the nonphosphorylatable mutant 35S-preSSU–M31/34-S/A (open diamonds) were fractionated in independent runs as before (see Figure 4) but with a fraction size of 250 μL. Only fractions of the relevant molecular size were collected. The positions of the molecular mass markers are indicated on top (open circles). Each fraction was precipitated by trichloroacetic acid, and precipitated proteins were separated by SDS-PAGE and subsequently transferred onto nitrocellulose membranes. Autoradiograms are shown below. Radioactivity present in each fraction was quantified by liquid scintillation counting after solubilizing nitrocellulose strips in dimethyl sulfoxide.
(B) Wheat germ lysate (∼130 μg protein) was used for coimmunoprecipitation by anti–14-3-3 and anti-Hsp70 antibodies. The immunoprecipitated polypeptides were separated by SDS-PAGE, blotted, and immunodecorated using 14-3-3 and Hsp70 antisera, as indicated by arrowheads. PIS, control experiment using preimmune serum.
To identify additional subunits of the precursor–14-3-3 complex, translationally active wheat germ lysate was subjected to coimmunoprecipitation analysis in the absence of exogenous mRNA by using either anti–14-3-3 or anti-Hsp70 antibodies (Figure 5B). It was possible to demonstrate that indeed anti–14-3-3 can coimmunoprecipitate Hsp70 and that anti-Hsp70 can coimmunoprecipitate 14-3-3 proteins, indicating a stable association between the two polypeptides. The 14-3-3–Hsp70 complex represents an easily detectable proportion but not a major amount of Hsp70 or 14-3-3 protein in the wheat germ lysate (Figure 5B). Our results suggest strongly that the 200-kD complex contains as minimal constituents the precursor protein, a 14-3-3 dimer (Aitken, 1996), and Hsp70.
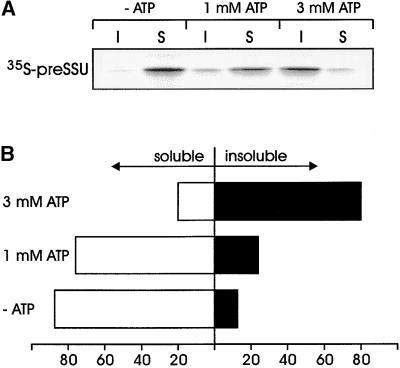
Earlier results indicated that the import of the heterologously expressed precursor of the light-harvesting chlorophyll a/b binding protein into chloroplasts was stimulated by cytosolic Hsp70 and another unidentified component, the activity of which depended on ATP (Waegemann et al., 1990). Therefore, we sought to determine whether ATP had an effect on the stability of the precursor complex. After size exclusion chromatography of freshly translated preSSU, fractions 11 to 13 (complex-containing fractions shown in Figure 4A) were combined and incubated in the absence or presence of 1 and 3 mM ATP, respectively. The addition of 3 mM ATP resulted in the dissociation of the complex (Figure 6A), and >80% of the preSSU was recovered as aggregated insoluble protein in a 165,000g pellet (Figure 6B). In the presence of 1 mM ATP, >75% of the preSSU remained basically in a soluble form (Figures 6A and 6B), whereas in the absence of ATP, only a very small amount of the preSSU (⩽10%) dissociated from the complex and was rendered insoluble. We were not able to determine the oligomeric state, that is, monomeric or oligomeric, of the preSSU after incubation with ATP due to detection limits, but we anticipated that the majority of soluble preSSU present in the supernatant remained in an oligomeric state with 14-3-3 and Hsp70.
Figure 6.
Precursor-Containing Complex Dissociates in an ATP-Dependent Manner.
(A) Freshly translated 35S-preSSU was fractionated by size exclusion chromatography (see Figure 4). Fractions 11 to 13 were combined, concentrated by ultrafiltration to a final volume of ∼150 μL, and divided into equal aliquots. Each aliquot was incubated for 15 min at 25°C in the absence (−ATP) or presence of different ATP concentrations as indicated. The incubation mixture was centrifuged at 165,000g for 30 min. Proteins present in the supernatant (S) or pellet (I) were separated by SDS-PAGE, transferred onto nitrocellulose, and subjected to autoradiography.
(B) Quantification of three independent results shown in (A). The sum of radioactivity in the soluble and the insoluble fractions, as determined by liquid scintillation counting, was taken as 100%. Values are given in percentages.
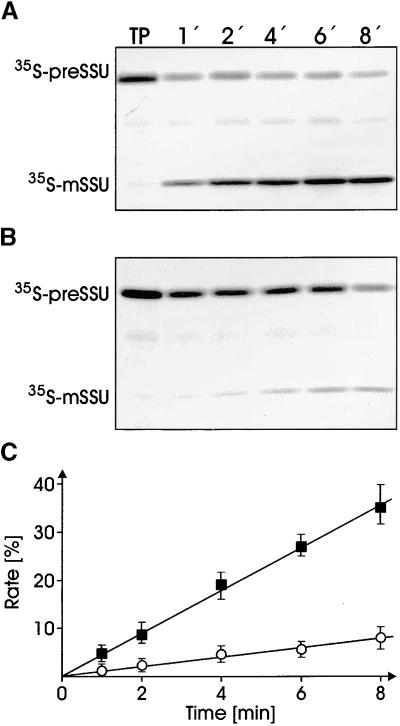
To answer the question of whether the oligomeric and monomeric forms of the preSSU differ in import competence, we separated the oligomeric preSSU from monomeric preSSU by size exclusion chromatography (as shown in Figure 4A) and used it in independent import experiments into purified chloroplasts (Figures 7A and 7B). Import experiments were performed under conditions in which import rates could be determined in the linear range (see Methods). The presence of 1 mM ATP in the import reaction prevents the formation of insoluble preSSU during the course of the experiment (cf. complex stabilty shown in Figure 6). Protein import, which is measured only by the appearance of processed mature SSU, was linear for both preSSU forms over the time range tested (Figure 7C). However, the import rate of the monomeric preSSU was low, that is, <10% of the amount of the monomeric preSSU added to an import reaction actually was translocated and processed within 8 min. In contrast, the preSSU present in the heterooligomeric complex was imported and processed with a three- to fourfold higher rate, that is, >35% of added preSSU became imported and processed after 8 min (Figure 7C). The import experiments demonstrate that efficient protein translocation into chloroplasts occurs with much higher rates when a precursor protein exists in an oligomeric complex with 14-3-3, Hsp70, and maybe additional components. We conclude that the cytosolic pretranslocation complex described here is also a bona fide intermediate in post-translational protein import into chloroplasts in vivo.
Figure 7.
Only Complexed but Not Free 35S-preSSU Is Imported into Isolated Chloroplasts with High Efficiency.
(A) and (B) Freshly translated 35S-preSSU was fractionated by size exclusion chromatography (see Figure 4). Fractions 11 to 13 (A) and 24 to 26 (B) from three independent runs were combined and concentrated, and equal aliquots (100 μL) were used in each import reaction. Chloroplasts equivalent to 7.5 μg of chlorophyll were incubated with preSSU at 25°C for the different times indicated. The import reaction was stopped by adding ice-cold EDTA solution to a final concentration of 37.5 mM. Chloroplasts were recovered from the import reaction by centrifugation. Chloroplast proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes, and radioactivity recovered in processed mature SSU was determined by autoradiography. A typical autoradiogram out of a set of five independent experiments is shown. TP, 10% of translation product used as internal standard; 1′, 1 min.
(C) Import efficiencies from complexed ([A], filled squares) or free ([B], open circles) preSSU as measured by the appearance of processed mature mSSU were quantified by liquid scintillation counting as above. The calculations were conducted from five independent experiments of five independent complex purifications. Values are given as percentages of the translation product supplied to each translocation reaction for the times indicated.
DISCUSSION
14-3-3 proteins represent a ubiquitous eukaryotic protein family with a wide range of regulatory and chaperone properties mediated by protein–protein interactions (Aitken et al., 1991). From analyses of the crystal structure of a 14-3-3 dimer and from peptide binding studies, it is predicted that the target proteins bind into a large groove formed by amphiphilic helices (Liu et al., 1995; Xiao et al., 1995). Each monomer is able to bind one target protein. Phosphorylated peptides bind with much higher affinity than do nonphosphorylated ones, which explains the involvement of 14-3-3 proteins in the regulation of protein kinases and other cellular events involving protein phosphorylation (Aitken, 1996; Palmgren et al., 1998). The preferred but not exclusive binding motif of 14-3-3 proteins is closely related to an amino acid motif in the targeting sequence of chloroplast precursor proteins (Waegemann and Soll, 1996). The phosphorylation site was determined experimentally in the precursor forms of SSU, OE23, and OE33, but it also can be deduced in many more chloroplast transit sequences, for example, heme oxygenases 1 and 2 (GenBank accession numbers AAD22108 and AAD22109), DNA repair protein RecA (Q39199), plastocyanin (P11490), ferredoxin (P16972), geranylgeranyl pyrophosophate synthetase (P34802), the 50S ribosomal protein L28 (O22795), acyl carrier protein 2 (P257010), glutathione reductase (P27456), glyceraldehyde 3-phosphate dehydrogenase (P09315 and P15985), and others (T. May and J. Soll, unpublished observations; Finnie et al., 1999).
The motif identified in preSSU is recognized specifically by wheat germ 14-3-3 proteins in vitro, even though it is not phosphorylated (Figure 1B), whereas interaction with preF1β could not be observed (Figure 1B). Phosphorylation of the transit sequence converts this peptide motif into a binding site that has much higher binding affinity for 14-3-3 (Muslin et al., 1996; Andrews et al., 1998). This is also supported by the observation that 14-3-3 dissociates from nonphosphor-ylated preSSU (as used in Figure 1) at ∼100 mM NaCl, whereas the phosphorylated precursor, that is, after translation in a wheat germ lysate, does not dissociate from 14-3-3 at 150 mM NaCl (as used in the coimmunoprecipitation experiments shown in Figure 2). Within this heterooligomeric complex, 14-3-3 cooperates with Hsp70 and perhaps with additional, as yet unidentified, components. The formation of the precursor guidance complex keeps the preprotein in a highly import-competent state, whereas the free, noncomplexed preprotein shows a much lower import competence. Although the binding of a phosphorylated precursor to the chloroplast surface is not disturbed, translocation into the chloroplast in a phosphorylated state is not possible. Dephosphorylation must occur before translocation can proceed, but it is most likely not a rate-limiting step in the overall translocation process, because dephosphorylation is only partially inhibited by the broad range phosphatase inhibitor NaF or by phosphorylating the precursor with ATPγS (MacKintosh, 1993; Waegemann and Soll, 1996).
It remains to be determined whether the precursor present in the guidance complex actually enters the chloroplast on an import route involving further unidentified import components, which subsequently might lead the precursor to a putative presequence receptor. Nevertheless, noncomplexed precursor proteins, that is, reticulocyte lysate–synthesized (preSSU–M31/34-S/A) or soluble overexpressed (preferredoxin; Pilon et al., 1995) precursor proteins, are imported into chloroplasts. However, it is not well established where these proteins enter the import pathway, because distinct precursor recognition sites have been suggested for all major Toc subunits, that is, Toc160, Toc75, and Toc34 (Perry and Keegstra, 1994; Ma et al., 1996; Hinnah et al., 1997; Kouranov and Schnell, 1997). Therefore, different precursor forms (states) might engage different subunits of the translocon machinery, which could lead to different import efficiencies. The highly efficient import rate for complexed precursor in comparison to the free precursor protein favors such an idea. In mitochondria from fungi, a preprotein–14-3-3 complex preferentially uses the TOM70–TOM37 receptor complex, whereas the precursor interacting only with Hsp70 binds to the TOM20–TOM22 receptor subcomplex (Hachiya et al., 1995; Haucke and Schatz, 1997; Komiya et al., 1997).
The plant system is faced with the challenge that precursor proteins must have the ability to discriminate between plastids and mitochondria in all cells. Our data show that only plastid precursor proteins interact with 14-3-3 proteins (Figures 1B and 3A). Even when these proteins are not phosphorylated, that is, as recombinant proteins, a low association with 14-3-3 proteins is detectable (Figure 1B). Thus, phosphorylation is not essential for interaction, although the affinity of interaction is greatly stimulated upon phosphorylation (Yaffe et al., 1997). Similarly, it might be expected that plant mitochondrial preproteins would be able to interact with the closely related plant 14-3-3 protein, but they did not (Figure 1B). Interaction of plant mitochondrial preproteins occurs to a detectable degree only in the heterologous mammalian translation system, the reticulocyte lysate. Consequently, in eukaryotic cells containing both plastids and mitochondria, the design of the plastid-targeting signal was predicated on guaranteeing a faithful targeting and translocation process. One event of this process could be represented by plastid precursor phosphorylation in conjunction with 14-3-3 binding. The 14-3-3 isoforms are closely related in all eukaryotic organisms analyzed thus far. Subtle changes unrecognized until now in the peptide binding regions of certain 14-3-3 isoforms in plants might be responsible for this. On the other hand, the estimated size of the precursor guidance complex suggests the presence of another component that could affect binding selectivity.
Size exclusion chromatography indicated the presence of three soluble precursor forms in a post-translational supernatant from wheat germ. One complex (∼200 kD) contains 14-3-3 most likely as a dimer (∼70 kD; Palmgren et al., 1998), with each monomer bound to a precursor molecule (∼40 kD; Aitken, 1996), and one Hsp70 (70 kD). At present, we cannot distiguish distinct 14-3-3 and Hsp70 isoforms recruited in the guidance complex. Another complex (∼170 kD) is made of precursor protein, Hsp70, and maybe other components but no 14-3-3 protein. The third contains the free precursor only. Precursor proteins are released from their complexed state by ATP. We do not know whether ATP is utilized by Hsp70 or by 14-3-3 protein, which has been suggested to have an intrinsic ATP-hydrolyzing capacity (Hachiya et al., 1993). In vitro translation is performed in the presence of 1 mM ATP, which is sufficient to promote slow dissociation of the complex, but it is not likely that in vivo import occurs such a long time after completion of translation (60 min in our study). We suggest that free precursor accumulates only in vitro and that preproteins exist in vivo in a cytosolic guidance complex. This idea is supported by our observations that (1) prolonged translation reactions (>60 min) as well as elevated temperatures (>25°C) lead to an increase of free and a decrease of complexed precursor; and (2) the import efficiency of free precursor is low compared with complexed precursor, indicating that the latter is the natural substrate for import in vivo.
Are the two distinguishable oligomeric precursor complexes formed sequentially or independently of each other? The following findings should be considered as an answer to this question. All proteins analyzed in this study, irrespective of their organellar location or the presence of a transit sequence, interact with Hsp70. The cotranslational formation of Hsp70 with the nascent polypeptide chain also has been described (Beckmann et al., 1990). The precursor–Hsp70 complex exists most likely very early after translation in the wheat germ system or maybe even cotranslationally. Binding of 14-3-3 proteins to the plastid-targeting signal then occurs, either in its nonphosphorylated form or in its phosphorylated form probably with higher affinity. The prebound Hsp70 might be replaced in this reaction by the preexisting 14-3-3–Hsp70 complex present in wheat germ (see Figure 5B). It seems to be possible that the Hsp70 precursor is an intermediate in the formation of a 14-3-3–containing precursor guidance complex.
In conclusion, increasing evidence accumulates in the literature (see citations above) and from our present study to demonstrate the significance of specific cytosolic protein components in post-translational protein import into mitochondria as well as into plastids. The use of experimental systems that resemble as closely as possible the homologous situation (de Castro Silva-Filho et al., 1997) will help to reveal the cytoplasmic route of the plastid precursor proteins and the consequences for recognition and translocation into the organelle.
METHODS
In Vitro Translation
Wheat germ lysate was prepared as described previously (Anderson et al., 1983). In vitro translation of preSSU, mature SSU (Klein and Salvucci, 1992), preOE23 (Cline et al., 1993), preSSU–M31/34-S/A, preOE23–M22-S/A (Waegemann and Soll, 1996), and preF1β (Boutry and Chua, 1985) was performed (1) in wheat germ lysate in 20 mM Hepes-KOH, pH 7.6, 1 mM ATP, 0.2 mM GTP, 2 mM DTT, 25 μM of each amino acid except methionine or leucine, 8 mM creatine phosphate, 40 μg/μL (w/v) creatine phosphokinase, 30 μg/μL (w/v) spermine, and 35% wheat germ lysate; and (2) in a reticulocyte lysate (kit type I; Roche, Mannheim, Germany), according to the manufacturer's protocol. In vitro translations for the various mRNAs contained 150 μCi 35S-methionine/cysteine or 30 μCi 3H-leucine, respectively, and were conducted for 60 min at 25°C. Postribosomal supernatants were prepared by centrifugation at 100,000g for 20 min.
Immunoprecipitation
Before immunoprecipitation, polyclonal antisera were coupled to protein A–Sepharose beads (Pharmacia, Uppsala, Sweden) for 45 min at room temperature. Coimmunoprecipitations were conducted in an immunoprecipitation (IP) buffer consisting of 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% (v/v) Nonidet P-40 for 90 min at room temperature. After washing the protein A–Sepharose material with IP buffer, the bound proteins were eluted with Laemmli solubilization buffer (Laemmli, 1970). The samples were analyzed by using SDS-PAGE, gel blot analysis, and immunostaining and/or autoradiography.
Chromatographical Methods
Affinity Chromatography
PreSSU, mature SSU, and preF1β were synthesized in Escherichia coli BL21 (DE3) with a C-terminal hexahistidine tag and isolated from inclusion bodies. Approximately 5 mg of the indicated proteins was denatured in a column buffer (20 mM sodium phosphate, pH 7.6, 50 mM NaCl, and 2.5 mM MgCl2) containing 8 M urea and bound to the Ni2+ metal chelate matrix Poros20MC (Perseptive Biosystems, Wiesbaden, Germany). To ensure equal amounts of binding partners attached on the different affinity matrices, we determined protein quantity by measuring the content of unbound protein in the flowthrough and the wash fractions obtained before the affinity column. The urea-containing buffer was displaced in a gradient (flow rate of 0.2 mL/min for 3 hr) by column buffer without urea. Translationally active wheat germ lysate with a protein content of ∼25 mg was applied to the column. After extensive washing in column buffer, bound proteins were eluted stepwise with column buffer containing 1 mM ATP and a gradient from 50 to 500 mM NaCl. Protein fractions of 1 mL were collected, precipitated by 10% (w/v) trichloroacetic acid, and analyzed by SDS-PAGE followed by immunoblot analysis.
Size Exclusion Chromatography
Postribosomal supernatants from in vitro translations were loaded on a size exclusion matrix (model G3000SW; Tosohaas, Stuttgart, Germany) equilibrated in 50 mM Tris-HCl, pH 7.4, and 100 mM NaCl, and the eluate was monitored by absorbance at 280 nm. Fractions of 0.5 or 0.25 mL were collected and analyzed by using SDS-PAGE, immunoblot analysis, and/or autoradiography or concentrated in an Ultrafree-4 Centrifugal Unit (Millipore, Eschborn, Germany) for further experiments.
Chloroplast Protein Import Assays
Chloroplasts were isolated as previously described (Waegemann and Soll, 1991; Bölter et al., 1998). Protein import assays were performed essentially as described previously (Waegemann and Soll, 1995), using chloroplasts equivalent to 7.5 μg of chlorophyll in a 100-μL import assay for the times indicated in the text. These conditions allowed us to measure import in the linear range of the reaction. Import reactions were stopped by the addition of ice-cold EDTA to a final concentration of 37.5 mM, and organelles were recovered by centrifugation before analysis by SDS-PAGE, blotting, and autoradiography. Processed, mature SSU was cut out from the nitrocellulose filter and dissolved in dimethyl sulfoxide, and radioactivity was quantitated by liquid scintillation counting. Import rates were calculated from the amount of radioactive precursor added to the import reaction and corrected for the loss of 35S-methionine due to cleavage of the transit sequence (the preSSU contains seven methionine residues versus three in the mature form).
Protein Gel Blotting
Proteins were analyzed by using SDS-PAGE and blotting onto nitrocellulose membranes followed by autoradiography and/or immunostaining with specific antisera according to published procedures (Waegemann and Soll, 1995).
Acknowledgments
We thank Bettina Bölter for excellent help in performing the import experiments. This investigation was supported by grants from the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie.
References
- Aitken, A. (1996). 14-3-3 and its possible role in coordinating multiple signaling pathways. Trends Cell Biol. 6 341–347. [DOI] [PubMed] [Google Scholar]
- Aitken, A., Collinge, D.B., van Heusden, B.P., Isobe, T., Roseboom, P.H., Rosenfeld, G., and Soll, J. (1991). 14-3-3 proteins: A highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17 498–501. [DOI] [PubMed] [Google Scholar]
- Anderson, C.W., Straus, J.W., and Dudock, B.S. (1983). Preparation of a cell-free protein-synthesizing system from wheat germ. Methods Enzymol. 101 635–644. [DOI] [PubMed] [Google Scholar]
- Andrews, R.K., Harris, S.J., McNally, T., and Berndt, M.C. (1998). Binding of purified 14-3-3 ζ signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry 37 638–647. [DOI] [PubMed] [Google Scholar]
- Beckmann, R.P., Mizzen, L.E., and Welch, W.J. (1990). Interaction of Hsp70 with newly synthesized proteins: Implications for protein folding and assembly. Science 248 850–854. [DOI] [PubMed] [Google Scholar]
- Bölter, B., May, T., and Soll, J. (1998). A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441 59–62. [DOI] [PubMed] [Google Scholar]
- Boutry, M., and Chua, N.-H. (1985). A nuclear gene encoding the β subunit of the mitochondrial ATP synthase in Nicotiana plumbaginifolia. EMBO J. 4 2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright, P., Beilharz, T., Hansen, P., Garrett, J., and Lithgow, T. (1997). Mft52, an acid-bristle protein in the cytosol that delivers precursor proteins to yeast mitochondria. J. Biol. Chem. 272 5320–5325. [DOI] [PubMed] [Google Scholar]
- Chen, X., and Schnell, D.J. (1999). Protein import into chloroplasts. Trends Cell Biol. 9 222–227. [DOI] [PubMed] [Google Scholar]
- Cline, K., Henry, R., Li, C., and Yuan, J. (1993). Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 12 4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro Silva-Filho, M., Wieërs, M.-C., Flügge, U.-I., Chaumont, F., and Boutry, M. (1997). Different in vitro and in vivo targeting properties of the transit peptide of a chloroplast envelope inner membrane protein. J. Biol. Chem. 272 15264–15269. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J., Koch, B.D., Werner-Washburne, M., Craig, E.A., and Schekman, R. (1988). A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332 800–805. [DOI] [PubMed] [Google Scholar]
- Ellis, R.J., and van der Vies, S.M. (1991). Molecular chaperones. Annu. Rev. Biochem. 60 321–347. [DOI] [PubMed] [Google Scholar]
- Ferl, R.J. (1996). 14-3-3 proteins and signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 49–73. [DOI] [PubMed] [Google Scholar]
- Finnie, C., Borch, J., and Collinge, D.B. (1999). 14-3-3 proteins: Eukaryotic regulatory proteins with many functions. Plant Mol. Biol. 40 545–554. [DOI] [PubMed] [Google Scholar]
- George, R., Beddoe, T., Landl, K., and Lithgow, T. (1998). The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. USA 95 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya, N., Alam, R., Sakasegawa, Y., Sakaguchi, M., Mihara, K., and Omura, T. (1993). A mitochondrial import factor purified from rat liver cytosol is an ATP-dependent conformational modulator for precursor proteins. EMBO J. 12 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya, N., Mihara, K., Suda, K., Horst, M., Schatz, G., and Lithgow, T. (1995). Reconstitution of the initial steps of mitochondrial protein import. Nature 376 705–709. [DOI] [PubMed] [Google Scholar]
- Haucke, V., and Schatz, G. (1997). Import of proteins into mitochondria and chloroplasts. Trends Cell Biol. 7 103–106. [DOI] [PubMed] [Google Scholar]
- Hennekes, H., Peter, M., Weber, K., and Nigg, E.A. (1993). Phosphorylation on protein kinase C sites inhibits nuclear import of lamin B2. J. Cell Biol. 120 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah, S.C., Hill, K., Wagner, R., Schlicher, T., and Soll, J. (1997). Reconstitution of a chloroplast protein import channel. EMBO J. 16 7351–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, S., Aitken, A., Bertsch, U., and Soll, J. (1992). A plant homologue to mammalian brain 14-3-3 protein and protein kinase C inhibitor. FEBS Lett. 296 222–224. [DOI] [PubMed] [Google Scholar]
- Jans, D.A., Ackermann, M.J., Bischoff, J.R., Beach, D.H., and Peters, R. (1991). p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 115 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., Olsen, L.J., and Theg, S.M. (1989). Chloroplastic precursors and their transport across the envelope membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 471–501. [Google Scholar]
- Klein, R.R., and Salvucci, M.E. (1992). Photoaffinity labeling of mature and precursor forms of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase after expression in Escherichia coli. Plant Physiol. 98 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya, T., Rospert, S., Schatz, G., and Mihara, K. (1997). Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 16 4267–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., and Schnell, D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the heads of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Liu, D., Bienkowska, J., Petosa, C., Collier, R.J., Fu, H., and Liddington, R. (1995). Crystal structure of the ζ isoform of the 14-3-3 protein. Nature 376 191–194. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Kouranov, A., LaSala, S.E., and Schnell, D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh, C. (1993). Assay and purification of protein (serine/threonine) phosphatases. In Protein Phosphorylation, D.G. Hardie, ed (New York: Oxford University Press), pp. 197–230.
- Moll, T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle–regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66 743–758. [DOI] [PubMed] [Google Scholar]
- Murakami, K., Tanase, S., Morino, Y., and Mori, M. (1992). Presequence binding factor-dependent and -independent import of proteins into mitochondria. J. Biol. Chem. 267 13119–13122. [PubMed] [Google Scholar]
- Muslin, A.J., Tanner, J.W., Allen, P.M., and Shaw, A.S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84 889–897. [DOI] [PubMed] [Google Scholar]
- Palmgren, M.G., Fuglsang, A.T., and Jahn, T. (1998). Deciphering the role of 14-3-3 proteins. Exp. Biol. Online 3 4. [Google Scholar]
- Perry, S.E., and Keegstra, K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon, M., Wienk, H., Sips, W., de Swaaf, M., Talboom, I., van't Hof, R., de Korte-Kool, G., Demel, R., Weisbeek, P., and de Kruijff, B. (1995). Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 270 3882–3893. [DOI] [PubMed] [Google Scholar]
- Rihs, H.-P., Jans, D.A., Fan, H., and Peters, R. (1991). The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 10 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf, M., Waegemann, K., and Soll, J. (1995). A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 7 401–411 [DOI] [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1991). Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1 149–158. [Google Scholar]
- Waegemann, K., and Soll, J. (1995). Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 50 255–267. [DOI] [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1996). Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 271 6545–6554. [DOI] [PubMed] [Google Scholar]
- Waegemann, K., Paulsen, H., and Soll, J. (1990). Translocation of proteins into chloroplasts requires cytosolic factors to obtain import competence. FEBS Lett. 261 89–92. [Google Scholar]
- Wu, K., Rooney, M.F., and Ferl, R.J. (1997). The Arabidopsis 14-3-3 multigene family. Plant Physiol. 114 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signaling pathways. Nature 376 188–191. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.B., Rittinger, K., Volinia, S., Caron, P.R., Aitken, A., Leffers, H., Gamblin, S.J., Smerdon, S.J., and Cantley, L.C. (1997). The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91 961–971. [DOI] [PubMed] [Google Scholar]
- Yang, J., Winkler, K., Yoshida, M., and Kornbluth, S. (1999). Maintenance of G2 arrest in the Xenopus oocyte: A role for 14-3-3–mediated inhibition of Cdc25 nuclear import. EMBO J. 18 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, R., Sagstetter, M., Lewis, M.J., and Pelham, H.R. (1988). Seventy-kilodalton heat shock proteins and an additional component from reticulocyte lysate stimulate import of M13 procoat protein into microsomes. EMBO J. 7 2875–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]