Abstract
Numerous cellular responses are reportedly regulated by blue light in gametophytes of lower plants; however, the molecular mechanisms of these responses are not known. Here, we report the isolation of two blue light photoreceptor genes, designated cryptochrome genes 4 and 5 (CRY4 and CRY5), from the fern Adiantum capillus-veneris. Because previously we identified three cryptochrome genes, this fern cryptochrome gene family of five members is the largest identified to date in plants. The deduced amino acid sequences of the five genes show remarkable similarities with previously identified cryptochromes as well as class I photolyases. Like the other plant cryptochromes, none of the cryptochromes of this fern possesses photolyase activity. RNA gel blot analysis and competitive polymerase chain reaction analysis indicate that the expression of the newly identified CRY4 and CRY5 genes is regulated by light and is under phytochrome control. The intracellular distribution of reporter β-glucuronidase (GUS)–CRY fusion proteins indicates that GUS–CRY3 and GUS–CRY4 localize in fern gametophyte nuclei. The nuclear localization of GUS–CRY3 is regulated in a light-dependent manner. Together with our physiological knowledge, these results suggest that CRY3, CRY4, or both might be the photoreceptor that mediates inhibition of spore germination by blue light.
INTRODUCTION
Blue light responses have been known to occur in various organisms, including plants, fungi, and bacteria, for many decades. In plants, phenomena such as phototropism, the inhibition of hypocotyl growth, flavonoid biosynthesis, and stomatal opening all are mediated by blue light photoreceptors. At least some of these photoreceptors are thought to contain a flavin chromophore (reviewed in Horwitz, 1994; Senger and Schmidt, 1994). One of the flavin chromophore class of photoreceptors, encoded by CRY1, was isolated from the Arabidopsis long hypocotyl mutant line hy4, which is defective in blue light inhibition of hypocotyl elongation (Ahmad and Cashmore, 1993). The deduced amino acid sequence of CRY1 shows substantial sequence similarity with class I photolyases, the repair enzymes that split cyclobutane pyrimidine dimers by using electrons obtained from blue light. CRY1 encodes a 75-kD protein that binds two cofactors, 5,10-methenyltetrahydrofolate and flavin adenine dinucleotide (FAD), as do the class I photolyases, but the CRY1 protein lacks DNA photorepair activity (Lin et al., 1995; Malhotra et al., 1995). To date, cryptochrome homologs have been identified from four different plant species: Arabidopsis CRY2 (AT-PHH1; Hoffman et al., 1996; Lin et al., 1996a), Sinapis alba SA-PHR1 (Batschauer, 1993), Chlamydomonas CPH1 (Small et al., 1995), and Adiantum capillus-veneris CRY1, CRY2, and CRY3 (Kanegae and Wada, 1998). The amino acid sequences deduced from these genes show remarkable similarity to CRY1 in their N-terminal domains but little similarity in their C-terminal domains.
Cryptochromes regulate many blue light responses in Arabidopsis. The physiological functions of CRY1 and CRY2 appear to overlap to some degree; for example, both CRY1 and CRY2 mediate inhibition of hypocotyl elongation and induction of anthocyanin synthesis (Lin et al., 1996b, 1998). Furthermore, functional analysis of plants overexpressing chimeric proteins comprising the N-terminal domain of CRY1 and the C-terminal domain of CRY2, or the N-terminal domain of CRY2 and the C-terminal domain of CRY1, indicates that the N-terminal domains and the C-terminal domains of CRY1 and CRY2 are interchangeable (Ahmad et al., 1998a). In addition to their common functions, both Arabidopsis CRY proteins have distinct functions. For example, CRY2 mediates cotyledon expansion and controls timing of flowering (Guo et al., 1998; Lin et al., 1998), whereas entrainment of the circadian clock by blue light is mediated by CRY1 (Somers et al., 1998).
Very recently, cryptochromes isolated from fruit flies and mice have been reported to play important roles in entraining and maintaining circadian rhythms in these organisms (Stanewsky et al., 1998; van der Horst et al., 1999). On the basis of amino acid sequence comparisons, cryptochromes are known to be ubiquitous photoreceptors in the plant and animal kingdoms, despite their distinct evolutionary histories (Cashmore et al., 1999). These findings raise the fascinating question of how individual cryptochromes evolved to perform diverse functions. To begin to answer this question, it is necessary to identify the functions of cryptochromes from a wide range of organisms. As discussed above, the only functions of plant cryptochromes known in any detail are those from Arabidopsis. However, numerous blue light responses have been characterized by focusing on the single cells and even on the single organelles in lower plants, particularly in mosses and ferns, because of the simple organization of their gametophytes. Thus, identifying the functions of individual lower plant cryptochromes is of particular interest.
Many physiological responses are induced by blue light in gametophytes of the fern A. capillus-veneris (reviewed in Wada and Sugai, 1994). Spore germination is inhibited by brief irradiation with blue light (Sugai and Furuya, 1985). Phototropism (Hayami et al., 1986), inhibition of tip growth (Kadota et al., 1979), apical swelling (Wada et al., 1978), and subsequent cell division (Wada and Furuya, 1972, 1978; Miyata et al., 1979) also are regulated by blue light in protonemata. In addition, blue light regulates organelle movements, including, for example, the orientational movements of chloroplasts (Yatsuhashi et al., 1985; Kagawa and Wada, 1994). Partial cell irradiation studies have further indicated that there are specific intracellular localizations for the blue light photoreceptors involved in each response (Kadota et al., 1986). For example, blue light photoreceptors involved in the inhibition of spore germination and cell cycle induction are shown to be localized in or close to the nuclear compartment (Wada and Furuya, 1978; Furuya et al., 1997). As a first step toward understanding the molecular mechanisms underlying these various blue light responses, we sought to clone and characterize the CRY gene family from A. capillus-veneris. Using the Arabidopsis CRY1 cDNA as a probe, we obtained nine clones from an A. capillus-veneris genomic library and have further classified these clones into five groups. Three of the five groups were characterized previously and designated A. capillus-veneris CRY1, CRY2, and CRY3, in accordance with their remarkable similarities with cryptochromes of higher plants (Kanegae and Wada, 1998).
Here, we report the molecular cloning of the two formerly unidentified genes, CRY4 and CRY5, and demonstrate both that cryptochromes in A. capillus-veneris are encoded by a small gene family comprising at least five members and that none of the CRY proteins has photoreactivating activity. In addition, we present the temporal and spatial expression patterns of individual A. capillus-veneris cryptochromes under various light conditions. This information provides important insights into cryptochrome function in A. capillus-veneris.
RESULTS
Isolation of Additional Cryptochrome Homologs: A. capillus-veneris CRY4 and CRY5 Genes
When three cryptochrome homologs (A. capillus-veneris CRY1, CRY2, and CRY3) were identified from the genomic library, two additional groups of unidentified clones were isolated. The partial sequences of these clones showed a similarity to the photolyase/blue light photoreceptor family (Kanegae and Wada, 1998). To determine whether these clones also encode cryptochrome homologs, we isolated the corresponding cDNAs and determined their nucleotide sequences by performing 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments using total RNA isolated from 1-day dark-imbibed spores as template. The resulting sequences confirm that these cDNAs encode cryptochrome homologs because their deduced amino acid sequences have a high degree of similarity to cryptochromes and share the features described below that are common to all cryptochromes. Thus, we designated these genes CRY4 and CRY5 (Figures 1A and 1B).
Figure 1.
Alignment of Amino Acid Sequences of Arabidopsis and A. capillus-veneris Cryptochromes.
The deduced amino acid sequences of A. capillus-veneris (Ac) CRY4 (AcCRY4; DDBJ accession number AB028928 for the cDNA and AB028930 for the gene) and CRY5 (AcCRY5; DDBJ accession number AB028929 for the cDNA and AB028931 for the gene) are compared with Arabidopsis CRY1 and CRY2 (AtCRY1; Ahmad and Cashmore, 1993; and AtCRY2; Lin et al., 1996a), and the three previously identified A. capillus-veneris cryptochromes, CRY1, CRY2, and CRY3 (AcCRY1, AcCRY2, and AcCRY3; Kanegae and Wada, 1998). Sequences were aligned by using the Clustal W program, version 1.74 (Thompson et al., 1994). The numbers at the ends of amino acid sequences indicate the positions of amino acids. The gaps are shown by dots.
(A) Residues present in four or more of the sequences are highlighted (BOXSHADE; http://ulrec3.unil.ch/software/BOX_form.html). The positions of 14 amino acids found in E. coli photolyase to interact with FAD by either direct H bonds (circles) or indirect H bonds (squares) are indicated below the alignment. The amino acids that are conserved between E. coli photolyase and cryptochromes are marked with closed circles and a closed square, and those that are not conserved are marked with open circles and an open square. The TGYP motif (asterisks) and the position of Trp-277 of E. coli photolyase (plus) are indicated below the alignment. STAESSSS motifs found in the C termini of cryptochromes are enclosed with boxes. The putative nuclear localization signals found in C termini of CRY3 and CRY4 are underlined. The N-terminal regions within the lines were used to construct the phylogenetic tree shown in Figure 8.
(B) The intron insertion positions of cryptochrome genes are indicated by the arrowheads on the partial amino acid alignment.
The longest CRY4 cDNA was 2892 bp in length and contained an open reading frame encoding a predicted protein of 699 amino acids. The longest CRY5 cDNA was 2828 bp long and contained an open reading frame of 487 amino acids. By comparing the cDNA and genomic sequences, we deduced that the CRY4 gene comprises five exons and the CRY5 gene comprises four exons. The intron positions in CRY4 and CRY5 are very similar to those in the previously characterized A. capillus-veneris and Arabidopsis CRY genes (Figure 1B). DNA gel blot analysis showed that both genes are present in a single copy in the haploid genome (data not shown). The N-terminal regions of A. capillus-veneris CRY4 and CRY5, which share similarities with photolyase (28.6 and 26.9% identities to Escherichia coli photolyase, respectively), are also well conserved with respect to those of plant cryptochromes (e.g., Figure 1A). Using BLAST (Altschul et al., 1997) and FASTA (Lipman and Pearson, 1985; Pearson and Lipman, 1988) searches, we could not find any sequences that showed notable similarity with any of the C termini. However, A. capillus-veneris CRY3 and CRY4 showed substantial similarity to each other at their C termini. The C-terminal conserved amino acid motifs DQMVP and STAESSSS found in other plant cryptochromes (Kanegae and Wada, 1998; Lin et al., 1998) were found in CRY4 (DQRVP and STAESSSS, respectively) but not in CRY5.
The cryptochrome apoproteins (Arabidopsis CRY1 and S. alba SA-PHR1) interact noncovalently with FAD chromophores (Lin et al., 1995; Malhotra et al., 1995). The amino acids binding to FAD have been deduced from the crystal structure of the E. coli photolyase (Park et al., 1995). In total, 10 of the 14 amino acids forming direct or indirect H bonds to FAD were found to be conserved in CRY4 and CRY5 (Figure 1A), which suggests that CRY4 and CRY5 might contain FAD as one of the two chromophores. In contrast, the amino acids binding to 5,10-methenyltetrahydrofolate in the E. coli photolyase were poorly conserved in CRY4 and CRY5. These chromophore binding site characteristics are similar in the other plant cryptochromes. Amino acids presumed to form a DNA binding site in the E. coli photolyase have been noted; some of these are also present in CRY4 and CRY5. However, Trp-277 in the E. coli protein, which is important for specific binding to pyrimidine dimers (Li and Sancar, 1990), is replaced by Phe in CRY4 and Leu in CRY5, as has been observed in the previously identified cryptochromes (Figure 1A). Moreover, CRY proteins from Arabidopsis and S. alba possess no photolyase activity (Lin et al., 1995; Malhotra et al., 1995; Hoffman et al., 1996).
A. capillus-veneris CRY Proteins Do Not Catalyze DNA Photorepair in a Photolyase-Deficient E. coli Strain
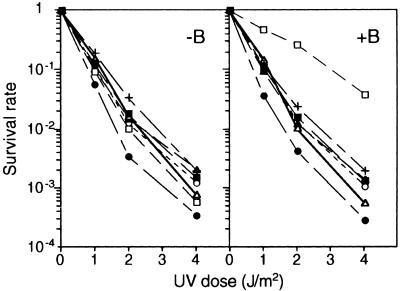
To investigate whether A. capillus-veneris CRY proteins can catalyze DNA repair, we cloned separately the full-length cDNAs encoding the five A. capillus-veneris CRY proteins and the E. coli phr gene (positive control) into an E. coli expression vector and transformed each plasmid into the photolyase-deficient strain SY2(DE3) of E. coli. These recombinant proteins also contain a calmodulin binding protein (CBP) tag at their N termini, and the expression vector that contained only the tag protein also was transformed as a negative control. The expression of the CBP-tagged recombinant CRY proteins was confirmed by protein gel blotting with biotinylated calmodulin and streptavidin–alkaline phosphatase (data not shown). After inducing synthesis of recombinant proteins, SY2(DE3) cells harboring each plasmid were exposed to UV light for specified times. These cells were plated in duplicate, and half the plates were irradiated with photoreactivating light (blue light) for 1 hr before dark incubation. As shown in Figure 2, blue light exposure did not change the survival rates of cells carrying either A. capillus-veneris CRY cDNAs or the negative control but did increase the survival rate of the cells carrying the E. coli phr gene. This result indicates that A. capillus-veneris cryptochromes do not function as photolyases in E. coli cells. Together with the characteristics of the primary structures of A. capillus-veneris CRY proteins as shown in Figure 1, this finding suggests that A. capillus-veneris CRYs are likely to be the cryptochromes of this fern.
Figure 2.
Comparison of Photoreactivation (PR) Effects of A. capillus-veneris CRY Genes with E. coli Photolyase on Survival Rates of the Photolyase-Deficient E. coli Strain.
The photolyase-deficient strain SY2(DE3) carrying each CRY cDNA in the pCAL-n-EK expression vector was exposed by UV light and plated in duplicate. Before overnight incubation in the dark, one plate of duplicates subsequently was irradiated with blue light (+B) for PR, but the others were not (−B). Each symbol represents the average of three independent survival rates of the cells carrying the following plasmids: pCALCRY1, closed circles; pCALCRY2, open circles; pCALCRY3, closed triangles; pCALCRY4, open triangles; pCALCRY5, closed squares; pCALPHR, open squares with positive control; and pCALSTOP, pluses with negative control.
Expression Patterns of A. capillus-veneris CRY Genes in Different Developmental Stages
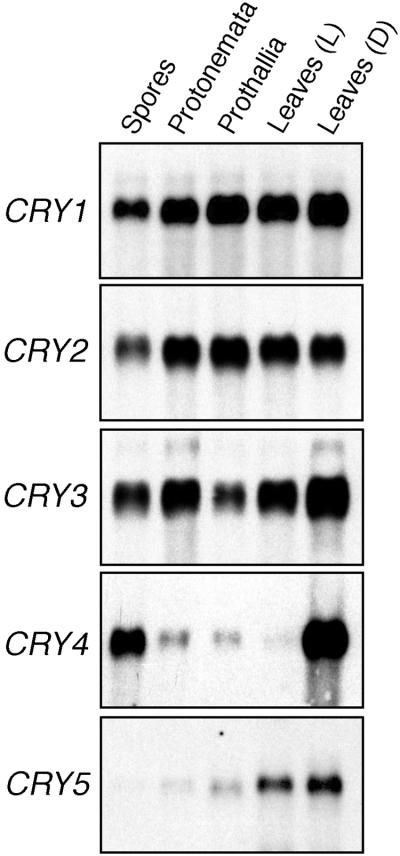
Because blue light responses have been observed in various developmental stages of A. capillus-veneris, we examined the expression patterns of the five CRY genes at different developmental stages by RNA gel blot analysis. All of the CRY genes are expressed in low amounts, so we prepared poly(A)+ RNA to detect the transcripts. As shown in Figure 3, all of the CRY mRNAs were expressed in both the sporophyte and gametophyte stages. The CRY1 and CRY2 mRNAs showed almost identical accumulation patterns. The amounts of these mRNAs rose a little after spore germination and stayed at the same level through the haploid and diploid phases. The amounts of CRY3 mRNA were relatively higher in protonemata and sporophyte tissues than in either spores or prothallia. The expression patterns of CRY4 and CRY5 genes were unique. The CRY4 mRNA was highly concentrated in the spores and in young leaves that had been grown in the dark but not in the other tissues examined. The CRY5 mRNA was observed mainly in sporophyte tissues, suggesting that expression of the CRY5 gene might depend on the nuclear phase. Both the protonemal and prothallial tissues examined were cultured under light, whereas the spores were prepared in the dark. In case the expression of the CRY4 gene might be downregulated by light, we further investigated the amount of CRY mRNAs that accumulated during light-dependent spore germination.
Figure 3.
Expression Patterns of Five CRY mRNAs in Different Developmental Stages.
Poly(A)+ RNA (4 μg) from spores, protonemata, prothallia, light-grown sporophytic leaves (Leaves [L]), and dark-grown leaves (Leaves [D]) was analyzed by RNA gel blotting by using specific probes corresponding to the C-terminal region of each CRY cDNA. The gametophyte tissues were obtained under the following culture conditions: imbibition for 1 day in darkness (Spores); imbibition for 4 days in darkness and incubation for an additional 3 days under red light (Protonemata); and incubation for 1 month under white light (Prothallia).
Both Red and Blue Light Altered the Expression of CRY Genes during Light-Dependent Spore Germination
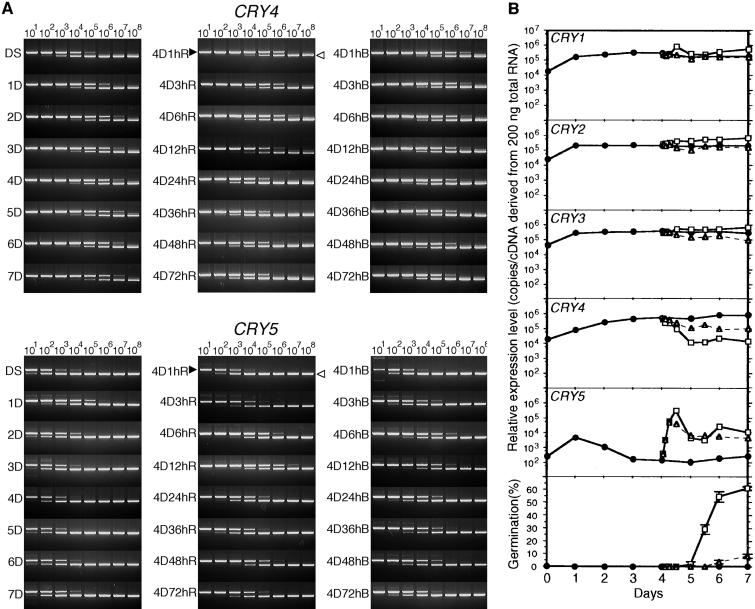
As was evident from the previous RNA gel blot analysis, the amounts of CRY transcripts were low in all the tissues investigated. Therefore, we used competitive polymerase chain reaction (PCR) to measure the changes in amount of individual CRY transcripts. The expression patterns of the CRY1, CRY2, and CRY3 genes were very similar (Figure 4B). The accumulated amounts of these mRNAs showed a 10-fold (CRY1 and CRY2) or sixfold (CRY3) increase in the first day after spore imbibition, but the amounts did not vary much during the following 6 days in the dark. When the spores were transferred to red light, which induces spore germination, the amounts of CRY1, CRY2, and CRY3 mRNAs increased two- to threefold within 12 hr and stayed at this level for as long as 72 hr. Blue light, which inhibits spore germination, had almost no effect on the amounts of either CRY1 or CRY2 mRNA expressed but did affect the amount of CRY3 mRNA, causing a fivefold decrease. Unexpectedly, it was found that not only the amount of CRY4 mRNA but also that of CRY5 mRNA was apparently regulated by light during spore germination. However, the ways in which mRNA accumulation was regulated by light included both inhibition and induction (Figures 4A and 4B). For CRY4, the amount of mRNA was reduced by ∼50-fold during the first 24 hr after the onset of red light irradiation; blue light reduced the level fivefold. Accumulation of CRY5 mRNA, however, was induced rapidly after the onset of exposure to both red and blue light, and it kept increasing to 300- to 400-fold the initial dark level during the first 12 hr. The mRNA amount then decreased for the next 12 hr, after which it remained at a level 20- to 40-fold higher than that of the dark control. All of the major changes in expression of the five CRY genes occurred before spore germination (Figure 4B).
Figure 4.
Competitive PCR Analysis of CRY mRNA Accumulation during Light-Dependent Spore Germination.
The amounts of each CRY transcript were determined by using competitive PCR. Spores imbibed in the dark were transferred to either red light or blue light. The amounts of CRY transcripts were measured at various times after transfer. This experiment was repeated twice in its entirety with very similar results in each case.
(A) Representative gel images of competitive PCR for CRY4 and CRY5. The PCR products derived from cDNA (closed arrowheads) and competitor (open arrowheads) are indicated on the gel image at each 1-hr red light irradiation sample after 4 days of incubation in the dark. The copy numbers of the competitors (from 101 to 108 copies) in each reaction tube are shown at the top of the sequential gel images. DS, dry spores; 1D, 1-day dark-imbibed spores; 4D1hB, 1-hr blue light irradiation samples after 4 days of incubation in the dark. The remaining samples are labeled similarly, with R indicating red light.
(B) Relative amounts of expression of five CRY genes and the germination rates in the same samples. Each point represents the average of the amounts of accumulation from two experiments. The symbols indicate the light conditions: circles, dark; squares, red light; triangles, blue light. The amounts of cDNA were determined from densitometric analysis of the gel images by using National Institutes of Health Image software.
Phytochrome Involvement in the Regulation of CRY4 and CRY5 mRNA Accumulation
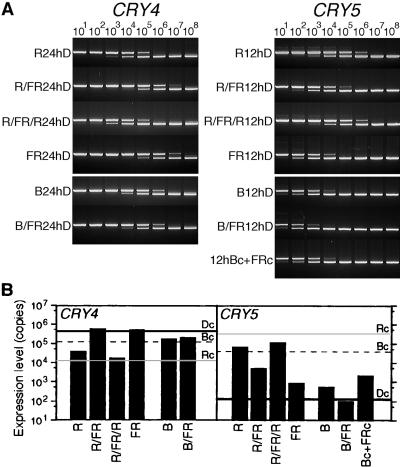
To determine which photoreceptor is involved in regulating CRY4 and CRY5 gene expression, we irradiated spores briefly in combinations of red, blue, or far-red light, and the expression patterns of both genes were measured by competitive PCR. Our results indicate that the amounts of both CRY4 and CRY5 mRNAs are regulated, at least in part, by phytochrome (Figures 5A and 5B). The accumulated amounts of CRY4 and CRY5 mRNAs were examined at 24 and 12 hr, respectively, after the light treatments, because the differences in the extents of their expression with or without light are clearly discernable at those times (see Figure 4A). The effect of red light on the accumulation of both mRNAs was cancelled by subsequent irradiation with far-red light and vice versa (Figures 5A and 5B). This red/far-red reversibility is a diagnostic characteristic of phytochrome regulation.
Figure 5.
Effects of Pulse Irradiation with Red, Far-Red, and Blue Light on the Regulation of CRY4 and CRY5 Gene Expression.
The effects of subsequent pulse irradiation (10 min each) with either red light (R) and far-red light (FR) or blue light (B) and far-red light (FR) on CRY4 and CRY5 transcript accumulation were measured using competitive PCR. Four-day dark-imbibed spores were irradiated in turn with red light and far-red light pulses or with blue light and far-red light. After 24- and 12-hr (h) dark incubation (for CRY4 and CRY5, respectively), the expression rates shown were observed. CRY5 was also simultaneously irradiated with continuous blue light (Bc) and far-red light (FRc).
(A) Representative gel images of competitive PCR for CRY4 and CRY5. (B) Relative amounts of CRY4 and CRY5 mRNAs. Each bar shows the average from two experiments. The gray, dotted, and black horizontal lines in each panel indicate the amounts of CRY4 and CRY5 cDNAs after the same lengths (24 and 12 hr) of continuous red (Rc) and blue (Bc) light exposure and of darkness (Dc), respectively (see Figure 4B).
A blue light pulse reduced the amount of CRY4 mRNA accumulation to that observed under continuous irradiation with blue light; subsequent exposure to far-red light did not alter the amount expressed. This indicates that CRY4 expression also was regulated by a blue light photoreceptor. In contrast, a blue light pulse was not sufficient to induce a large increase in CRY5 transcript abundance, and subsequent irradiation with far-red light decreased the amount even further. Moreover, the amount of mRNA present after simultaneous irradiation with continuous blue and far-red light was 20 times less than that measured after continuous blue light irradiation. Together, these results suggest that continuous blue light induction of CRY5 mRNA occurred mainly by way of a phytochrome.
Intracellular Localization of CRYs under Different Light Conditions
Physiological observations suggest that the blue light photoreceptor that mediates inhibition of spore germination and induction of the cell cycle is localized in or close to the nucleus (Kadota et al., 1986; Furuya et al., 1997). To determine whether any cryptochromes are located in the nucleus, we transiently expressed in fern gametophytes fusion genes containing each CRY cDNA inserted as a translational fusion to β-glucuronidase (GUS). Because both red and blue light are known to cause various responses during the early stages of gametophyte development, the transfected cells were incubated under either red light or blue light or in the dark to examine the effects of light on the intracellular distribution of CRY proteins. Representative intracellular staining patterns of GUS–CRY fusion proteins are shown in Figure 6. We observed the same distribution patterns of each GUS fusion protein in both the basal cells and the tip cells. The intracellular localization of GUS activity varied slightly among individual cells, even those expressing a single fusion construct, probably because of damage from particle bombardment. Therefore, we show the tendency of the intracellular distribution pattern of each GUS–CRY fusion protein in Figure 7.
Figure 6.
Representative Images of the Intracellular Distribution of GUS–CRY Fusion Proteins in Germinating Cells.
Intracellular distribution of GUS–CRY fusion proteins under different light conditions is shown.
(A) to (C) GUS–CRY1.
(D) to (F) GUS–CRY2.
(G) to (I) GUS–CRY3.
(J) to (L) GUS–CRY4.
(M) to (O) GUS–CRY5.
(P) to (R) GUS.
(S) to (U) GUS–CRY3C (CRY3 C terminus).
(V) to (X) GUS–CRY4C.
Protonemal cells expressing various GUS–CRY fusion proteins were incubated under red light ([A], [D], [G], [J], [M], [P], [S], and [V]), blue light ([B], [E], [H], [K], [N], [Q], [T], and [W]), or in the dark ([C], [F], [I], [L], [O], [R], [U], and [X]) for 16 hr and stained under the same light conditions. The cells showing GUS activity were photographed by using Nomarski optics (left panels); the fluorescence micrographs show the position of the nuclei in the same cells after staining with 4′,6-diamidino-2-phenylindole (right panels). Note that under fluorescence, chlorophyll autofluoresces red and spore coats appear bluish white.  .
.
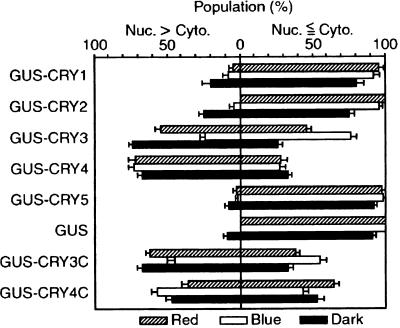
Figure 7.
Intracellular Distribution of GUS–CRY Fusion Proteins under Different Light Conditions.
Distribution patterns of GUS activities in individual transformed cells were classified into two groups: (1) nuclear enrichment of GUS activity was seen (nucleus [Nuc.] > cytoplasm [Cyto.]) or (2) nuclear enrichment of GUS activity was not seen (Nuc. ⩽ Cyto.). These cells expressed each GUS–CRY fusion protein under red light (striped bars), blue light (open bars), or in the dark (black bars). From protonemal cells prepared under the same experimental conditions as in Figure 6, we randomly selected cells that showed GUS activities. The first group (Nuc. > Cyto.) contained cells that showed clear nuclear enrichment with various degrees of cytoplasmic GUS staining; and the second group (Nuc. ⩽ Cyto.) contained cells that showed nuclear staining indistinguishable from or less than cytoplasmic staining. The data were obtained from between three and seven independent experiments. In total, we examined 45, 119, and 104 cells (red light, blue light, and dark treatment, respectively) for GUS–CRY1; 30, 76, and 117 cells for GUS–CRY2; 117, 129, and 114 cells for GUS–CRY3; 120, 120, and 124 cells for GUS–CRY4; 105, 139, and 139 cells for GUS–CRY5; 144, 113, and 130 cells for GUS; 120, 136, and 111 cells for GUS–CRY3C; and 141, 128, and 119 cells for GUS–CRY4C. For each experiment, we calculated the percentage of population of each group, dividing the number of cells belonging to each group by the total number of cells examined in that experiment. The bars show the average percentages of populations derived from all experiments. Standard errors of the mean are indicated.
The GUS–CRY3 and GUS–CRY4 proteins showed clear nuclear localization, but GUS–CRY1, GUS–CRY2, and GUS–CRY5 did not. The GUS–CRY3 protein tended to accumulate in the nucleus in the dark and in red light but not in blue light. The GUS–CRY4 protein was predominantly nuclear under all the light conditions examined. The degree of nuclear enrichment of GUS staining in GUS–CRY3—expressing cells under red light seemed to be less than that in dark-incubated cells (see Figure 6). In the dark, a few of the cells showed nuclear enrichment in GUS–CRY1 and GUS–CRY2; in those cells, GUS activity also was observed in the cytoplasm. In onion epidermal cells, we also observed nuclear localization of GUS–CRY3 and GUS–CRY4 but not of GUS–CRY1, GUS–CRY2, and GUS–CRY5 (data not shown). The expression rates of all GUS–CRY fusion proteins in A. capillus-veneris—especially those of GUS–CRY1 and GUS–CRY2—under red or blue light were much lower than their expression rates in the dark. This observation suggests that a light-dependent protein degradation mechanism similar to that seen for Arabidopsis CRY2 (Lin et al., 1998) and Drosophila CRY (Emery et al., 1998) might also operate in A. capillus-veneris.
The N-terminal regions of cryptochromes, especially of the A. capillus-veneris CRYs, are highly conserved with respect to each other (see Figure 1A). Nonetheless, our results indicate that the subcellular distributions of these cryptochromes differ, with only GUS–CRY3 and GUS–CRY4 showing clear nuclear localization. This implies that the divergent C-terminal regions of these cryptochromes might play important roles in the determination of subcellular distribution. In addition, small basic amino acid clusters (e.g., KRKAK), which might function as monopartite nuclear localization signals, are present in the conserved regions found at the CRY3 and CRY4 C termini (Figure 1A). To test this possibility, we generated fusion genes encoding GUS at the N-terminal half and CRY3 or CRY4 C-terminal regions at the C-terminal half (GUS–CRY3C and GUS–CRY4C) and transformed these into A. capillus-veneris cells. The results show that the light-dependent intracellular localization patterns of GUS–CRY3C and the nuclear localization pattern of GUS–CRY4C are similar to those of GUS–CRY3 and GUS–CRY4 (Figures 6 and 7). This finding suggests that the C-terminal regions of both CRY3 and CRY4 are required to import these proteins into the nucleus.
DISCUSSION
Including the two additional cryptochrome genes reported here, the A. capillus-veneris cryptochrome gene family comprises at least five members, all of which are expressed during the haploid and diploid phases of the life cycle (see Figure 3). Until now, the two Arabidopsis CRY genes represented the only completely characterized family (Ahmad and Cashmore, 1993; Lin et al., 1998). The five-member gene family that we report here for the fern A. capillus-veneris is the largest cryptochrome gene family characterized to date. Considering the complexity of blue light–induced phenomena found in this fern, it is not surprising that this fern has a large number of blue light photoreceptors.
The N termini of these five cryptochromes are highly conserved, and the amino acid residues necessary for interaction with FAD and presumptive DNA binding are found mostly in the same positions as in the E. coli photolyase. However, as we have shown, none of these cryptochromes has photolyase activity. Their characteristics, therefore, are the same as in previously identified cryptochromes, suggesting that the same redox electron transfer mechanism from blue light photons by way of FAD also functions in A. capillus-veneris CRY proteins. The STAESSSS-related amino acid motif also is found at the C termini of CRY2, CRY3, and CRY4 (STAESTS, PMTESSSS, and STAESSSS, respectively). Although the function of this motif is unknown, it has been reported to be important for the phosphorylation of cryptochrome 1 by phytochrome A in vitro (Ahmad et al., 1998b). Physiological responses in A. capillus-veneris that are regulated antagonistically or cooperatively by phytochrome and blue light photoreceptors include spore germination, phototropism, the cell cycle, and chloroplast movement (Wada and Sugai, 1994). We have identified three phytochrome genes from A. capillus-veneris (Okamoto et al., 1993; Nozue et al., 1998a, 1998b). Recently, Yeh and Lagarias (1998) showed that phytochromes from oat and alga expressed in yeast have serine/threonine kinase activity. Thus, in the signal transduction cascades upstream of some photoresponses in A. capillus-veneris, direct interaction of phytochromes and cryptochromes might happen by way of the STAESSSS motif by phosphorylation. However, the A. capillus-veneris CRY1, CRY5, Chlamydomonas CPH1, and S. alba SA-PHR1 sequences do not possess the STAESSSS motif, which suggests other blue light signal transduction cascades that do not involve phosphorylation of this motif are present.
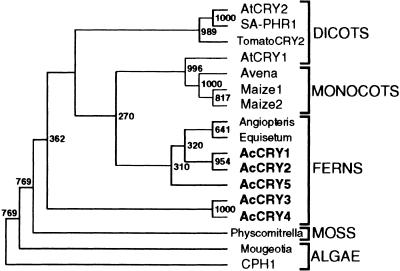
Cryptochromes have been found in divergent species across the biological kingdom (Cashmore et al., 1999). Of particular interest is how cryptochromes evolved in plants. To address this question, we performed a phylogenetic tree analysis using 10 full-length cryptochrome cDNA sequences (alga, Chlamydomonas [Small et al., 1995]; moss, Physcomitrella patens [Imaizumi et al., 1999]; fern, A. capillus-veneris [Kanegae and Wada, 1998; this article]; dicots, Arabidopsis Arabidopsis [Ahmad and Cashmore, 1993][Lin et al., 1996a] and S. alba [Batschauer, 1993]) and related partial sequences found in the database. The regions of each N terminus indicated in Figure 1 were used for construction of the tree, and the bootstrap values from 1000 replicates were calculated by using the neighbor-joining method. The tree was rooted by using Chlamydomonas CPH1 as the outgroup (Figure 8). Although some branching points are not reliable because of the low bootstrap values, the branching order of cryptochromes and the related sequences closely correspond to the organismal phylogeny. As in the phylogenetic analysis of another plant photoreceptor, phytochrome (Mathews and Sharrock, 1997), multiple cryptochrome lineages in cryptogams are clearly separable from those in seed plant clusters.
Figure 8.
Phylogenetic Analysis of Plant Cryptochromes.
The regions of partial amino acid sequences used to generate the phylogenetic tree are shown in Figure 1. The neighbor-joining method was used to calculate bootstrap values from 1000 replicates (http://www.genome.ad.jp/) and to draw the tree by using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The numbers located at branch points are the bootstrap values. GenBank/DDBJ/EMBL accession numbers for each cryptochrome DNA sequence, except for those given in the legend of Figure 1, are as follows: Angiopteris for Angiopteris evecta, X99261; Avena for Avena sativa, X99262; CPH1 for Chlamydomonas CPH1, L07561; Equisetum for Equisetum arvense, X99263; Maize1 and Maize2 for Zea mays, X99265 and X99266; Mougeotia for Mougeotia scalaris, AJ000694; Physcomitrella for P. patens, AB027528; SA-PHR1 for S. alba SA-PHR1, X72019; and TomatoCRY2 for Lycopersicon esculentum, AJ000695.
Consistent with the amino acid sequence alignment, A. capillus-veneris CRY1 and CRY2, and CRY3 and CRY4 are in each cluster. Three recent independent gene duplications might have occurred to yield CRY3 and CRY4, CRY1/CRY2 and CRY5, and CRY1 and CRY2. This information, together with the mRNA accumulation pattern of CRY1 and CRY2 (Figure 3) and the intracellular distribution of CRY1/CRY2 and CRY3/CRY4 (Figures 6 and 7), indicates that CRY1/CRY2 and CRY3/CRY4 might comprise subfamilies of A. capillus-veneris cryptochromes. Because the diversity of cryptogam phytochrome genes is thought to result from taxon-specific diversification, it is of great interest to learn whether the diversification of A. capillus-veneris CRY genes also occurred taxon specifically, as in the case of the cryptogam phytochromes, or whether cryptogams have more than one cryptochrome subfamily, as has been suggested for the phytochromes of seed plants. In seed plants, cryptochrome-related partial sequences from monocots are closely related to Arabidopsis CRY1, whereas sequences from dicots are in the same cluster as Arabidopsis CRY2. It is also of interest to know whether seed plant cryptochromes diverged before or after the diversification of dicots and monocots. To address these questions requires identifying additional cryptochrome genes from other plant species.
In A. capillus-veneris, the previous physiological observations indicated that blue light photoreceptors were involved in phototropism (Hayami et al., 1992), apical swelling (Wada et al., 1978), and chloroplast orientational movement (Yatsuhashi et al., 1987) localized on or close to the plasma membrane in protonemata. Figures 6 and 7 show that CRY1, CRY2, and CRY5 proteins are localized in the cytoplasm. We could not find the putative membrane-spanning domains in CRY1, CRY2, or CRY5 proteins. However, some data showed that Arabidopsis CRY1 protein, which was not noted to contain any membrane-spanning domains, was enriched in the membrane fractions (Ahmad et al., 1998c). Possibly, therefore, some portions of CRY1, CRY2, or CRY5 proteins may attach to the plasma membrane where they might be involved in blue light–induced phototropism, apical swelling, chloroplast orientational movement in protonemata, or some combination of these responses.
Blue light suppresses the entry into the S phase of the first mitosis leading to spore germination (Uchida and Furuya, 1997). In protonemata, blue light photoreceptors also control shortening of the G1 phase (Miyata et al., 1979). The blue light photoreceptors involved in these responses are localized in or close to the nucleus (Wada and Furuya, 1978; Furuya et al., 1997). Such physiological observations indicate that the blue light photoreceptor mediating inhibition of spore germination must be present in or close to the nucleus before the onset of blue light irradiation. Although whether these blue light photoreceptors are derived from the maternal cytoplasm or are newly translated during imbibition in the dark is unknown, CRY1, CRY2, CRY3, and CRY4 genes would be the candidates for such receptors, given our results showing that these mRNAs accumulate in dark-imbibed spores (Figure 4). In addition, we demonstrated that GUS–CRY3 and GUS–CRY4 apparently accumulate in the nucleus in dark-incubated protonemal cells (Figures 6 and 7).
These results are consistent with the notion that CRY3 and/or CRY4 genes could encode the blue light photoreceptors regulating spore germination. Under natural light conditions, the ratio between blue light and red light may differ, depending on the surrounding environment, but both are always present. Even after the steps leading to spore germination are activated by light signals by way of a phytochrome, any blue light photoreceptors working antagonistically would interfere with germination. Although we could not find any amino acid sequences that fit the consensus sequence of the nuclear export signal in the C-terminal region of CRY3 (Bogerd et al., 1996; Kim et al., 1996), our data showed the apparent blue light–dependent export of GUS–CRY3 fusion protein from the nuclei of dark-grown cells to the cytoplasm (Figures 6 and 7). If CRY3 is the blue light photoreceptor of the inhibition of spore germination, the nuclear export of CRY3 might be one of the important mechanisms to cause the inhibition of spore germination. CRY4 seems to localize in the nucleus under all light conditions examined, but the accumulation of CRY4 mRNA is regulated by light quality. If CRY4 is the blue light photoreceptor that inhibits spore germination, the suppression of CRY4 mRNA accumulation observed under red light (Figures 4 and 5) might reduce the interruption of the germination processes by blue light, allowing uninterrupted completion of germination. Currently, the answers to these questions remain obscure, but abundant physiological data suggest that cryptochromes might work as negative photoswitches for cell cycle promotion in germination of A. capillus-veneris spores.
METHODS
Plant Materials and Light Sources
Spores of Adiantum capillus-veneris were harvested in either 1994 or 1995 in a greenhouse at Tokyo Metropolitan University. Spores were sterilized with 10% (v/v) antiformin (NaClO) containing 0.1% (v/v) Triton X-100. After three rinses with sterile distilled water, the spores were suspended in 1:10 strength of modified Murashige and Skoog culture medium (Wada and Furuya, 1970) and sown on a cellophane sheet that covered the same Murashige and Skoog medium solidified with 0.5% (w/v) agar. The spores were incubated under different light conditions at 25 ± 1°C. The gametophytes were collected on filter paper by using vacuum filtration; they were then frozen immediately in liquid nitrogen and stored at −80°C before use. All manipulations after sowing were performed under a dim green safelight (Kadota et al., 1984). Sporophytes were cultivated in the greenhouse under natural light conditions. After all leaves were cut, young light-grown leaf tissue was collected as emerging croziers. Some sporophytes were transferred into complete darkness after removal of all the leaves. Young dark-grown leaves (croziers) were collected in the dark for 3 to 4 weeks under a safelight. This tissue, too, was frozen immediately and stored at −80°C.
White light (25.7 μmol m−2 sec −1) was obtained from broad range–emitting fluorescent tubes (model FL40SD; Toshiba Ltd., Tokyo, Japan). Red light (9.9 μmol m−2 sec −1) was obtained from either red-emitting fluorescent tubes (model FL20S-Re-66; Toshiba Ltd.) or fluorescent tubes (model FL40SD; Toshiba Ltd.) filtered through a red plastic sheet (model Acrylite 102; Mitsubishi Rayon, Tokyo, Japan). Far-red light (9.2 μmol m−2 sec −1) was provided by far-red-emitting fluorescent tubes (model FL20S-FR-74; Toshiba Ltd.) and filtered through the same red plastic sheet (model Acylite 102; Mitsubishi Rayon) and a far-red filter (IR-1; Koto Electronic Company Ltd., Urawa, Japan). Blue light (4.6 μmol m−2 sec −1) was obtained from blue-emitting fluorescent tubes (models FL20S-B and FL20S-BW; Toshiba Ltd.) filtered through a blue plastic sheet (model Acrylite 302; Mitsubishi Rayon). The light conditions described above were used throughout all experiments.
DNA and RNA Isolation
Genomic DNA and total RNA derived from either gametophytes or sporophytes were isolated by using the cetyltrimethylammonium bromide (CTAB) method (Kanegae and Wada, 1998), with some modifications. When gametophyte tissue was ground with a mortar and pestle, an amount of sterilized quartz sand equal to 10 times the volume (w/w) was added to each sample to help break the cell walls in liquid nitrogen. Frozen tissue was incubated with an amount of 2 × CTAB solution (1 × CTAB solution is 2% [w/v] CTAB, 0.1 M Tris-HCl, pH 8.0, 20 mM EDTA, and 1.4 M NaCl) equal to 10 times the volume (w/v) containing 5% (v/v) 2-mercaptoethanol at 60°C. The chloroform treatments were repeated three times. DNA was precipitated with 2-propanol. RNA was precipitated twice with LiCl. The precipitate was dissolved in Tris-EDTA or diethyl pyrocarbonate–treated water. To purify poly(A)+ RNA, we used mRNA purification kits (Amersham Pharmacia Biotech, Buckinghamshire, UK) according to the manufacturer's protocol.
Cloning of A. capillus-veneris Cryptochrome 4 and 5 Genes
Three clones (λ-1, λ-3, and λ-5) contained the same gene (CRY4). The other clone (λ-2), corresponding to CRY5, was isolated from the A. capillus-veneris genomic λ phage library by using the full-length Arabidopsis CRY1 cDNA as a probe (Kanegae and Wada, 1998). These clones were subcloned into the plasmid vector pBluescript II SK+ (Stratagene, La Jolla, CA). Both genomic clones and cDNA clones were sequenced by using either fluorescent-labeled primers or dye terminators (BigDye terminator cycle sequencing ready reaction; Applied Biosystems, Foster City, CA). For isolation of CRY4 and CRY5 cDNAs, a 3′ rapid amplification of cDNA ends (RACE) kit (Life Technologies, Rockville, MD) and a 5′ RACE kit (Life Technologies) were used according to the system protocols. In 3′ RACE, cDNA synthesized from 1 μg of total RNA derived from 1-day dark-imbibed spores was amplified by gene-specific primers: 5′-CCAAGCTTGGGGAGGAGAG-3′ for CRY4 and 5′-AGGGAAGCCAGATGAAGAGC-3′ for CRY5. Both primary polymerase chain reaction (PCR) products were amplified with the same nested primer (5′-CCYYTDGTKGATGCHGGVATG-3′). In 5′ RACE, first-strand cDNAs were synthesized from the same total RNA used in 3′ RACE by using gene-specific primers: 5′-AAGCCTCAGCATCAGT-3′ for CRY4 and 5′-AACTAATGGTCGTGGA-3′ for CRY5. The cDNAs were then amplified by PCR with nested gene-specific primers: 5′-CCCAAGCACATCAGACTCCAAA-3′ for CRY4 and 5′-TCATCCATCCGTTCGAGTTC-3′ for CRY5. Primary PCR products that were between 1 and 2.5 kb long were recovered and reamplified by using another nested gene-specific primer: 5′-AGACTATGACTCGCACTCGGTT-3′ for CRY4 and 5′-CCCATGTCCAAGGTAGCTGC-3′ for CRY5. Afterward, 3′- and 5′ RACE products were cloned into the plasmid vector pGEM-T Easy (Promega, Madison, WI) and sequenced.
Photoreactivation Analysis
Full-length coding regions of five CRYs and an Escherichia coli photolyase (phr) gene (pRT2; kindly provided by T. Todo, Kyoto University, Kyoto, Japan) were cloned into the expression vector pCAL-n-EK (affinity LIC cloning and protein purification kit; Stratagene) according to the manufacturer's procedure. The primers used for PCR cloning were the following: 5′-GACGACGACAAGGCCTGCACAATTGTGTGGTT-3′ and 5′-GGAACAAGACCCGTGAGTTTCTGAGAC-AATCT-3′ for pCALCRY1; 5′-GACGACGACAAGGCGGCACAC-ACAATTGTGGC-3′ and 5′-GGAACAAGACCCGTCTGACCCTTAACT-TCAAC-3′ for pCALCRY2; 5′-GACGACGACAAGGCAAAATCATGT-ACCGTTGT-3′ and 5′-GGAACAAGACCCGTCACGTCAGTTTAAACA-AC-3′ for pCALCRY3; 5′-GACGACGACAAGGCAAAACCTTGTACA-ATAGT-3′ and 5′-GGAACAAGACCCGTCACCGATGCATTTTTTCG-3′ for pCALCRY4; 5′-GACGACGACAAGACCACCTCTACAACCATT-GT-3′ and 5′-GGAACAAGACCCGTAGCTGCAGACTAATCAAC-3′ for pCALCRY5; and 5′-GACGACGACAAGACTACCCATCTGGTCTGG-TT-3′ and 5′-GGAACAAGACCCGTATTGCCTGACGCGTCTGT-3′ for pCALPHR. These recombinant proteins possess a calmodulin binding protein (CBP) tag in the N terminus, so a vector expressing only the CBP was generated to insert synthetic oligonucleotides consisting of three stop codons (TGATAATAG) into the cloning site as a negative control vector (pCALSTOP). The resulting clones were sequenced to confirm that they contained the correct inserts. A photolyase-deficient E. coli strain SY2 (JM107; phr::Cmr uvrA::Kmr recA::Tetr; Yasuhira and Yasui, 1992; kindly provided by T. Todo) was lysogenized with λDE3 prophage (Novagen, Madison, WI) to enable the expression system to use the T7 RNA polymerase promoter. Plasmids pCALCRY1, pCALCRY2, pCALCRY3, pCALCRY4, pCALCRY5, pCALPHR, and pCALSTOP were transformed into the resulting SY2(DE3) cells. The SY2(DE3) cells harboring each plasmid were cultured until the OD600 reached 0.6; then isopropylthiogalactopyranoside was added to a final concentration of 1 mM. After an additional 3.5-hr incubation at 37°C in the dark, the cells were collected and washed with phosphate buffer (pH 6.8; Yamamoto, 1985). The cells were resuspended in phosphate buffer at a concentration of 1 to 2 × 107 cells per mL. The cell suspensions, in 30-mm-diameter plastic dishes, were exposed to UV light (0.2 μmol m−2 sec −1) obtained from a germicidal light tube (model GL-20; Toshiba Ltd.) and then plated in duplicate in two to three dilution series. For photoreactivation, half the group of Luria-Bertani plates were irradiated with blue light for 1 hr before overnight incubation in the dark. The number of surviving colonies was independently counted three times to calculate the survival rates. After resuspension of the cells, all manipulations were performed under red light (model National FL20S.R-F; Matsusita Electric Co., Osaka, Japan).
RNA Gel Blot Analysis
Poly(A)+ RNA (4 μg per lane) was electrophoresed on 1.2% formaldehyde-agarose gels and transferred to nylon membranes (Hybond N+; Amersham Pharmacia Biotech) over a period of 14 to 16 hr with 20 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate). For hybridization and detection, a chemifluorescence-labeled gene detection kit (Gene Images; Amersham Pharmacia Biotech) was used according to the manufacturer's instruction. The 3′ regions of each CRY cDNA (nucleotide sequence positions 1451 to 2457 for the CRY1 cDNA, 1691 to 2754 for the CRY2 cDNA, 1448 to 2440 for the CRY3 cDNA, 1634 to 2857 for the CRY4 cDNA, and 1328 to 2444 for the CRY5 cDNA) were amplified by PCR to be used as probes. The blots were hybridized with the probes at 65°C. The membranes were washed in 1 × SSC, 0.1% (w/v) SDS, and then in 0.1 × SSC, 0.1% (w/v) SDS, for 15 min each time at 65°C. Cross-hybridization did not occur when we performed DNA gel blot analyses with the same probes at the lower stringency condition (0.5 × SSC, 0.1% [w/v] SDS at 60°C for the second wash; data not shown).
Competitive Reverse Transcription–PCR Analysis
DNA competitors were designed to have different sequences on both 5′ and 3′ ends in which each set of CRY-specific primers could be annealed but with the same sequences derived from λ phage DNA in between (Competitive DNA construction kit; Takara Shuzo Co., Kyoto, Japan). Five primer sets were used for PCR synthesis of each competitor: 5′-CAGAATTTATGCCCGAGTCCGCGTGAGTATTAC-GAAGGTG-3′ and 5′-TATGGAAACAGGAAGACCAGTGAAGACGACGCGAAATTCA-3′ for CRY1 competitor; 5′-GCACATTCACAC-AACTCTCCGCGTGAGTATTACGAAGGTG-3′ and 5′-CTAATGTTGGTGTAAGGAGGTGAAGACGACGCGAAATTCA-3′ for CRY2 competitor; 5′-GCCCATTGAGAGCCATTAAGGCGTGAGTATTACGAA-GGTG-3′ and 5′-AGCTCACGACGGGATCAAATTGAAGACGACGCG-AAATTCA-3′ for CRY3 competitor; 5′-CTCTGACAATGGTGACTT-CTGCGTGAGTATTACGAAGGTG-3′ and 5′-AGACAGCAGTGGCAG-GAACATGAAGACGACGCGAAATTCA-3′ for CRY4 competitor; and 5′-ACAATCCACCAATGAGCCCAGCGTGAGTATTACGAAGGTG-3′ and 5′-GAACTCGAACGGATGGATGATGAAGACGACGCGAAATTCA-3′ for CRY5 competitor.
A gradient series from 101 to 108 copies per μL of competitors was used for the analysis. Complementary DNA was synthesized from 2 μg of each total RNA pretreated with DNase I (Stratagene) by using an oligo(dT) primer according to the supplied procedure (SUPERSCRIPT preamplification system for first-strand cDNA synthesis; Life Technologies). One-tenth volume of the reaction solution of cDNA and 1 μL of each concentration of competitors were put into the same reaction tube. The locations of the primers were designed to contain the last intron of each CRY gene so that we could distinguish whether the amplified fragments were derived from cDNA or from contaminated genomic DNA. PCR was performed with the following primer sets: 5′-CAGAATTTATGCCCGAGTCC-3′ and 5′-TATGGAAAC-AGGAAGACCAG-3′ for CRY1; 5′-GCACATTCACACAACTCTCC-3′ and 5′-CTAATGTTGGTGTAAGGAGG-3′ for CRY2; 5′-GCCCATTGAGAG-CCATTAAG-3′ and 5′-AGCTCACGACGGGATCAAAT-3′ for CRY3; 5′-CTCTGACAATGGTGACTTCT-3′ and 5′-AGACAGCAGTGGCAGGAACA-3′ for CRY4; and 5′-GAACTCGAACGGATGGATGA-3′ and 5′-ACAATCCACCAATGAGCCCA-3′ for CRY5. The PCR schedule was 1 min at 94°C; 40 cycles of 10 sec at 98°C, 30 sec at 55°C, and 30 sec at 72°C; and then 3 min at 72°C. Equal volumes of PCR products were separated in 3% (w/v) agarose gels. The expected lengths of amplified fragments derived either from each CRY cDNA or from each competitor were 0.5 and 0.4 kb, respectively. The gel images were captured and analyzed by using the public domain National Institutes of Health Image program (http://rsb.info.nih.gov/nih-image/).
β-Glucuronidase–CRY Plasmid Construction
A PCR-based cloning method (seamless cloning kit; Stratagene) was used to construct vectors containing β-glucuronidase (GUS)–CRY fusion genes. pBI221 plasmid (Clontech, Palo Alto, CA) that contained a cauliflower mosaic virus 35S promoter–driven GUS gene was used as a template. The primers used for making GUS–CRY constructs were as follows (forward primer and reverse primer, respectively): 5′-AGTTACTCTTCAGCTACCGAGCTCGAATTTCCCCGA-3′ and 5′-AGTTACTCTTCATTGTTTGCCTCCCTGCTGCGGTTT-3′ for amplifying the pBI221 vector that has a specific in-frame cloning site; 5′-AGTTACTCTTCACAAGCCTGCACAATTGTGTGGTTTCGG-3′ and 5′-AGTTACTCTTCAAGCGAGAACCAACCAACCGCCACATTC-3′ for CRY1 insert; 5′-AGTTACTCTTCACAAGCGGCACACACAATT-GTGGCACAC-3′ and 5′-AGTTACTCTTCAAGCCCAGGCTTTGATCCT-AGTGGGTGT-3′ for CRY2 insert; 5′-AGTTACTCTTCACAAGCAAAA-TCATGTACCGTTGTGTGG-3′ and 5′-AGTTACTCTTCAAGCCAGTGC-AACTGCACGTCAGTTTAAAC-3′ for CRY3 insert; 5′-AGTTACTCTTCACAAGCAAAACCTTGTACAATAGTGTGG-3′ and 5′-AGTTACTCT-TCAAGCCGTCACCAGCCCCACTCATGAAGA-3′ for CRY4 insert; 5′-AGTTACTCTTCACAAACCACCTCTACAACCATTGTCTGGC-3′ and 5′-AGTTACTCTTCAAGCGCTGGGGCTCTGAGACATCCTTTC-3′ for CRY5 insert; 5′-AGTTACTCTTCACAAGTGGGCGATTCAATTGCG-AAGGTC-3′ for CRY3C insert; and 5′-AGTTACTCTTCACAACCA-CCACTGGTTTCAACTCACTCC-3′ for CRY4C insert. To amplify CRY3C and CRY4C inserts, we used the same reverse primers as for the CRY3 and CRY4 inserts. Both junctions between vector and amplified CRY inserts and CRY inserts themselves were sequenced to confirm whether or not these fragments were inserted in the same frame correctly.
Analysis of GUS Intracellular Localization in Protonemal Cells
Gold particles (1.6 μm in diameter) coated with GUS–CRY or pBI221 plasmids were introduced into the two-celled protonemata, which were incubated for 4 days in the dark and for an additional 3 days under red light, by using a biolistic gun transformation system (DuPont, Wilmington, DE). To bombard gold particles, we used 1350-psi rupture discs according to the manufacturer's procedure. Transfected cells were incubated in red light, blue light, or in the dark for 16 hr at 25°C and then stained with 100 mM sodium phosphate, pH 7.0, 1 mM EDTA, 0.3% (v/v) Triton X-100, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1 μg/mL 4′,6-diamidino-2-phenylindole, and 1 mM X-gluc. Staining was performed under the same light conditions at 37°C overnight. Cells showing GUS staining were selected under low magnifying power (×10 to 25) through a stereomicroscope, and the intracellular localization of GUS products was observed under a microscope (×400). The optic images of GUS activity and 4′,6-diamidino-2-phenylindole staining were obtained by using a microscope equipped with Nomarski optics. A dim green safelight was used in every step of the transfections and staining. Transfections using each recombinant plasmid were repeated at least five times.
Acknowledgments
We thank Dr. Takeshi Todo for providing the SY2 strain and the pRT2 plasmid. We are grateful to Dr. Jane Silverthorne (University of California, Santa Cruz) for critical reading of the manuscript. We thank Hyun-Sook Park for 3′ RACE analysis and Kayoko Hara, Eitetsu Sugiyama, and Takako Yasuki for their technical assistance. This work was supported in part by the Grant-in-Aid for International Scientific Research (Joint Research) (Grant Nos. 07044206 and 09044232) and for Scientific Research (B) (Grant Nos. 07458196 and 09440270) from the Ministry of Education, Science, Sports, and Culture of Japan; by grants from the Mitsubishi Foundation, PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences), and the NOVARTIS Foundation to M.W.; by the Grant-in-Aid for Encouragement of Young Scientists (Grant No. 10740373) from the Ministry of Education, Science, Sports, and Culture of Japan to T.K.; and also by a grant from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (Grant No. 10639301) to T.I.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., and Cashmore, A.R. (1998. a). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998. b). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1 939–948. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998. c). Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392 720–723. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschauer, A. (1993). A plant gene for photolyase: An enzyme catalyzing the repair of UV-light-induced DNA damage. Plant J. 4 705–709. [DOI] [PubMed] [Google Scholar]
- Bogerd, H.P., Fridell, R.A., Benson, R.E., Hua, J., and Cullen, B.R. (1996). Protein sequence requirements for function of the human T-cell leukemia virus type1 Rex nuclear export signal delineated by a novel in vitro randomization-selection assay. Mol. Cell. Biol. 16 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- Emery, P., So, W.V., Kaneko, M., Hall, J.C., and Rosbash, M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95 669–679. [DOI] [PubMed] [Google Scholar]
- Furuya, M., Kanno, M., Okamoto, H., Fukuda, S., and Wada, M. (1997). Control of mitosis by phytochrome and a blue-light receptor in fern spores. Plant Physiol. 113 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hayami, J., Kadota, A., and Wada, M. (1986). Blue light–induced phototropic response and the intracellular photoreceptive site in Adiantum protonemata. Plant Cell Physiol. 27 1571–1577. [Google Scholar]
- Hayami, J., Kadota, A., and Wada, M. (1992). Intracellular dichroic orientation of the blue light–absorbing pigment and the blue-absorption band of the red-absorbing form of phytochrome responsible for phototropism of the fern Adiantum protonemata. Photochem. Photobiol. 56 661–666. [Google Scholar]
- Hoffman, P.D., Batschauer, A., and Hays, J.B. (1996). PHH1, a novel gene from Arabidopsis thaliana that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol. Gen. Genet. 253 259–265. [DOI] [PubMed] [Google Scholar]
- Horwitz, B.A. (1994). Properties and transduction chains of the UV and blue light photoreceptors. In Photomorphogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 327–350.
- Imaizumi, T., Kiyosue, T., Kanegae, T., and Wada, M. (1999). Cloning of the cDNA encoding the blue-light photoreceptor (cryptochrome) from the moss Physcomitrella patens. Plant Physiol. 120 1205.10490396 [Google Scholar]
- Kadota, A., Wada, M., and Furuya, M. (1979). Apical growth of protonemata in Adiantum capillus-veneris. III. Action spectra for the light effect on dark cessation of apical growth and the intracellular photoreceptive site. Plant Sci. Lett. 15 193–201. [Google Scholar]
- Kadota, A., Koyama, M., Wada, M., and Furuya, M. (1984). Action spectra for polarotropism and phototropism in protonemata of the fern Adiantum capillus-veneris. Physiol. Plant. 61 327–330. [Google Scholar]
- Kadota, A., Fushimi, Y., and Wada, M. (1986). Intracellular photoreceptive site for blue light–induced cell division in protonemata of the fern Adiantum—Further analyses by polarized light irradiation and cell centrifugation. Plant Cell Physiol. 27 989–995. [Google Scholar]
- Kagawa, T., and Wada, M. (1994). Brief irradiation with red or blue light induces orientational movement of chloroplasts in dark-adapted prothallial cells of the fern Adiantum. J. Plant Res. 107 389–398. [Google Scholar]
- Kanegae, T., and Wada, M. (1998). Isolation and characterization of homologues of plant blue-light photoreceptor (cryptochrome) genes from the fern Adiantum capillus-veneris. Mol. Gen. Genet. 259 345–353. [DOI] [PubMed] [Google Scholar]
- Kim, F.J., Beeche, A.A., Hunter, J.J., Chin, D.J., and Hope, T.J. (1996). Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol. Cell. Biol. 16 5147–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.F., and Sancar, A. (1990). Active site of Escherichia coli DNA photolyase: Mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry 29 5698–5706. [DOI] [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., and Cashmore, A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, C., Ahmad, M., Chan, J., and Cashmore, A.R. (1996. a). CRY2: A second member of the Arabidopsis cryptochrome gene family. Plant Physiol. 110 1047.8819875 [Google Scholar]
- Lin, C., Ahmad, M., and Cashmore, A.R. (1996. b). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light–dependent regulation of plant growth and development. Plant J. 10 893–902. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman, D.J., and Pearson, W.R. (1985). Rapid and sensitive protein similarity searches. Science 227 1435–1441. [DOI] [PubMed] [Google Scholar]
- Malhotra, K., Kim, S.-T., Batschauer, A., Dawut, L., and Sancar, A. (1995). Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34 6892–6899. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and Sharrock, R.A. (1997). Phytochrome gene diversity. Plant Cell Environ. 20 666–671. [Google Scholar]
- Miyata, M., Wada, M., and Furuya, M. (1979). Effects of phytochrome and blue-near ultraviolet light-absorbing pigment on duration of component phases of the cell cycle in Adiantum gametophytes. Dev. Growth Differ. 21 577–584. [DOI] [PubMed] [Google Scholar]
- Nozue, K., Fukuda, S., Kanegae, T., and Wada, M. (1998. a). Isolation of a second phytochrome cDNA from Adiantum capillus-veneris. Plant Physiol. 118 711.9867602 [Google Scholar]
- Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K.-C., Lagarias, J.C., and Wada, M. (1998. b). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, H., Hirano, Y., Abe, H., Tomizawa, K., Furuya, M., and Wada, M. (1993). The deduced amino acid sequence of phytochrome from Adiantum includes consensus motifs present in phytochrome B from seed plants. Plant Cell Physiol. 34 1329–1334. [Google Scholar]
- Park, H.-W., Kim, S.-T., Sancar, A., and Deisenhofer, J. (1995). Crystal structure of DNA photolyase from Escherichia coli. Science 268 1866–1872. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., and Lipman, D.J. (1988). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger, H., and Schmidt, W. (1994). Diversity of photoreceptors. In Photomorphogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 301–325.
- Small, G.D., Min, B., and Lefebvre, P.A. (1995). Characterization of a Chlamydomonas reinhardtii gene encoding a protein of the DNA photolyase/blue light photoreceptor family. Plant Mol. Biol. 28 443–454. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S.A., Rosbash, M., and Hall, J.C. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95 681–692. [DOI] [PubMed] [Google Scholar]
- Sugai, M., and Furuya, M. (1985). Action spectrum in ultraviolet and blue light region for the inhibition of red-light-induced spore germination in Adiantum capillus-veneris L. Plant Cell Physiol. 26 953–956. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, K., and Furuya, M. (1997). Control of the entry into S phase by phytochrome and blue light receptor in the first cell cycle of fern spores. Plant Cell Physiol. 38 1075–1079. [Google Scholar]
- van der Horst, G.T.J., et al. (1999). Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398 627–630. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Furuya, M. (1970). Photocontrol of the orientation of cell division in Adiantum. I. Effects of the dark and red periods in the apical cell of gametophytes. Dev. Growth Differ. 12 109–118. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Furuya, M. (1972). Phytochrome action on the timing of cell division in Adiantum gametophytes. Plant Physiol. 49 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., and Furuya, M. (1978). Effects of narrow-beam irradiations with blue and far-red light on the timing of cell division in Adiantum gametophytes. Planta 138 85–90. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Sugai, M. (1994). Photobiology of ferns. In Photomorphogenesis in Plants, 2nd ed. R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 783–802.
- Wada, M., Kadota, A., and Furuya, M. (1978). Apical growth of protonemata in Adiantum capillus-veneris. II. Action spectra for the induction of apical swelling and the intracellular photoreceptive site. Bot. Mag. Tokyo 91 113–120. [Google Scholar]
- Yamamoto, K. (1985). Photoreactivation reverses ultraviolet radiation induced premutagenic lesions leading to frameshift mutations in Escherichia coli. Mol. Gen. Genet. 201 141–145. [DOI] [PubMed] [Google Scholar]
- Yasuhira, S., and Yasui, A. (1992). Visible light-inducible photolyase gene from the goldfish Carassius auratus. J. Biol. Chem. 267 25644–25647. [PubMed] [Google Scholar]
- Yatsuhashi, H., Kadota, A., and Wada, M. (1985). Blue- and red-light action in photoorientation of chloroplasts in Adiantum protonemata. Planta 165 43–50. [DOI] [PubMed] [Google Scholar]
- Yatsuhashi, H., Hashimoto, T., and Wada, M. (1987). Dichroic orientation of photoreceptors for chloroplast movement in Adiantum protonemata. Non-helical orientation. Plant Sci. 51 165–170. [Google Scholar]
- Yeh, K.-C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]