Abstract
To determine the scope of gene expression controlled by the maize transcription factors C1/R and P, which are responsible for activating flavonoid synthesis, we used GeneCalling, an open-ended, gel-based, mRNA-profiling technology, to analyze cell suspension lines of the maize inbred Black Mexican Sweet (BMS) that harbored estradiol-inducible versions of these factors. BMS cells were transformed with a continually expressed estrogen receptor/maize C1 activator domain fusion gene (ER–C1) and either a fusion of C1 and R (CRC), P, or luciferase genes regulated by a promoter containing four repeats of an estrogen receptor binding site. Increasing amounts of luciferase activity, anthocyanins, and flavan-4-ols were detected in the respective cell lines after the addition of estradiol. The expression of both known and novel genes was detected simultaneously in these BMS lines by profiling the mRNA isolated from replicate samples at 0, 6, and 24 hr after estradiol treatment. Numerous cDNA fragments were identified that showed a twofold or greater difference in abundance at 6 and 24 hr than at 0 hr. The cDNA fragments from the known flavonoid genes, except chalcone isomerase (chi1), were induced in the CRC-expressing line after hormone induction, whereas only the chalcone synthase (c2) and flavanone/dihydroflavonol reductase (a1) genes were induced in the P-expressing line, as was expected. Many novel cDNA fragments were also induced or repressed by lines expressing CRC alone, P alone, or both transcription factors in unique temporal patterns. The temporal differences and the evidence of repression indicate a more diverse set of regulatory controls by CRC or P than originally expected. GeneCalling analysis was successful in detecting members of complex metabolic pathways and uncovering novel genes that were either coincidentally regulated or directly involved in such pathways.
INTRODUCTION
Flavonoids, a diverse group of low molecular weight secondary metabolites found throughout the plant kingdom, play key roles in a variety of developmental programs, biochemical processes, and environmental responses. Examples include the 3-hydroxy flavonoids or anthocyanins, which constitute flower pigments that attract pollinating insects and shield DNA from UV-B damage (Koes et al., 1994; Stapleton and Walbot, 1994). The flavonol aglycones are essential for pollen viability in some plant species (Koes et al., 1994; Deboo et al., 1995), and the silk-localized C-glycosylflavone, maysin, confers host resistance to the insect pest corn earworm (Helicoverpa zea; Lee et al., 1998). Luteolin (3′,4′,5,7-tetrahydroxyflavone) helps to establish beneficial symbiotic plant–microbe interactions between Rhizobium spp and members of the legume family (Peters et al., 1986). Interest in the impact of flavonoids on human health is growing, because of their common occurrence in food and their reputed but as yet unconfirmed nutritional and pharmaceutical effects (Hollman and Katan, 1998; Wang et al., 1998).
The first committed step of flavonoid biosynthesis is catalyzed by the enzyme chalcone synthase, which is encoded in maize by the genetic loci c2 and white pollen (whp; Wienand et al., 1986; Dooner et al., 1991; Franken et al., 1991). Chalcone synthase produces the aglycone flavonoid naringenin chalcone from malonyl-CoA and coumaroyl-CoA precursors (reviewed in Heller and Forkmann, 1994). In maize, anthocyanin synthesis requires chalcone isomerase (chi; Grotewold and Peterson, 1994; Grotewold et al., 1998), flavanone 3-hydroxylase (f3h; Deboo et al., 1995), and the well-documented genetic loci a1, a2, bronze1 (bz1), and bz2 (Reddy et al., 1987; Furtek et al., 1988; Menssen et al., 1990; Marrs et al., 1995). After the final genetically defined step, which involves a glutathione S-transferase, the cyanidin 3-O-glucoside is transferred into the vacuole by an as yet unknown pump (Alfenito et al., 1998). In a second major branch of the maize flavonoid pathway, 3-deoxyflavones, C-glycosylflavones, and the pericarp/seedling cell wall–associated phlobaphene pigment are produced from flavanone through the conversion to flavan-4-ol by the a1 gene product, which is followed by several poorly characterized steps (Grotewold et al., 1998).
Our knowledge of the complex regulatory mechanisms controlling flavonoid synthesis is mainly limited to the anthocyanin pathways in maize (Dooner et al., 1991; Mol et al., 1998), petunia (Avila et al., 1993), and Antirrhinum (Martin et al., 1991). In maize, the Myb transcription factor homolog C1 and the basic helix-loop-helix–containing factors R or B activate several genes in the anthocyanin pathway, whereas a single Myb homolog, P, can activate the subset of the known flavonoid genes that directs phlobaphene synthesis (reviewed in Koes et al., 1994; Mol et al., 1998). These factors activate certain flavonoid genes through interactions with Myb-related cis-elements (Grotewold et al., 1994; Sainz et al., 1997; Lesnick and Chandler, 1998).
Expressing the C1/R or P transcription factors in transgenic maize Black Mexican Sweet (BMS) cell lines stimulates expression of the genes of the flavonoid biosynthetic pathway, resulting in accumulation of anthocyanins and 3-deoxy flavonoids (Grotewold et al., 1998). In the P-expressing cell line, additional compounds are produced, including three types of C-glycosylflavones and the two phenylpropanoids, chlorogenic acid and ferulic acid. Because the C1/R– and P-expressing cell lines also exhibit marked changes in the subcellular localization of the synthesized compounds and subcellular structures relative to the untransformed control cell line, genes other than those of the known flavonoid pathway—such as those that synthesize coumaroyl-CoA as one of the precursors to flavonoid synthesis—are probably also regulated by these transcription factors. A key step in the phenylpropanoid pathway is the conversion of phenylalanine to cinnamic acid, catalyzed by the enzyme phenylalanine ammonia-lyase (PAL). The biochemistry of PAL is well understood, and several pal genes have been cloned and characterized (Logemann et al., 1995; Wanner et al., 1995; Rosler et al., 1997). Although the regulation of pal expression by C1/R or P has not been directly demonstrated, PAL activity in maize aleurones is controlled by viviparous 1 (Dooner, 1985), which also regulates C1 (Hattori et al., 1992).
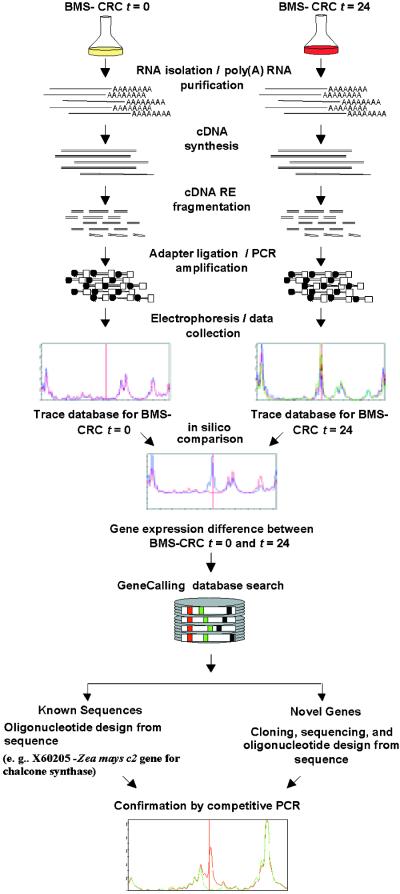
Several methods of searching for the mRNA of differentially expressed genes have been developed, including transcript sampling–based, hybridization-based, or cDNA-amplified/imaging–based technologies. Both the transcript sampling methods, such as serial analysis of gene expression (Velculescu et al., 1995) and representational difference analysis (Hubank and Schatz, 1994), and the cDNA-amplified/imaging methods, such as differential display (Liang and Pardee, 1992) and amplified fragment length polymorphism (Bachem et al., 1996), have proved successful in identifying differentially expressed genes. Yet these methods are both time consuming and resource intensive, especially when used to confirm the sequence identity of the target genes. Genomic-scale hybridization-based technologies, such as oligonucleotide arrays (Lockhart et al., 1996) and cDNA microarrays (Schena et al., 1995), have improved throughput, sensitivity, and versatility for identifying differentially expressed genes but are limited to use only with those clones and sequences that are already available. In this study we used a recently developed method that comprehensively samples cDNA populations in a highly sensitive manner (Shimkets et al., 1999) to identify differential gene expression patterns of transgenic maize cell lines that produce inducible forms of either a C1 and R fusion gene or the P gene. This method, termed GeneCalling analysis, is an open-architecture, gel-based assay that reproducibly measures changes in RNA amounts of both known and novel genes. The method is described schematically in Figure 1. Using this method, we reproducibly measured the induction of the known maize flavonoid genes and detected the expression modulation of other known and newly discovered genes.
Figure 1.
Schematic of the GeneCalling Analysis.
Total RNA is extracted from the cell lines, and poly(A)+ RNA is purified and converted to cDNA, which is fragmented by using pairs of restriction enzymes (RE). Adapters are then ligated to the ends of the fragments, which are amplified by polymerase chain reaction (PCR). Because one of the PCR primers is labeled with a fluorescent tag (fluorescamine; FAM), amplified fragments can be visualized during electrophoresis. For each sample and restriction enzyme pair, electronic images of the gel lane traces are collected and kept in a sample trace database. Comparisons of the trace databases reveal specific expression differences that are characterized by the length of the amplified fragment and by restriction enzyme sequence information. The identity of each differentially expressed gene fragment can be established either by performing a GeneCalling search in a sequence database or by cloning and sequencing the desired cDNA fragment. The identity of the cDNA fragment is confirmed by competitive PCR in which the original PCR reaction is reamplified in the presence or absence of an excess of an unlabeled, gene-specific PCR primer. Further characterization of known and newly discovered sequences that are identified by GeneCalling analysis as differentially expressed can be obtained by BLASTX and BLASTN analyses against public and proprietary databases. t, time in hours.
RESULTS
Induction of the Flavonoid Pathway
BMS cell cultures were stably transformed with both the estrogen receptor/maize C1 activator domain fusion gene (ER–C1) and one of the following constructs: the C1–R fusion gene (CRC), P, or the luciferase genes under the control of a promoter containing four repeats of an estrogen receptor binding site (4X-ERE). Upon addition of estradiol, the production of CRC and P activated the anthocyanin and phlobaphene/C-glycosylflavone pathways, respectively, in maize BMS cells as schematically shown in Figure 2. Figure 3A shows the induction of luciferase activity from the 4X-ERE::luciferase–containing control BMS cell line. Luciferase activity was not detectable at 0 hr or in BMS cells lacking the ER–C1 receptor, thus showing that in the absence of estradiol or of the modified estrogen receptor, the 4X-ERE constructs were essentially silent (data not shown; Figure 3). Following a short lag after the addition of estradiol, luciferase activity was detectable, increasing linearly for 48 hr, the longest duration of sampling. BMS cells harboring the 4X-ERE::CRC fusion gene or the 4X-ERE::P gene produced no detectable anthocyanins or flavan-4-ols, respectively (Figure 3), before induction. After estradiol was added, increases in anthocyanins and flavan-4-ols were detected by spectral absorbance after lags of 12 and 24 hr, respectively (Figure 3). In addition, the red pigmentation of the anthocyanins and the slightly brown pigmentation of the downstream metabolites of the flavan-4-ols (phlobaphenes and C-glycosylflavones; Grotewold et al., 1998) were visible 24 to 48 hr after treatment with estradiol.
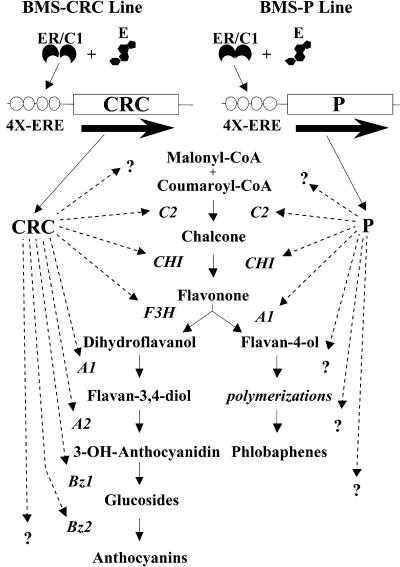
Figure 2.
Schematic of the Estradiol-Inducible Anthocyanin and Phlobaphene Pathways.
BMS cell lines harbor a constitutively expressed estrogen receptor–maize C1 activator domain fusion gene (ER/C1) that binds to estradiol (E) and in turn activates the expression of the P or CRC genes by way of 4X-ERE promoter. Members of the flavonoid pathway are then activated, generating anthocyanins or phlobaphenes. Other genes suspected to be regulated by P or CRC are designated by question marks. Genes known to be involved in the flavonoid pathway are designated as follows: C2, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; A1, flavanone/dihydroflavanonol NADPH-dependent reductase; A2, proanthocyanidin synthase; Bz1, UDP-glucose:flavonoid 3-O-glucosyltransferase; Bz2, a glutathione S-transferase.
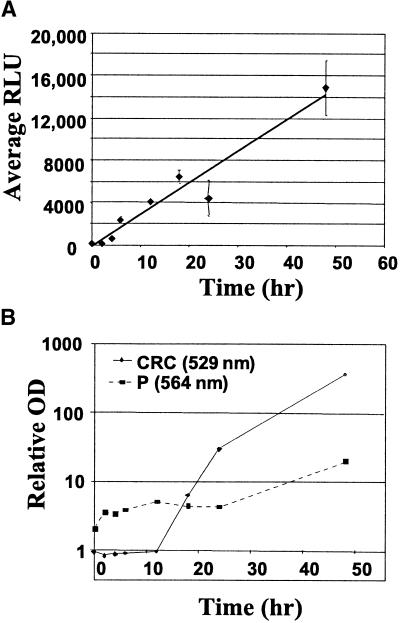
Figure 3.
Estradiol-Induced Luciferase Activity and Accumulation of Anthocyanins and Flavan-4-ols in Transformed BMS Cell Lines.
(A) Luciferase data are expressed as the mean relative light units (RLU) of three sample replicates from three independent samples for each time point, with error bars representing ±sd.
(B) Accumulation of anthocyanins and flavan-4-ols was measured by spectral absorption at 529 and 564 nm, respectively, of extracts from three replicate samples. Note the log scale of absorption values. Standard deviation for each absorbance value was <15% of the mean value, except for the 4-hr samples from the CRC-expressing line, in which the standard deviation was 39% of the mean absorbance value.
RNA Profiling
Poly(A)+ RNA was purified from three replicates of the control BMS lines and of the P- and CRC-expressing lines at 0, 6, and 24 hr after the addition of estradiol. The experimental treatments were designated CRC0, CRC6, CRC24, P0, P6, and P24. Using the GeneCalling process, as outlined in Figure 1, we focused on the gel traces of cDNA fragments ranging from 50 to 500 bp from the reactions of 68 restriction enzyme pairs for the subsequent comparisons. The trace database was analyzed by measuring the relatedness of the expression profiles among replicates within and between the cell lines, as shown in Figure 4. Cluster analysis of the trace profiles from samples between the cell lines revealed more differences than between the sampling times after treatment within a cell line. For example, computer-based comparisons of the trace data at 0 hr from the CRC- and P-expressing lines (CRC0 and P0, respectively) with those from the control line at 0 hr generated 2.83 and 1.04% of cDNA fragment differences, respectively, from >19,000 fragments detected. Although the nature of these differences is not clear, the results suggest that the expression patterns of the BMS lines may have diverged after transformations with the recombinant plasmids and subsequent culturing. We maintained the CRC- and P-expressing lines on selection media by using both the bialaphos resistance (Bar; Kumada et al., 1988) and acetolactate synthase (ALS; Fang et al., 1992) genes, because these lines were transformed with multiple templates, whereas the control line was maintained on selection media with use of only the Bar gene. The difference in selection regimes may account for the differences in cDNA expression between the lines. We therefore restricted our comparisons to a time series by analyzing the results of the treatments at 6 and 24 hr in comparison with the results at 0 hr within each cell line. The data are represented as the N-fold difference ratio of the average peak-height values for the cDNA fragments at 6 or 24 hr to that at 0 hr. The cDNA fragments that showed a significant (P < 0.1) expression difference of at least twofold had very little overlap with the fragments from the time series from the comparisons between the cell lines.
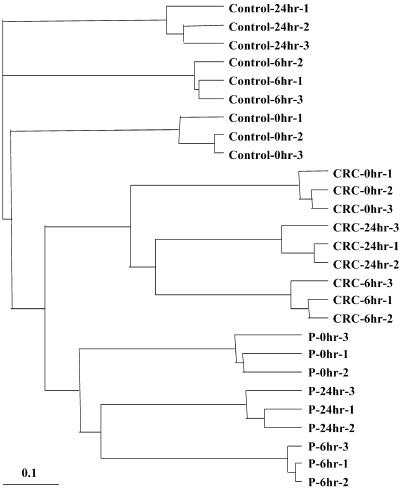
Figure 4.
Hierachical Cluster Analysis of Gene Expression Profiles of BMS Cell Line Treatments and Replicates.
Trace profiles for each replicate were compared with those for all the other replicates. The distances for clustered replicates were based on the Pearson's correlation coefficients for peak intensities of all the cDNA fragments within the corresponding traces. The dendogram showing the relatedness of samples was produced using PHYLIP (Felsenstein, 1989) and visualized using TREEVIEW (Page, 1996).  .
.
As shown in Table 1, >19,000 cDNA fragments were detected for each of the primary comparisons. The differentially expressed cDNA fragments ranged from 0.50 to 2.18% of all the gene fragments detected, resulting in a total of 919 unique cDNA fragments showing at least a twofold change in average band intensities on the gel. Ninety-two of the 919 cDNA fragments induced or repressed in the P- or CRC-expressing line (or both) were similarly induced or repressed in the control line. These fragments may have arisen from genes affected by the estradiol or the ethanol-carrier treatment directly and probably represent genes not controlled by CRC or P. Therefore, the remaining 827 fragments were expected to correspond directly to the induction of CRC and P and were analyzed further. The CRC gene product apparently affected the expression of nearly three times the number of cDNA fragments affected by P, suggesting a greater complexity of gene expression evoked by CRC than P. For the CRC6, CRC24, and P6 samples, approximately half of the cDNA fragments were upregulated and the other half were downregulated. For P24, however, almost 73% were upregulated and only 27% were downregulated. These data not only portray the potentially large numbers and complexity of the differentially regulated genes affected by CRC and P, but they also show differences in the regulated profiles between the CRC- and P-expressing lines after induction.
Table 1.
Results of Differentially Expressed cDNA Fragments Derived from the Three Maize BMS Cell Lines at Various Time Points after Estradiol Induction
| Number and Percentages of cDNA Fragments Showing Expression Differencesc
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Total
|
Unique to CRC or P
|
|||||||
| Primary Comparisonsa |
Assayed cDNA Fragmentsb |
No. | %d | Common to Control-6 or Control-24 |
No. | %d | Upregulated(No.) | Downregulated(No.) |
| Control-6 | 19,606 | 165 | 0.84 | 165 | ||||
| Control-24 | 19,406 | 172 | 0.89 | 172 | ||||
| CRC6 | 19,670 | 428 | 2.18 | 34 | 394 | 2.00 | 224 | 170 |
| CRC24 | 19,562 | 420 | 2.15 | 49 | 371 | 1.90 | 217 | 154 |
| P6 | 19,365 | 236 | 1.22 | 58 | 178 | 0.92 | 94 | 84 |
| P24 | 19,117 | 95 | 0.50 | 26 | 69 | 0.36 | 50 | 19 |
Comparisons were made against the same cell line at 0 hr.
Based on data from 68 pairs of restriction enzymes.
At least a twofold (P < 0.1) difference.
Differences as a percentage of the total cDNA fragments assayed.
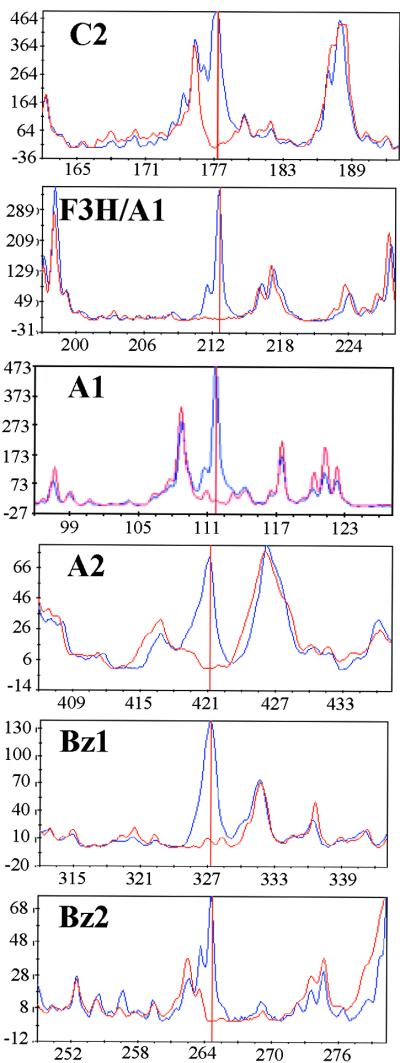
To identify the genes corresponding to the differentially expressed cDNA fragments, we first used the gene fragment length and the restriction enzyme sequence at each end of the fragment to search the public and the Pioneer/DuPont proprietary sequence databases. The cDNA fragments corresponding to known genes were subsequently cloned, sequenced, and confirmed by competitive polymerase chain reaction (PCR), as described by Shimkets et al. (1999). The differentially expressed cDNA fragments corresponding to the flavonoid pathway genes were detected in CRC24 and are shown in Figure 5 as an average trace from six to nine replicated gel lanes from the 24- and 0-hr samples. The gel band corresponding to the F3H/A1 trace (Figure 5) consisted of cDNA fragments, from both genes, that had identical band size and restriction enzyme ends, as confirmed by competitive PCR reactions. Although we detected this gel band only for the f3h gene, two other bands for the a1 gene were detected that exhibited the appropriate expression regulation (data not shown). As expected, the cDNA fragments corresponding to c2 and a1 genes were detected as induced bands in the P24 sample (data not shown). The gel traces for most of the cDNA fragments showed little variance among the samples taken at different times or among the replicates (Figure 5; data not shown).
Figure 5.
Induction of the Flavonoid Gene Fragments by CRC Measured at 24 Hr after Estradiol Treatment.
Each panel shows the average gel trace based on six to nine replicate gels of digested gene fragments from the CRC-expressing BMS line at 24 hr (blue) and 0 hr (red) after estradiol treatment. Three replicates of three independent samples were analyzed by GeneCalling. The vertical red line indicates the gene fragment that corresponds to the confirmed gene product. The designations C2, A2, Bz1, and Bz2 are described in the legend to Figure 2. The designation F3H/A1 refers to cDNA fragments from the F3H and A1 genes with identical fragment size and restriction sites at the fragment ends. The y axis is in arbitrary fluorescence units, whereas the x axis is in base pairs of nucleotides.
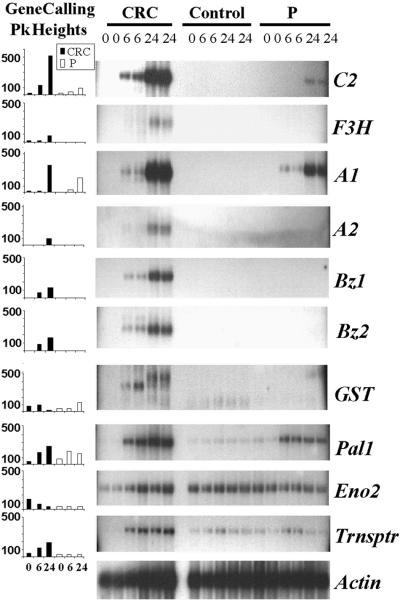
The relative change in abundance of cDNA fragments for each of the flavonoid-related genes corresponded closely with the RNA gel blot analysis, as depicted in Figure 6. The cDNA fragments corresponding to chi1 were not detected in our BMS cell lines, which is similar to the results of Grotewold et al. (1998). Other CRC- or P-regulated cDNA fragments that were identified by GeneCalling and confirmed by RNA gel blot analysis (Figure 6) included maize pal1 (Rosler et al., 1997), a multidrug-resistant protein (MRP)–type transporter (Buchler et al., 1996), and a gene for glutathione S-transferase (GST). The GeneCalling analysis of both the MRP transporter cDNA fragment and the pal1 cDNA fragment were in good agreement with the RNA gel blot analysis (Figure 6). The newly discovered GST gene showed a more complex banding pattern on the RNA gel blot, suggesting the presence of multiple mRNA species from a large gene family that hybridized with the probe. However, we detected an mRNA band that was upregulated in CRC6 and downregulated in CRC24, which was similar to the data exhibited by the profiling analysis (Figure 6). We did not detect a band of the same size for GST in the RNA gel lanes corresponding to the P-expressing line. We also detected downregulation of the enolase (eno2) (Lal et al., 1998) gene in the P-expressing line when we used GeneCalling but not in RNA gel blot analysis (Figure 6). This result may reflect cross-hybridization to RNA species from the second maize enolase gene, eno1 (Lal et al., 1998). In general, the RNA gel blot analysis reflected the results of the GeneCalling analysis.
Figure 6.
Comparison of GeneCalling and RNA Gel Blot Analyses for Several CRC- and P-Regulated Genes.
The GeneCalling peak (Pk) height graphs indicate the maximal average peak height in arbitrary fluorescence units (y axis) for the cDNA fragment corresponding to each of the analyzed genes from each sampling time. The adjacent RNA gel blots show the signals obtained in hybridization by using ∼2 μg of poly(A)+ RNA. RNA gel blot analysis was performed on two independent replicates of the three BMS cell lines (CRC, Control, and P). The blots were hybridized with probes corresponding to C2, A1, A2, Bz1, Bz2, GST, pal1 (Rosler et al., 1997), eno2 (Lal et al., 1998), and Trnsptr (MRP transporter; Buchler et al., 1996). A maize actin cDNA clone acted as an RNA loading control. Hours after estradiol treatment are indicated for the x axis of the graphs and for each lane of the gel.
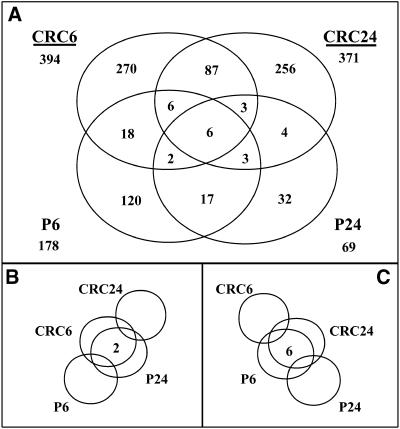
The expression levels of 663 unique cDNA fragments were shown to change specifically when CRC was present, whereas those of 217 unique cDNA fragments changed in response to P. The number of unique and shared cDNA fragments for all possible combinations of treatments that were differentially expressed are shown in Figure 7. Most of the differentially expressed cDNA fragments were specific to one treatment (Figure 7). We observed only six fragments that were induced or repressed by both CRC and P for both time points (Figure 7), among which was pal1. The c2 cDNA fragment was differentially expressed in CRC24, CRC6, and P24, whereas a1 showed a delay in expression in both CRC- and P-expressing lines and thus was shared between CRC24 and P24. The other flavonoid-related cDNA fragments were upregulated either in both CRC6 and CRC24 or in the CRC24 sample alone. A few cDNA fragments were differentially expressed either early in the CRC-expressing line and late in the P-expressing line or vice versa after estradiol treatment (Figures 7B and 7C). As these data show, all of the flavonoid-related cDNA fragments that exhibited differential gene expression were present in the expected compartments of the Venn diagram (Figure 7), demonstrating the ability of this technology to identify the gene members in the proper context of complex metabolic pathways.
Figure 7.
A Venn Diagram of Shared and Specific Differentially Expressed cDNA Fragments for All Treatments Analyzed.
(A) The numbers of unique cDNA fragments for the majority of comparisons are shown. The cDNA fragments are either induced or repressed, with a more than twofold difference detected between 6 or 24 hr and 0 hr after initiating treatment, in the cell lines indicated. These fragments are either shared or unique to particular treatments. The values shown under each treatment name are the total number of unique cDNA fragments that showed differential expression for that treatment.
(B) The number of cDNA fragments showing differences at 6 hr after treatment for CRC-expressing cells and at 24 hr after treatment for P-expressing cells versus their corresponding 0-hr values.
(C) The number of cDNA fragments showing differences at 24 hr after treatment for CRC-expressing cells and at 6 hr after treatment for P-expressing cells versus their corresponding 0-hr values. The number of cDNA fragments with responses in agreement with that of the control BMS line was excluded.
Temporal Correlation of CRC- and P-Regulated Genes
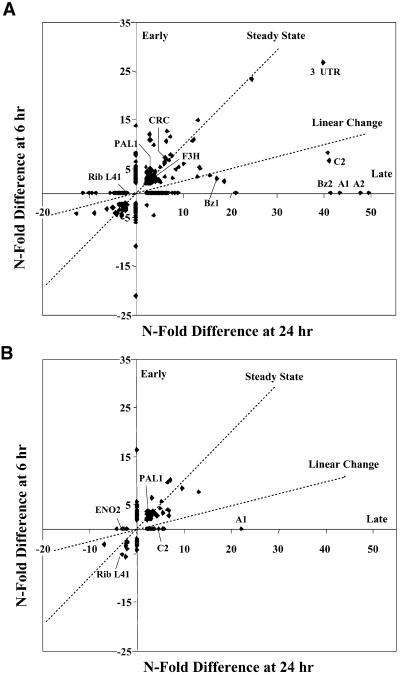
To more easily visualize the change in population of bands over time, we plotted the N-fold difference values for the cDNA fragments from the 6-hr sample against those for the 24-hr time point, as shown in Figure 8. Although transient changes in mRNA amounts may have occurred between 6 and 24 hr, these changes did not significantly affect the interpretations described here. For the CRC-expressing line, four basic clusters were observed (Figure 8A). Most of the bands were in clusters that could be designated as early (6 hr only), steady state (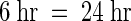 ), or late (24 hr only); a fourth, much smaller cluster of bands displayed a fourfold linear change between 6 and 24 hr. In the P-expressing line, the majority of fragments were observed as early or steady state, with fewer fragments in the late cluster and only one fragment in the linear change cluster (Figure 8B). The CRC cDNA fragment showed an increase by 6 hr but apparently reached steady state because the N-fold difference at CRC24 was nearly the same as that at CRC6. As confirmed by the RNA gel blot analysis in Figure 6, the c2 cDNA fragment increased at a nearly linear rate in the CRC-expressing line, whereas the a1 and a2 cDNA fragments were induced relatively late after the addition of estradiol (Figure 8A). A cDNA fragment for bz1 showed a nearly linear increase, whereas the cDNA fragment for bz2 was shown to be exclusively late (Figure 8A), despite the detection of the bz2-hybridizing band at 6 hr on the RNA gel blot analysis. The apparent difference between the plotted GeneCalling analysis (Figure 8A) and the RNA gel blot analysis (Figure 6) for bz2 reflected our assignment of the value zero (Figure 8A) to any change of less than twofold for the bz2 cDNA fragment at 6 hr.
), or late (24 hr only); a fourth, much smaller cluster of bands displayed a fourfold linear change between 6 and 24 hr. In the P-expressing line, the majority of fragments were observed as early or steady state, with fewer fragments in the late cluster and only one fragment in the linear change cluster (Figure 8B). The CRC cDNA fragment showed an increase by 6 hr but apparently reached steady state because the N-fold difference at CRC24 was nearly the same as that at CRC6. As confirmed by the RNA gel blot analysis in Figure 6, the c2 cDNA fragment increased at a nearly linear rate in the CRC-expressing line, whereas the a1 and a2 cDNA fragments were induced relatively late after the addition of estradiol (Figure 8A). A cDNA fragment for bz1 showed a nearly linear increase, whereas the cDNA fragment for bz2 was shown to be exclusively late (Figure 8A), despite the detection of the bz2-hybridizing band at 6 hr on the RNA gel blot analysis. The apparent difference between the plotted GeneCalling analysis (Figure 8A) and the RNA gel blot analysis (Figure 6) for bz2 reflected our assignment of the value zero (Figure 8A) to any change of less than twofold for the bz2 cDNA fragment at 6 hr.
Figure 8.
Temporal Correlation of cDNA Fragment N-Fold Difference Values for the CRC- and P-Expressing Lines.
(A) Individual cDNA fragments from the CRC-expressing BMS line were plotted as their N-fold difference between the results for the 6-hr and 24-hr samples. The N-fold differences are the ratios of the average peak height values at either 24 or 6 hr after treatment to those at 0 hr for each cDNA fragment. Results for differences of less than twofold are not shown.
(B) Individual cDNA fragments from the P-expressing BMS line were plotted as the N-fold difference in results between 6 hr and 24 hr after treatment.
Early and Late refer to genes that are induced or repressed at either the 6- or 24-hr times only, respectively. Steady State refers to genes for which the differential expression at 6 hr remains the same at 24 hr, whereas Linear Change refers to those that are either induced or repressed in a linear manner from 6 hr to 24 hr. Some of the cDNA fragments corresponding to genes regulated by CRC or P are indicated as follows: CRC, the C1–R fusion gene; C2, A1, A2, Bz1, Bz2, and F3H (as in the legend to Figure 2); Pal1 and Eno2 (as in the legend to Figure 6); Rib L41, ribosomal protein L41; 3′ UTR, the poly(A) addition sequence from the potato PinII gene present in all of the clones introduced into the BMS cells.
Identification of cDNA Fragments
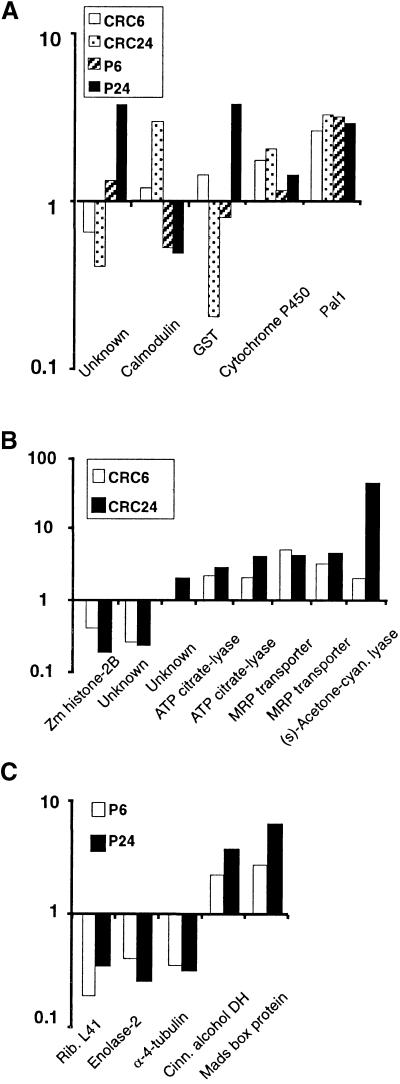
Twenty-eight cDNA fragments that were induced or repressed by CRC, P, or both were cloned, sequenced, and confirmed by competitive PCR analysis. Ten corresponded to the six known maize flavonoid genes. Of the remaining fragments described in Table 2, 15 were assigned to known genes on the basis of sequence similarity; the sequences of the remaining three cDNA fragments were not similar to entries in any database. Of the 15, some of the fragments did not show any marked resemblance to the public database entries. Nonetheless, these were assigned gene names on the basis of sequence identity with expressed sequence tag (EST) clones in the Pioneer/DuPont maize EST database that showed a strong similarity to entries in public databases entry. Figures 9A and 9B show the N-fold differences between the cell lines expressing CRC and P for the cDNA fragments described in Table 2.
Table 2.
GeneCalling cDNA Fragments Related to Genes in GenBank
| GenBank Accession No.
|
||||
|---|---|---|---|---|
| Band Namea | Gene Similarity | BLASTXb | Public Match | cDNA Fragment |
| Both CRC and P | ||||
| CG02-94.1 | Unknown | AF204888 | ||
| CG04-231.6 | Calmodulin | 6.50E-23 | L19359c | AF204891 |
| CG02-243.4 | Glutathione S-transferase | 1.00E-24 | AF004358c | AF204887 |
| CG43-398.3 | Cytochrome P450 | 8.60E-27 | Q42700c | AF204893 |
| CG22-161.8 | Phenylalanine ammonia-lyase 1 | 0.00E+00d | L77912 | AF204900 |
| CRC specific | ||||
| CG66-135.1 | Maize histone 2B | 3.00E-14 | P30755 | AF204897 |
| CG71-261.9 | Unknown | AF204886 | ||
| CG10-213.9 | Unknown | AF204894 | ||
| CG28-291.1 | ATP citrate-lyase | 9.30E-18 | P16638 | AF204892 |
| CG31-165.4 | ATP citrate-lyase | 2.00E-05 | P16638c | AF204885 |
| CG10-267.2 | MRP transporter | 1.80E-11 | U92650c | AF204895 |
| CG47-259.8 | MRP transporter | 1.20E-64 | NM_000392 | AF204901 |
| CG36.437.3 | (s)-Acetone-cyanohydrin lyase | 8.00E-14 | AC003040 | AF204896 |
| P specific | ||||
| CG45.139.0 | Ribosomal protein L41 | 3.90E-06 | U26255 | AF204898 |
| CG69-64.3 | Enolase 2 | 3.60E-54 | U17973c | AF204899 |
| CG97-163.0 | Maize α-4 tubulin | 0.00E+00 | P33626c | AF204884 |
| CG04-168.2 | Cinnamoyl alcohol dehydrogenase | 5.60E-19 | T02767 | AF204890 |
| CG25-383.8 | MADS box protein | 2.20E-09 | Q41828 | AF204889 |
Band name is designated by an arbitrary labeling convention that includes the cDNA fragment size in base pairs.
BLASTX output is the probability value of 1.0E-5 or less of the translated cDNA fragment randomly matching a public database entry. Values of 0.00E + 00 indicate an exact amino acid match.
The corresponding GeneCalling cDNA fragments did not show a direct match to an entry in GenBank but were essentially identical to EST clones from the Pioneer/DuPont maize database that matched the GenBank accessions indicated with probability value of 1.0E-5 or less.
The fragment CG22-161.8 was based on only competitive PCR using the matching public entry.
Figure 9.
Differential Expression of Known and Newly Discovered Gene Fragments after Induction of CRC and P.
(A) The N-fold differences for cDNA fragments affected by both CRC and P. The data are shown as the ratio of the average peak-height values from cDNA fragments at 6 or 24 hr after treatment initiation to the 0-hr value.
(B) N-fold differences for cDNA fragments affected by CRC alone.
(C) N-fold differences for cDNA fragments affected by P alone. The average peak-height values showed <15% variance over the mean of six to nine replicate gel analyses. Shown are the gene names from GenBank matching most closely the amino acid sequences of the corresponding cDNA fragments.
Several of the fragments corresponded to genes that were expected to impact the flavonoid pathway directly. For example, a newly discovered GST sequence with 58% amino acid identity to bz2 was repressed by CRC but induced by P at 24 hr, in contrast with the regulation of the bz2 gene. An MRP transporter fragment, also determined to be induced by CRC alone, is possibly the pump necessary for transporting flavonoid compounds across the tonoplast (Alfenito et al., 1998). Fragments that arose from genes involved in producing precursors to the flavonoid pathway included pal1 (Rosler et al., 1997) and ATP citrate-lyase (Elshourbagy et al., 1990); fragments that arose from genes affecting the precursor pools included the P-induced cinnamoyl alcohol dehydrogenase (cad, which is identical to bm1; Halpin et al., 1998). A cDNA fragment that was homologous to a cytochrome P450 gene was shown to be upregulated by both CRC and P and may correspond to a previously unidentified flavonoid oxidoreductase. Interestingly, the cDNA fragments for some of the downregulated genes included those that affect general cell function, namely, eno2 and the genes for ribosomal protein L41, histone 2B, and α-4 tubulin (Figure 9).
DISCUSSION
mRNA Profiling Analysis
Using GeneCalling technology, we profiled gene expression of maize BMS cell lines that had been induced to generate either the P or the CRC transcription activators and measured the differences in amounts of cDNA fragments corresponding to most of the known genes in the maize flavonoid pathway. We found differential expression of genes that were not known to be affected by P- and C1/R–directed regulation. We also identified differential expression of cDNA fragments with no sequence similarities to entries in either the public databases or the Pioneer/DuPont proprietary maize EST database. Therefore, we found this open architecture, gel-based method to be quite useful in the discovery of known and unknown genes and in placing the genes in an appropriate biological context.
GeneCalling profiling allows identification of differentially expressed gene fragments by pairwise comparisons of expression profiles. The extent of the expressed gene coverage with 72 restriction enzyme pairs was estimated to be >90% for various genomes, including maize (Shimkets et al., 1999). We detected >19,000 cDNA fragments in each of the nine treatments (three cell lines and three time points) and estimated their relative abundance by using six to nine trace profiles per treatment. Because approximately two to three cDNA fragments can be detected per gene by using 72 restriction enzyme pairs with maize sequences (Shimkets et al., 1999), the number of unique genes corresponding to the detected cDNA fragments per BMS cell line treatment performed with 68 restriction pairs was extrapolated to be between 6000 and 8000. Computer-based treatment comparisons identified 827 unique cDNA fragments that were differentially expressed in the CRC- or P-expressing lines but not in the control line (Table 1 and Figure 7). A hierarchical cluster analysis of the cDNA fragment profiles showed that the replicates within a cell line were related, whereas those between cell lines diverged, which reaffirmed the strategy of examining a time series to generate an appropriate set of differentially expressed genes (Figure 4).
The range of N-fold differences for each treatment was generally proportional to that observed by RNA gel blot analysis (Figure 6). Because cDNA fragments are generated by restriction enzyme digestion, the GeneCalling analysis is sensitive to gene- or allele-specific polymorphisms and is more likely to provide information at a gene-specific level than does RNA gel blot analysis. Although efforts were made to achieve gene-specific hybridizations for the RNA gel blots, results for some samples did not correlate with the mRNA profiling, possibly because probes may have cross-hybridized with transcripts corresponding to other gene family members. Both the GST and eno2 genes have known paralogs in maize (Marrs et al., 1995; Lal et al., 1998; W. Bruce, unpublished results), which may have affected the RNA gel blot analysis relative to the regulation observed by the GeneCalling method.
chi1 was the only known gene of the flavonoid pathway for which we did not detect expression in these BMS cell lines. In a previous study, expression of chi1 was not detected in BMS cells in which P and C1/R were continually expressed at relatively high amounts (Grotewold et al., 1998). Perhaps the continual presence of these transcription factors negatively affected chi1 expression. However, we found that the induction of P or CRC from near background amounts also did not stimulate chi1 expression as determined by the gel-based profiling or RNA gel blot analysis (data not shown). This confirmed the previously reported observation that the chi1 gene either is not actively inducible or is nonfunctional in BMS cell lines. Because chalcone can undergo spontaneous isomerization (Moustafa and Wong, 1967) and no known maize mutants of chi have been identified to date, chi1 may be unnecessary for anthocyanin production in these cell lines.
Temporal and Induction/Repression Controls by CRC and P
Because hundreds of gene fragments displayed differential expression over time, clustering and temporal correlation analyses were used to understand more clearly the relationship between gene expression and the biochemical changes that occurred after the induction of CRC and P. Comparisons of the expression profiles between the BMS lines showed both common and P- or CRC-specific cDNA fragments that were induced or repressed. The temporal correlation analysis revealed unexpected regulation either as a direct influence of expression of CRC or P or as an indirect consequence resulting from the overall changes to the cell function caused by activating the flavonoid pathways (Figure 8). In the CRC-expressing line, cDNA fragments corresponding to the gene products at the beginning of the flavonoid pathway, before the branch point between flavonoid- and phlobaphene-specific pathways (i.e., the early flavonoid biosynthetic genes; Jackson et al., 1992), were detected generally before those at the end of the pathway. This pathway-dependent temporal control is somewhat similar to the expression control of the Antirrhinum anthocyanin pathway described by Jackson et al. (1992).
In addition to increased amounts of transcripts after the induction of CRC or P, the amounts of numerous cDNA fragments were downregulated, indicating inhibition of gene expression. Many of the fragments showed repression as early as 6 hr after the synthesis of these transcription factors, suggesting a more direct effect on transcriptional or post-transcriptional control not previously attributed to these factors. Additional examination of the effects of P or CRC on gene expression, such as mRNA profiling of CRC-, C1-, R-, or P-expressing lines with and without protein translation inhibitors (e.g., cycloheximide) or profiling samples collected at earlier times, may be necessary before drawing more concrete conclusions. Other Myb-related factors such as the human c-Myb factor are known to function both as activators and repressors (Nakagoshi et al., 1989; Mizuguchi et al., 1995; Oelgeschlager et al., 1996; Ganter and Lipsick, 1997; Salomoni et al., 1997). Perhaps the CRC and P Myb-related factors also function in both regulatory capacities.
cDNA Fragments Affected by Both CRC and P
We further analyzed a small number of cDNA fragments from the hundreds showing differential expression affected by CRC, P, or both. Several of the cDNA fragments corresponded to gene members of the flavonoid pathway (Figure 5). The remaining cDNA fragments were grouped into three expression regulation categories: CRC specific, P specific, and those affected by both CRC and P (Table 2 and Figure 9). Of those affected by both transcription factors, a diverse set of expression controls was evident and included examples of induction or repression by both factors as well as contrasting regulation between the two factors.
One cDNA fragment induced by both CRC and P, CG22-161.8, was upregulated by both transcription factors and corresponded to the maize pal1 gene (Rosler et al., 1997), which initiates the pathway to produce 4-coumaroyl-CoA, one of the precursors of flavonoid synthesis. Although pal regulation has been linked to flavonoid regulation in many plant species, including maize (Dooner, 1985; Christie et al., 1994; Koes et al., 1994), no direct evidence of C1/R or P regulating the amounts of pal1 mRNA in maize has been previously documented. Another fragment induced by both factors, CG43-398.3, was similar to a soybean cytochrome P450 and could potentially function as another flavonoid hydroxylase for both pathways. Of those that showed contrasting regulation by CRC and P, the fragment CG02-243.4 is similar to both the wheat GST TSI-1 (Riechers et al., 1997) and the maize bz2 genes (Marrs et al., 1995). This CRC-repressed, P-induced gene may encode an enzyme that adds glutathione to metabolites of the phlobaphene pathway analogous to the function of bz2 in the anthocyanin pathway.
cDNA Fragments Regulated Specifically by CRC or P
We cloned and sequenced several cDNA fragments that were regulated by either CRC or P specifically (Figure 9 and Table 2). The CRC-induced fragments CG28-291.1 and CG31-165.4 both corresponded to an ATP citrate-lyase (Elshourbagy et al., 1990). This enzyme generates acetyl-CoA that could be converted to the flavonoid precursor malonyl-CoA by a cytosolic version of acetyl-CoA carboxylase (Ashton et al., 1994; Podkowinski et al., 1996). The fragment CG47-259.8 was induced by CRC specifically and its sequence highly resembled that of a human MRP transporter gene (Buchler et al., 1996). We think it very likely that this MRP transporter homolog functions as the tonoplast glutathione pump necessary for transporting cyanidin 3-O-glucosides into the vacuole after the last genetically defined step in anthocyanin synthesis (Alfenito et al., 1998). Two other fragments induced by either CRC or P are CG36-437.3, which shows marked identity to an Arabidopsis open reading frame matching an acetone–cyanohydrin lyase (Trummler and Wajant, 1997), and CG04-168.2, which exhibits strong identity to the maize cad gene. Halpin et al. (1998) reported that maize cad is probably the same as that encoded by bm1, which contributes to lignin synthesis and shares precursors with the known flavonoid pathways. We also identified three different cDNA fragments affected by CRC and/or P that lacked any noticeable sequence identity to known genes or ESTs at either the amino acid or nucleotide level.
Conclusion
Overall, the mRNA-profiling method provided new information about the regulatory effects of C1/R and P on gene expression and revealed genes that were not known to be regulated by these transcription factors. Grotewold et al. (1998) previously demonstrated that when the flavonoid pathway is active, BMS cells undergo detectable changes involving numerous cellular processes and structures. The rate of cell growth in cell lines producing anthocyanins, for example, slowed markedly (Grotewold et al., 1998), concomitant with the apparent repression of cDNA fragments corresponding to eno2 and the genes for histone 2B, ribosomal L41, and α-4 tubulin, as detected by the mRNA profiling method (Figure 9 and Table 2). We would also expect that various genes involved in signal transduction, such as the cDNA fragments identified as calmodulin and MADS box protein (Table 2), might play an important role in such modifications at the cellular level. C1/R and P apparently affected the expression of genes from an extensive set of pathways that provide the necessary components for synthesizing the flavonoids, suggesting a more complexly regulated pathway than previously known. We analyzed only a small portion of the cDNA fragments that showed differential regulation; analysis of additional cDNA fragments should provide a more complete understanding of how the flavonoid pathway and basic cellular functions are integrated. Determining how the known genes, shown to be associated with the flavonoid pathway, and newly identified genes under apparent control of CRC or P, are expressed and regulated in intact plants will also be important. Continuing analysis of the expression of the gene fragments identified in this work, in various P- and C1/R–containing genotypes, will further our understanding of the potentially broad repertoire of gene expression control attributed to these transcription factors.
METHODS
Plant/DNA Materials and Transformation
The Zea mays inbred Black Mexican Sweet (BMS) cells, P2 No. 10, were maintained on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 2,4-D at 1.5 mg/L as a suspension culture shaken at 250 rpm and kept in the dark at 27°C. The genotype of BMS is P-ww A1 A2 C1 C2 R-g b pl (Grotewold et al., 1998). These cells, derived from cambial tissue in the mesocotyl region, do not ordinarily express any of the known transcription factors controlling the flavonoid pathway, including B, C1, P, Pl, or R (see references in Grotewold et al., 1998).
Transformation of BMS cells by microprojectile bombardment was conducted essentially as described (Grotewold and Peterson, 1994). The control BMS line was transformed with the following plasmids: PHP610 (35S::Bar), PHP5947 (4X-ERE::LUC), and PHP6417 (−59_35S::ER–C1). The CRC-expressing BMS line was transformed with the plasmids PHP6417 (−59_35S::ER–C1) and PHP6680 (UBI::ALS and 4X-ERE::CRC). Plasmid PHP6417 involved the modification of the estrogen receptor from the MCF-7 cell line (Greene et al., 1986) as follows. Between amino acids 71 and 78 of the estrogen receptor, the acidic activation domain of the maize C1 transcription activator (amino acids 146 to 269; Paz-Ares et al., 1987) was inserted in frame. This ER–C1 fusion gene was under the control of the truncated −59-bp cauliflower mosaic virus (CaMV) 35S promoter similar to vectors described in Unger et al. (1993). Plasmid PHP6680 included both the maize Ubi1 promoter and its first exon and intron (Bruce et al., 1989) fused to the maize acetolactate synthase coding sequence (UBI::ALS; L. Tagliani, unpublished results) and the four repeats of the ERE element, 5′-GGCCGCTCGAGTCCAAAGTCA-GGTCACAGTGACCTGATCAAAGTGTCCAAAGTCAGGTCACAGTGA-CCTGATCAAAGTTGTCA-3′ (Klein-Hitpass et al., 1988), upstream of a −59-nucleotide 5′ deletion of the CaMV 35S core promoter (4X-ERE) fused to a chimeric C1 and R-paralog fusion gene, designated CRC. The 4X-ERE::CRC (PHP6680) construct contains a transactivator fusion of Lc, a member of the maize R gene family (amino acids 1 to 610; Ludwig et al., 1989), inserted into the maize C1 gene (Paz-Ares et al., 1987) between the C1 Myb and transcription activation domains at amino acid 125. To create the P-containing BMS cell line, the control BMS line was transformed with an additional plasmid, PHP6682 (UBI::ALS and 4X-ERE::P). All fusion genes were terminated with the poly(A) addition site from the potato PinII gene (Unger et al., 1993).
Estradiol Induction and RNA Isolation of BMS Cell Lines
Each transformed BMS cell line was grown as three replicate cultures in 1-liter flasks with 300 mL of MS medium plus 2,4-D at 1.5 mg/L. After 4 days, 15 g of cells was transferred with 150 mL of conditioned medium to a sterile 1-liter flask containing an additional 150 mL of fresh MS medium and 2,4-D at 1.5 mg/L. Ethanol (100 μL) was added to each culture cell line 24 hr before the induction with estradiol. The next day, 280 μL of 10 mM 17α-ethylnylestradiol (Sigma) in ethanol was added to each culture to a final concentration of 10 μM. Approximately 20 mL of each cell suspension was collected at 0, 6, and 24 hr after the estradiol addition and was transferred into individual preweighed 50-mL conical centrifuge tubes. After centrifugation and aspiration of liquid medium, the fresh weight of the remaining cells was recorded, and the cells were frozen in liquid nitrogen and stored at −80°C. Frozen tissues were ground, and total RNA was extracted with Tripure isolation reagent (Boehringer Mannheim) according to the manufacturer's instructions. Total RNA samples were used both for the RNA gel blot and the GeneCalling analyses (Shimkets et al., 1999).
Flavonoid and LUC Detection
At 0, 2, 4, 6, 12, 18, 24, and 48 hr after estradiol addition, 1 mL of each cell suspension (ranging in fresh weight from 40 to 70 mg) was transferred to 1.5-mL microcentrifuge tubes and centrifuged at 1300g. Cells from the CRC BMS and P BMS lines were lyophilized and used for analysis of anthocyanin or flavan-4-ols essentially as described (Grotewold et al., 1998), except that 564 nm instead of 559 nm was used for the cell line expressing P. For comparisons between samples, absorbance values were adjusted according to the mass of lyophilized tissue. Cells from the control BMS line were pulverized in 200 μL of luciferase grinding buffer (Unger et al., 1993), centrifuged for 5 min at 16,000g, and then stored at −80°C. Luciferase assays were performed as previously described (Unger et al., 1993), and data were normalized to the amount of soluble protein present, as determined by the Bradford assay (BioRad).
RNA Gel Blot Analysis
Isolated from total RNA with the use of a Poly-A-Tract kit (Promega), ∼2 μg of poly(A)-enriched RNA was separated on a 1.2% SeaKem gel containing MOPS (Ambion, Inc.) and 4% formaldehyde. After electrophoresis, the gel was washed twice in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and blotted overnight onto a Nytran membrane by using the TurboBlotter (Schleicher and Schuell) system and protocol. The blot was air-dried for 15 min and UV cross-linked in a Stratalinker (Stratagene) at 1200 μJ/cm2. The RNA gel blot was prehybridized and hybridized in ExpressHyb buffer (Clontech), according to the manufacturer's protocol, with the following randomly primed 32P-labeled probes from our maize expressed sequence tag (EST) library or Pioneer plasmid collections: c2 (CHWAR86), a1 (CZAAZ08), a2 (CR1.pk002.C12), bz1 (pPHP2012), bz2 (CKDAF37), GST (CFBBF17), eno2 (CLDCX48), pal1 (CCMAV49), MRP transporter (CGSNW75), chi1 (CPICB71), and actin (provided by M. Cigan and E. Unger, Pioneer Hi-Bred). The blot was successively probed and stripped by using the Strip-EZ kit (Ambion, Inc.), according to the manufacturer's protocol.
GeneCalling Analysis of RNA from BMS Lines
All steps in GeneCalling analysis (Figure 1) were performed according to Shimkets et al. (1999), except that poly(A)+ RNA was isolated from 50 μg of total RNA. Additional details of this method are also described by Rothberg et al. (1999). Briefly, poly(A)+ RNA was converted to double-stranded cDNA and digested with each of the 68 restriction enzyme pairs. The resulting fragments were ligated to a pair of restriction enzyme-based oligonucleotide adapters, one of which was labeled with fluorescamine (FAM) and the other with biotin. After a 20-cycle polymerase chain reaction (PCR), the fragments were isolated with streptavidin-coated beads, separated on an ultrathin gel, and visualized by using the FAM label to generate gel lane trace data, which were stored electronically in a database.
For each BMS cell line, the GeneCalling data for the 24- and 6-hr samples were compared with those for their respective 0-hr samples. The cDNA fragments from the control BMS line that showed twofold or greater differences (either induced or repressed after the addition of estradiol to the liquid medium) were removed from the data sets of differentially expressed gene fragments of the CRC- and P-expressing lines. The remaining data represented gene fragments that showed changes specifically resulting from the induction of CRC or P. Known genes were identified by a GeneCalling-mediated search in a proprietary maize database, consisting of all publicly available maize sequences and maize ESTs from the proprietary Pioneer/DuPont database. Competitive PCR (described below) was used to confirm that a known gene did indeed correspond to the differentially expressed cDNA fragment, although this procedure cannot delineate the difference between highly homologous gene family members. Differentially expressed fragments that did not have a match in the GeneCalling maize sequence database were cloned from a reamplification of the original PCR reaction into a TA cloning vector (InVitrogen) by using standard protocols.
Competitive PCR confirmation involved using the sequence of the known genes or novel clones to design a gene-specific primer. The gene-specific PCR primer was synthesized such that it overlapped with the adapter sequence restriction enzyme site and extended into the putative gene sequence identified in the database search. The original PCR reaction containing the FAM-labeled, adapter-specific primer was repeated in the presence or absence of excess unlabeled gene-specific primer. If, in the presence of the competing primer, the differentially expressed fragment was ablated without affecting the amplification of neighboring peaks, the identification of the differentially expressed gene fragment was considered to be correct.
The cloned gene fragments were sequenced by conventional methods, and sequences were subjected initially to BLASTX searches of public databases. Those with little or no match to existing database entries (with P > 1.0 × 10−5) were subjected to BLAST 2.0 searches of the Pioneer/DuPont maize EST database for matches that showed significant sequence similarities to an EST. The corresponding EST sequence was then used in BLASTX searches, and any matches with a probability ⩽ 1.0 × 10−5 were entered into Table 2.
Hierarchical cluster analysis was performed to evaluate the relatedness in RNA profiling across all replicates by using the expression profiles of all the samples and their replicates. The dendogram showing the relatedness of samples was produced by using the PHYLIP program (Felsenstein, 1989) and visualized by using the TREEVIEW program (Page, 1996). The distances (Dij) for the dendogram were calculated as follows:
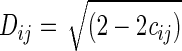 |
where cij is the Pearson's correlation coefficient between any two replicates and was calculated from the peak intensities of gene fragments that were significantly different between the composite trace profiles of any two replicates.
Acknowledgments
We thank Pete Solan and Dr. Cory Brouwer for producing and maintaining the CRC- and P- expressing cell lines, respectively. We also extend our thanks to Sandi Meyer and Joni Heller for assistance in the luciferase and protein assays, respectively. Our gratitude is also extended to Drs. Joe Felsenstein (University of Washington, Seattle) and Roderic Page (University of Glasgow, Scotland) for access to the programs PHYLIP and TREEVIEW, respectively, and to Dr. Joel Bader (CuraGen Corp.) for providing the RNA-profiling clustering algorithms. We especially thank the GeneCalling and Confirmation teams at CuraGen Corporation under the supervision of Dr. Michael McKenna for their excellent contributions. We also thank Drs. Erich Grotewold and Gerard Irzyk for critical evaluation of the manuscript.
References
- Alfenito, M.R., Souer, E., Goodman, C.D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton, A.R., Jenkins, C.L.D., and Whitfeld, P.R. (1994). Molecular cloning of two different cDNAs for maize acetyl-CoA carboxylase. Plant Mol. Biol. 24 35–49. [DOI] [PubMed] [Google Scholar]
- Avila, J., Nieto, C., Canas, L., Benito, M.J., and Paz-Ares, J. (1993). Petunia hybrida genes related to the maize regulatory C1 gene and to animal Myb proto-oncogenes. Plant J. 3 553–562. [DOI] [PubMed] [Google Scholar]
- Bachem, C.W., van der Hoeven, R.S., de Bruijn, S.M., Vreugdenhil, D., Zabeau, M., and Visser, R.G. (1996). Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J. 9 745–753. [DOI] [PubMed] [Google Scholar]
- Bruce, W.B., Christensen, A.H., Klein, T., Fromm, M., and Quail, P.H. (1989). Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc. Natl. Acad. Sci. USA 86 9692–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler, M., Konig, J., Brom, M., Kartenbeck, J., Spring, H., Horie, T., and Keppler, D. (1996). cDNA cloning of the hepatocyte canalicular isoform of the multidrug resistance protein, cMrp, reveals a novel conjugate export pump deficient in hyperbilirubinemic mutant rats. J. Biol. Chem. 271 15091–15098. [DOI] [PubMed] [Google Scholar]
- Christie, P.J., Alfenito, M.R., and Walbot, V. (1994). Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194 541–549. [Google Scholar]
- Deboo, G.B., Albertsen, M.C., and Taylor, L.P. (1995). Flavanone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinated in maize anthers. Plant J. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Dooner, H.K. (1985). Viviparous-1 mutation in maize conditions pleiotropic enzyme deficiencies in the aleurone. Plant Physiol. 77 486–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25 173–199. [DOI] [PubMed] [Google Scholar]
- Elshourbagy, N.A., Near, J.C., Kmetz, P.J., Sathe, G.M., Southan, C., Strickler, J.E., Gross, M., Young, J.F., Wells, T.N., and Groot, P.H. (1990). Rat ATP citrate-lyase. Molecular cloning and sequence analysis of a full-length cDNA and mRNA abundance as a function of diet, organ, and age. J. Biol. Chem. 265 1430–1435. [PubMed] [Google Scholar]
- Fang, L.Y., Gross, P.R., Chen, C.H., and Lillis, M. (1992). Sequence of two acetohydroxyacid synthase genes from Zea mays. Plant Mol. Biol. 18 1185–1187. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5 164–166. [Google Scholar]
- Franken, P., Niesbach-Klosgen, U., Weydemann, U., Marechal-Drouard, L., Saedler, H., and Wienand, U. (1991). The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene In. EMBO J. 10 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtek, D., Schiefelbein, J., Johnston, F., and Nelson, O.E. (1988). Sequence comparisons of three wild-type Bronze-1 alleles from Zea mays. Plant Mol. Biol. 11 473–481. [DOI] [PubMed] [Google Scholar]
- Ganter, B., and Lipsick, J.S. (1997). Myb binding sites within the N-ras promoter repress transcription. Oncogene 15 193–202. [DOI] [PubMed] [Google Scholar]
- Greene, G.L., Gilna, P., Waterfield, M., Baker, A., Hort, Y., and Shine, J. (1986). Sequence and expression of human estrogen receptor complementary DNA. Science 231 1150–1154. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., and Peterson, T. (1994). Isolation and characterization of a maize gene encoding chalcone flavanone isomerase. Mol. Gen. Genet. 242 1–8. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The Myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., Clair, G.S., and Bowen, B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10 721–740. [PMC free article] [PubMed] [Google Scholar]
- Halpin, C., Holt, K., Chojecki, J., Oliver, D., Chabbert, B., Monties, B., Edwards, K., Barakate, A., and Foxon, G.A. (1998). Brown-midrib maize (bm1)—A mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14 545–553. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Vasil, V., Rosenkrans, L., Hannah, L.C., McCarty, D.R., and Vasil, I.K. (1992). The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev. 6 609–618. [DOI] [PubMed] [Google Scholar]
- Heller, W., and Forkmann, G. (1994). Biosynthesis of flavonoids. In The Flavonoids, J.B. Harborne, ed (New York: Chapman and Hall), pp. 499–535.
- Hollman, P.C., and Katan, M.B. (1998). Bioavailability and health effects of dietary flavonols in man. Arch. Toxicol. Suppl. 20 237–248. [DOI] [PubMed] [Google Scholar]
- Hubank, M., and Schatz, D.G. (1994). Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22 5640–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D., Roberts, K., and Martin, C. (1992). Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinum majus. Plant J. 2 425–434. [Google Scholar]
- Klein-Hitpass, L., Ryffel, G.U., Heitlinger, E., and Cato, A.C. (1988). A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 16 647–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, R.E., Quattrocchio, F., and Mol, J.N.M. (1994). The flavonoid biosynthetic pathway in plants—Function and evolution. Bioessays 16 123–132. [Google Scholar]
- Kumada, Y., Anzai, H., Takano, E., Murakami, T., Hara, O., Itoh, R., Imai, S., Satoh, A., and Nagaoka, K. (1988). The bialaphos resistance gene (bar) plays a role in both self-defense and bialaphos biosynthesis in Streptomyces hygroscopicus. J. Antibiot. 41 1838–1845. [DOI] [PubMed] [Google Scholar]
- Lal, S.K., Lee, C., and Sachs, M.M. (1998). Differential regulation of enolase during anaerobiosis in maize. Plant Physiol. 118 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.A., Byrne, P.F., McMullen, M.D., Snook, M.E., Wiseman, B.R., Widstrom, N.W., and Coe, E.H. (1998). Genetic mechanisms underlying apimaysin and maysin synthesis and corn earworm antibiosis in maize (Zea mays L.). Genetics 149 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, P., and Pardee, A.B. (1992). Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257 967–971. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J., Dong, H., Byrne, M.C., Follettie, M.T., Gallo, M.V., Chee, M.S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., and Brown, E.L. (1996). Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14 1675–1680. [DOI] [PubMed] [Google Scholar]
- Logemann, E., Wu, S.C., Schroder, J., Schmelzer, E., Somssich, I.E., and Hahlbrock, K. (1995). Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle–related genes. Plant J. 8 865–876. [DOI] [PubMed] [Google Scholar]
- Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcription activators and contains the Myc-homology region. Proc. Natl. Acad. Sci. USA 86 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, K.A., Alfenito, M.R., Lloyd, A.M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375 397–400. [DOI] [PubMed] [Google Scholar]
- Martin, C., Prescott, A., Mackay, S., Bartlett, J., and Vrijlandt, E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1 37–49. [DOI] [PubMed] [Google Scholar]
- Menssen, A., Hohmann, S., Martin, W., Schnable, P.S., Peterson, P.A., Saedler, H., and Gierl, A. (1990). The En/Spm transposable element of Zea mays contains splice sites at the termini generating a novel intron from a dSpm element in the A2 gene. EMBO J. 9 3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., Kanei-Ishii, C., Takahashi, T., Yasukawa, T., Nagase, T., Horikoshi, M., Yamamoto, T., and Ishii, S. (1995). c-Myb repression of c-erbB-2 transcription by direct binding to the c-erbB-2 promoter. J. Biol. Chem. 270 9384–9389. [DOI] [PubMed] [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3 212–217. [Google Scholar]
- Moustafa, E., and Wong, E. (1967). Purification and properties of chalcone-flavanone isomerase from soya bean seeds. Phytochemistry 6 625–632. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakagoshi, H., Nagase, T., Ueno, Y., and Ishii, S. (1989). Transcriptional trans-repression by the c-Myb proto-oncogene product. Nucleic Acids Res. 17 7315–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager, M., Janknecht, R., Krieg, J., Schreek, S., and Luscher, B. (1996). Interaction of the co-activator CBP with Myb proteins: Effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 15 2771–2780. [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P.A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, N.K., Frost, J.W., and Long, S.R. (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233 977–980. [DOI] [PubMed] [Google Scholar]
- Podkowinski, J., Sroga, G.E., Haselkorn, R., and Gornicki, P. (1996). Structure of a gene encoding a cytosolic acetyl-CoA carboxylase of hexaploid wheat. Proc. Natl. Acad. Sci. USA 93 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.R., Britsch, L., Salamini, F., Saedler, H., and Rohde, W. (1987). The A1 (anthocyanin-1) locus in Zea mays encodes dihydroquercetin reductase. Plant Sci. Lett. 52 7–14. [Google Scholar]
- Riechers, D.E., Irzyk, G.P., Jones, S.S., and Fuerst, E.P. (1997). Partial characterization of glutathione S-transferases from wheat (Triticum spp.) and purification of a safener-induced glutathione S-transferase from Triticum tauschii. Plant Physiol. 114 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosler, J., Krekel, F., Amrhein, N., and Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, J.M., Deem, M.W., and Simpson, J.W. (1999). Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing. United States Patent No. 5,871,697. Issued February 16, 1999.
- Sainz, M.B., Grotewold, E., and Chandler, V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni, P., Perrotti, D., Martinez, R., Franceschi, C., and Calabretta, B. (1997). Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc. Natl. Acad. Sci. USA 94 3296–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270 467–470. [DOI] [PubMed] [Google Scholar]
- Shimkets, R.A., et al. (1999). Gene expression analysis by transcript profiling coupled to a gene database query. Nat. Biotechnol. 17 798–803. [DOI] [PubMed] [Google Scholar]
- Stapleton, A.E., and Walbot, V. (1994). Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol. 105 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trummler, K., and Wajant, H. (1997). Molecular cloning of acetone cyanohydrin lyase from flax (Linum usitatissimum). Definition of a novel class of hydroxynitrile lyases. J. Biol. Chem. 272 4770–4774. [DOI] [PubMed] [Google Scholar]
- Unger, E., Parsons, R.L., Schmidt, R.J., Bowen, B., and Roth, B.A. (1993). Dominant negative mutants of opaque2 suppress transactivation of a 22-kD zein promoter by opaque2 in maize endosperm cells. Plant Cell 5 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. (1995). Serial analysis of gene expression. Science 270 484–487. [DOI] [PubMed] [Google Scholar]
- Wang, H.K., Xia, Y., Yang, Z.Y., Natschke, S.L., and Lee, K.H. (1998). Recent advances in the discovery and development of flavonoids and their analogues as antitumor and anti-HIV agents. Adv. Exp. Med. Biol. 439 191–225. [DOI] [PubMed] [Google Scholar]
- Wanner, L.A., Li, G., Ware, D., Somssich, I.E., and Davis, K.R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27 327–338. [DOI] [PubMed] [Google Scholar]
- Wienand, U., Weydemann, U., Niesbach-Klosgen, U., Peterson, P.A., and Saedler, H. (1986). Molecular cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol. Gen. Genet. 203 202–207. [Google Scholar]