Abstract
Phospholipids play an important role in many signaling pathways in animal cells. Signaling cascades are triggered by the activation of phospholipid cleaving enzymes such as phospholipases C, D (PLD), and A2. Their activities result in the formation of second messengers and amplification of the initial signal. In this study, we provide experimental evidence that PLD is involved in the early events of dehydration in the resurrection plant Craterostigma plantagineum. The enzymatic activity of the PLD protein was activated within minutes after the onset of dehydration, and although it was not inducible by abscisic acid, PLD activity did increase in response to mastoparan, which suggests a role for heterotrimeric G proteins in PLD regulation. Two cDNA clones encoding PLDs, CpPLD-1 and CpPLD-2, were isolated. The CpPLD-1 transcript was constitutively expressed, whereas CpPLD-2 was induced by dehydration and abscisic acid. Immunological studies revealed changes in the subcellular localization of the PLD protein in response to dehydration. Taken together, the data on enzymatic activity as well as transcript and protein distributions allowed us to propose a role for PLD in the events leading to desiccation tolerance in C. plantagineum.
INTRODUCTION
Craterostigma plantagineum (Scrophulariaceae) belongs to the small group of angiosperms referred to as resurrection plants. Extremely tolerant to dehydration (Gaff, 1971), their vegetative tissue can tolerate loss of as much as 98% of its water content, yet return to active metabolism and growth within hours after rehydration. For this reason, C. plantagineum has been used to investigate the molecular mechanisms underlying desiccation tolerance (Ingram and Bartels, 1996). Many transcripts are synthesized de novo during dehydration, and they are presumed to encode gene products with protective functions (Bartels et al., 1990; Piatkowski et al., 1990). Most of the genes can also be induced by exogenous abscisic acid (ABA), suggesting that ABA is at least partly involved in dehydration-activated signal transduction. Induction of most of these genes requires de novo protein synthesis, an indication that other cellular responses must precede transcription of these genes.
Despite the characterization of many dehydration-induced genes, little information is currently available on the early events elicited by desiccation. One interesting gene identified in C. plantagineum, CDT-1, acts in response to ABA and is able to activate a pathway that leads to desiccation tolerance (Furini et al., 1997). Two genes for transcription factors, C. plantagineum homeodomain–leucine zipper gene-1 (CPHB-1) and CPHB-2, encode interacting homeodomain–leucine zipper proteins, which may interconnect ABA-dependent and ABA-independent pathways (Frank et al., 1998). Given their differential expression in response to ABA, they may act in different branches of the dehydration-induced signaling network.
Phospholipid signaling plays an important role in diverse early signaling cascades in animal cells (Liscovitch and Cantley, 1994, 1995). In plants, phospholipid signaling has been suggested to play a role in several developmental processes and environmental adaptations involving phospholipases A2, C, and D (PLD) (reviewed in Chapman, 1998; Munnik et al., 1998a). Initial evidence that phospholipid signaling may also play a role in dehydration came from the isolation of Arabidopsis dehydration-responsive genes encoding a phosphatidylinositol-specific phospholipase C (AtPLC1; Hirayama et al., 1995) and a phosphatidylinositol-4-phosphate 5-kinase (PIP5K; Mikami et al., 1998). Although the expression patterns of these genes imply a role in dehydration-responsive phospholipid signaling, no direct evidence based on measurements of enzyme activity has been found.
Here, we report that PLD enzyme activation is an early event in the response of C. plantagineum to dehydration. PLD cleaves phospholipids within membranes, resulting in the formation of a polar head group and a phosphorylated lipid moiety, termed phosphatidic acid (PtdOH). PtdOH is a second messenger in animal cells, activating targets such as phospholipase C and protein kinase C (Liscovitch and Cantley, 1994, 1995; English, 1996; English et al., 1996). Genes encoding PLD have been isolated from various plant species (Wang, 1997), and their molecular characterization has suggested involvement in different signaling pathways. The genes are important in phospholipid catabolism and initiate a lipolytic cascade in membrane disintegration during germination, senescence, and stress injuries (Paliyath and Droillard, 1992; Chapman, 1998; Munnik et al., 1998a). Recent studies also suggest that they play a role in early events of various signal transduction pathways. PLD and its product PtdOH have been found to be involved in wound- and pathogen-induced signaling pathways (Ryu and Wang, 1996; Young et al., 1996; Lee et al., 1997) and in the response of barley aleurone cells to ABA (Ritchie and Gilroy, 1998).
An understanding of the molecular mechanisms underlying the activation of PLD was derived from studies with the G protein–activating peptide mastoparan. Treatment of Chlamydomonas cells and carnation petals with mastoparan resulted in a time- and dose-dependent activation of PLD measured in vivo (Munnik et al., 1995; de Vrije and Munnik, 1997), indicating that the activation of the enzyme is G protein mediated. Using an in vivo assay, we measured PLD activity during dehydration in C. plantagineum leaf discs. Here, we report on the induction of PLD by dehydration and provide molecular evidence for a differential regulation of PLD-encoding genes in response to water stress.
RESULTS
PLD Activity Is Rapidly Induced during Dehydration
To analyze the role of PLD in dehydration-activated signal transduction in C. plantagineum, we measured PLD activity in response to dehydration and ABA. PLD activity in vivo was determined by an assay using leaf discs prelabeled with 32Pi and n-butanol as reporter alcohol (Munnik et al., 1995). n-Butanol is converted by PLD to phosphatidylbutanol (PtdButOH), a specific measure for PLD activity. In addition, PtdOH was quantified, both because it is the natural product of PLD and because it may also be generated by way of the phospholipase C pathway.
PLD activity was measured in two series of dehydration experiments. Lipids were extracted and separated by thin-layer chromatography (TLC), and PtdButOH and PtdOH were quantified as a percentage of total incorporated radioactivity. In one set of experiments, leaf discs were dried for 30 min or for 1, 2, 3, or 4 hr (Figures 1A to 1C). PLD activity increased rapidly after the onset of dehydration, as demonstrated by a fivefold increase in PtdButOH and a 17-fold increase of PtdOH, representing ∼40% of total labeled phospholipids. The PtdButOH concentration reached its maximum after 2 hr of dehydration, whereas the concentration of PtdOH stayed constant after 2 hr.
Figure 1.
Dehydration Stimulates PLD Activity.
(A) Leaf discs from C. plantagineum were prelabeled with 32Pi for 16 hr, removed from the labeling buffer, and dried for the times indicated. To measure PLD activity, we again incubated them in buffer that included <0.75% n-butanol (ButOH). After 5 min of vacuum infiltration, the discs were incubated for 25 min at room temperature.
(B) Autoradiography of a TLC plate with the separated phospholipids extracted from one leaf disc, showing the extent of PtdButOH and PtdOH formation in response to dehydration. The positions of PtdButOH and PtdOH are indicated. C, control disc.
(C) Quantification of PtdButOH and PtdOH by using a PhosphorImager. The amount of each phospholipid is expressed as fold stimulation relative to that in the control. The horizontal lines in the graph indicate the amount of PtdButOH (top) and PtdOH (bottom) measured in the control discs. The values are means of two independent experiments. The error bars indicate the standard deviations.
(D) Leaf discs of C. plantagineum were treated as described in (A) but were sampled after different durations of dehydration.
(E) Autoradiography of the TLC plate after separation of the phospholipids (cf. [B]).
(F) Quantification of PtdButOH and PtdOH by using a PhosphorImager (cf. [C]).
In a second series of experiments, the time of dehydration was reduced to 10 min. Prelabeled leaf discs were dried for 10, 20, or 30 min or for 1 or 2 hr; an increase in PLD activity was measured within the first 10 min after the onset of dehydration (Figures 1D to 1F). This activity continued to increase for 2 hr. The formation of PtdButOH was stimulated fourfold, and PtdOH levels increased 14-fold. These experiments demonstrate that the stimulation of PLD activity by dehydration is a rapid response to the environmental stimulus, producing the potential second messenger PtdOH.
PLD Stimulation Is Mediated by G Proteins but Not by ABA
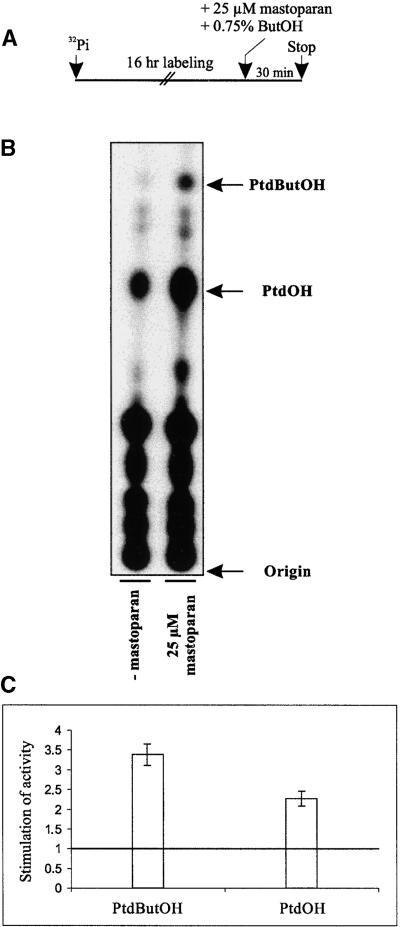
Experiments using the G protein–activating peptide mastoparan revealed a time- and dose-dependent stimulation of PLD activity in plants (Munnik et al., 1995; de Vrije and Munnik, 1997). Prelabeled C. plantagineum leaf discs were incubated for 30 min in the presence of 25 μM mastoparan and 0.75% butanol. Compared with the control, PtdButOH formation increased 3.4-fold and PtdOH 2.3-fold after mastoparan treatment (Figures 2A to 2C). Mastoparan was thus able to mimic the dehydration-related response of PLD, suggesting that heterotrimeric G proteins are involved in PLD regulation. When the inactive mastoparan analog mas17 was used instead of mastoparan, no particular increase in PtdOH or PtButOH was observed (data not shown). This finding points to the specific activation of PLD by G proteins rather than by perturbation of the membranes by mastoparan.
Figure 2.
PLD Stimulation Is Mediated by Heterotrimeric G Proteins.
(A) Leaf discs of C. plantagineum were prelabeled with 32Pi for 16 hr and incubated in the presence of 25 μM mastoparan and 0.75% n-butanol (ButOH) for 30 min at room temperature. Control discs were incubated in the absence of mastoparan.
(B) Autoradiography of a TLC plate after separation of the phospholipids from mastoparan-treated discs (25 μM mastoparan) and control leaf discs not treated with mastoparan (− mastoparan) (cf. Figure 1B).
(C) Quantification of PtdButOH and PtdOH by using a PhosphorImager. The values are means of two independent experiments. The horizontal line in the graphs indicates the amount of PtdButOH and PtdOH measured in the control experiments (no mastoparan added). The error bars indicate the standard deviations (cf. Figure 1C).
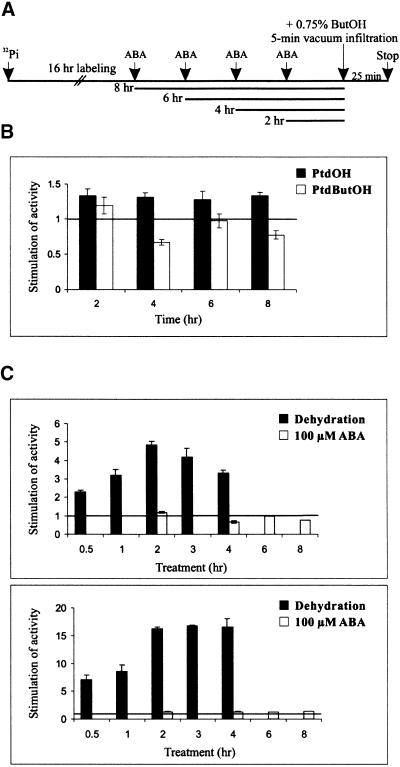
To test whether treatment with ABA induced PLD activity, we measured PLD activities after incubating leaf discs, prelabeled with 32Pi, in 100 μM ABA for 2, 4, 6, or 8 hr (Figures 3A to 3C). Control leaf discs were incubated in 0.1% ethanol, which was the same concentration used in the ABA treatment. Application of ABA did not lead to increased formation of PtdButOH, indicating that ABA is not able to stimulate PLD activity. Also, the 1.3-fold increase in PtdOH observed after treatment with ABA is negligible compared with the 17-fold induction of PtdOH triggered by dehydration (Figure 3C). This experiment shows that the activation of PLD is triggered by dehydration but not by exogenous ABA.
Figure 3.
ABA Does Not Stimulate PLD Activity.
(A) Leaf discs were prelabeled with 32Pi for 16 hr and treated with 100 μM ABA or 0.1% ethanol (control) for the times indicated. To measure PLD activity, they were incubated in the presence of 0.75% n-butanol (ButOH). After 5 min of vacuum infiltration, the discs were incubated for 25 min at room temperature.
(B) Formation of PtdButOH and PtdOH in response to ABA. After separation of phospholipids by TLC, the TLC plate was exposed to a PhosphorImager screen; the amount of each phospholipid was quantified and is expressed as fold stimulation relative to the amount in the control. The values are means of four different experiments. The error bars indicate the standard deviations.
(C) Comparison of PLD activity in response to dehydration and ABA. The values for PtdButOH (top) and PtdOH (bottom), shown in Figures 1C and 3B, are compared. They demonstrate that the activation of PLD by dehydration is independent of ABA.
The horizontal lines in the graphs indicate the amount of PtdButOH and PtdOH measured in the control discs. Error bars indicate the standard deviation.
Isolation of Two cDNAs for PLD
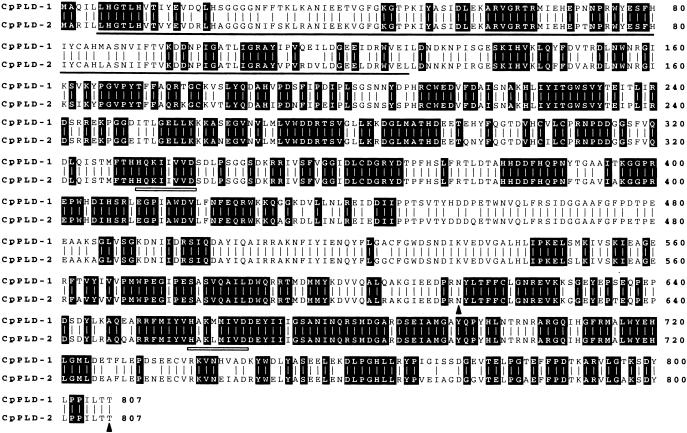
The effect of dehydration on the induction of PLD activity prompted us to isolate genes encoding PLD from C. plantagineum. A differential display–reverse transcription–polymerase chain reaction (DD-RT-PCR) approach led to the isolation of a 355-bp cDNA fragment exhibiting >88% similarity to genes encoding PLD from plants. Using this cDNA fragment as a hybridization probe led to isolation of two full-length cDNAs, each encoding a different PLD. The cDNAs have been designated CpPLD-1 and CpPLD-2 (for C. plantagineum phospholipase D-1 and D-2). CpPLD-2 contains the 355-bp DD-RT-PCR fragment at its 3′ end; CpPLD-1 represents a second member of the PLD family, its prod-uct being 90.8% identical and 94.7% similar to CpPLD-2 (Figure 4). Both cDNAs encode 807–amino acid proteins with a molecular mass of 92 kD. Based on size and acidic pI (5.18 for CpPLD-1 and 5.46 for CpPLD-2), both PLDs can be grouped into the α class of plant PLDs (Wang, 1997). CpPLD-1 and CpPLD-2 are very similar to 11 other plant PLD proteins of the α class and share 388 identical amino acid residues. CpPLD-1 and CpPLD-2 also contain the C2 or calcium binding domain near their N termini. This domain, first identified in Ca2+-dependent protein kinase C isoforms, is present in all plant PLD sequences and is involved in the binding of Ca2+ and phospholipids (Sutton et al., 1995). Notably, the C2 domain is present in several different proteins involved in signal transduction and membrane trafficking, including phospholipase A2 and PIP2 phospholipase C isoforms (Ponting and Parker, 1996). Both PLDs from C. plantagineum also contain the duplicated HxKxxxxD motif (HKD motif, where x stands for any amino acid), which is found twice within all PLD amino acid sequences and is thought to represent the catalytic domain. The strict conservation of these amino acids suggests that the His, Lys, and Asp residues at these positions are functionally important. When gene-specific CpPLD-1 and CpPLD-2 probes were used for DNA gel blot analyses, distinct hybridization patterns were obtained (Figure 5). The number of hybridizing bands indicates that both CpPLD-1 and CpPLD-2 are present in at least two copies in the C. plantagineum genome.
Figure 4.
Amino Acid Sequence Comparison of CpPLD-1 and CpPLD-2.
In the aligned amino acid sequences deduced from the cDNA clones CpPLD-1 and CpPLD-2, vertical lines denote identical amino acids present in both sequences. Invariant amino acid residues found in 11 other plant α-class PLD proteins are indicated by black boxes. The C2 (CalB) domain, which is involved in Ca2+/phospholipid binding, is indicated by a thick black line. The duplicated HKD catalytic domains are indicated by open bars. The region of CpPLD-1 used for antiserum production is indicated by an arrowhead. The GenBank accession numbers for CpPLD-1 and CpPLD-2 are AJ133001 (CpPLD-1) and AJ133000 (CpPLD-2).
Figure 5.
DNA Gel Blot Analyses.
C. plantagineum DNA (10 μg per lane) was digested with the indicated restriction enzymes. Identical membranes were probed at high stringency with the gene-specific fragments of CpPLD-1 and CpPLD-2.
CpPLD-1 and CpPLD-2 Are Differentially Expressed
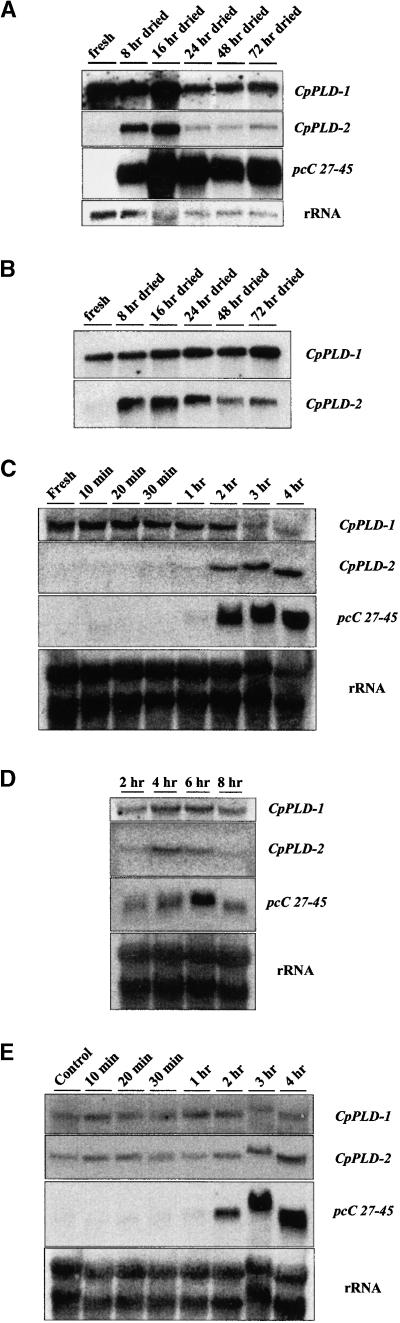
RNA gel blot analysis was used to determine CpPLD-1 and CpPLD-2 transcript levels during dehydration and ABA treatment. CpPLD-1 and CpPLD-2 were differently regulated in leaves and roots (Figures 6A and 6B). The CpPLD-1 transcript was constitutively expressed in both tissues; the CpPLD-2 transcript, in contrast, was absent from both in well-watered plants but was induced after 8 hr of dehydration, declining after 24 hr of dehydration in leaves and after 48 hr in roots.
Figure 6.
Expression Pattern of CpPLD-1 and CpPLD-2 Transcripts.
(A) RNA gel blot loaded with 3 μg of poly(A)+ RNA per lane extracted from leaves harvested from dried plants at the times indicated. The membrane was probed with the gene-specific fragments of CpPLD-1 and CpPLD-2. The same membrane was used for control hybridizations with the LEA-type cDNA pcC27-45 and an rDNA gene from wheat.
(B) RNA gel blot loaded with 3 μg of poly(A)+ RNA per lane from roots harvested from dried plants at the times indicated. The membrane was probed with the gene-specific fragments of CpPLD-1 and CpPLD-2.
(C) RNA gel blot loaded with 40 μg of total RNA per lane from detached leaves, which were dried for the times indicated, and hybridized with the same probes as described in (A).
(D) RNA gel blot loaded with 40 μg of total RNA per lane from detached leaves, which were incubated in 100 μM ABA for the times indicated, and hybridized with the same probes as described in (A).
(E) RNA gel blot with 40 μg of total RNA per lane from leaf discs, which were dried for the times indicated, and hybridized with the same probes as described in (A). Control discs were left in buffer for 4 hr.
The transcript levels of both genes were also investigated in detached leaves that had been dried or treated with exogenous ABA (Figures 6C and 6D). Again, the CpPLD-1 transcript was constitutively expressed, whereas the CpPLD-2 mRNA was induced after 2 hr of dehydration. Treatment with ABA induced a signal from CpPLD-2 mRNA after 4 hr that declined after 6 hr.
CpPLD-1 and CpPLD-2 mRNAs were also analyzed in excised leaf discs (Figure 6E), because excised leaf discs had been used to determine PLD activity. Both the CpPLD-1 and CpPLD-2 transcripts were detected in all samples, including control discs left in buffer. As in previous experiments, CpPLD-1 showed a constitutive expression pattern. Conversely, the amount of CpPLD-2 transcript was found to increase in leaf discs dehydrated for >2 hr, even though its expression was also induced by wounding. In all dehydration experiments, the timing of induction of CpPLD-2 transcript accumulation correlated with the onset of expression of the late embryogenesis abundant (LEA)–type gene C. plantagineum pcC27-45 (Bartels et al., 1990), which was used to monitor the dehydration treatment.
Anti-CpPLD Antiserum Recognizes Different PLD Isoforms
An antiserum raised against a 202–amino acid fragment (see Figure 4 and Methods) of CpPLD-1 not only detected CpPLD-1 but also recognized other members of the PLD family, because the antigen used contained a conserved protein domain. For immunodetection of PLD, total protein extracts and soluble protein extracts from leaves and roots harvested from fresh and dehydrated plants were used. A 92-kD protein was detected on immunoblots in all samples of total proteins (Figure 7A). After the total protein fractions were further fractionated and the soluble proteins were purified, differentially expressed PLD isoforms were found. When proteins were separated under denaturing conditions, PLD was detected after 16 hr of dehydration in roots and after 48 hr of dehydration in leaves. In both cases, the protein was still present after 72 hr of desiccation. The dehydration treatment was monitored by reprobing the membranes with a polyclonal antibody raised against the dehydration-inducible cytosolic LEA-type protein pcC27-45. The soluble proteins were further analyzed by one-dimensional isoelectric focusing (Figure 7B). PLD was detected after 48 hr of dehydration in leaves and after 16 hr of desiccation in roots, but the complexity of the resolved bands differed in leaves and roots. A single protein band was recognized in leaves, and at least three different PLD isoforms were observed in the root extracts. These results indicate a complex regulation of the expression and distribution of PLD proteins, which depend on the water content and the tissue of the plant.
Figure 7.
Protein Gel Blot Analyses with the Anti–CpPLD-1 Antiserum.
(A) One hundred micrograms of total and soluble proteins per lane from leaves and roots harvested from plants treated as indicated was separated by 10% SDS-PAGE and blotted onto a polyvinyl difluoride membrane. Immunodetection was performed by using the polyclonal anti–CpPLD-1 antiserum. The membrane with the separated soluble proteins was reprobed with a polyclonal anti–pcC27-45 antiserum.
(B) Fifty micrograms of the soluble protein extracts described in (A) was separated by one-dimensional isoelectric focusing (pH 3.5 to 9.5) and blotted onto a polyvinyl difluoride membrane. Immunodetection was performed as described in (A). The pH values of 1-cm gel slices were determined and are marked by horizontal lines.
DISCUSSION
Early Activation of PLD Enzyme Activity
Our studies have shown that the activity of PLD, an enzyme involved in phospholipid signaling, is stimulated by dehydration. PLD activity was monitored by its ability to transfer the phosphatidyl moiety of a structural phospholipid to a primary alcohol (transphosphatidylation). In C. plantagineum, PLD responds within 10 min of the onset of dehydration, and PtdOH reaches a maximum after 3 hr. Because this response is much faster than any other changes observed during dehydration of C. plantagineum plants (Bockel et al., 1998), we propose that the stimulation of PLD activity opens a very early signaling pathway.
During dehydration treatment, PtdOH accounts for as much as 40% of the total amount of labeled phospholipids, which represents a 20-fold increase over that in control samples. This large increase is consistent with a role for PtdOH as a second messenger that interacts rapidly with downstream targets. Interestingly, the amount of dehydration-induced PtdOH exceeds that of PtdButOH, probably because PtdButOH is a selective relative measure for PLD, whereas PtdOH is a relative measure of several possible reactions that involve PLD, phospholipase C, or formation of glycerolipids (Munnik et al., 1998b).
The increased PLD activity most likely results from the stimulation of preexisting PLD isoforms rather than de novo synthesis of the enzyme. This form of activation could be a general mechanism in processes involving PLD, because similar observations have been made for wounding stress (Ryu and Wang, 1996; Lee et al., 1997). In these studies, increased concentrations of PtdOH were measured within 5 min at the wound site and in neighboring leaves, suggesting that PLD activation is part of the systemic response to wounding. PLD was also activated after treatment of plants with red light and red/far-red light, indicating that the enzyme also is involved in phytochrome signaling (Park et al., 1996).
Activation of PLD involves heterotrimeric G proteins, as demonstrated by the effects of G protein activators, such as alcohols or mastoparan (Munnik et al., 1995; de Vrije and Munnik, 1997). Indeed, treating Chlamydomonas cells with G protein activators stimulated PLD activity within minutes in a dose-dependent manner. The stimulation of PLD in C. plantagineum was also mediated through G proteins, as shown by the application of mastoparan. Although many targets of PtdOH, including protein kinase C and phospholipase C, have been identified in animal cells (Liscovitch and Cantley, 1994, 1995; English, 1996; English et al., 1996), their precise targets in plants are still mainly unknown.
Activation of the Enzymatic Activity of PLD Is Independent of ABA
Studies of mutants and molecular analysis of dehydration-responsive genes have suggested that ABA plays a major role in cellular responses to water deficit (Ingram and Bartels, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1997; Bonitta and McCourt, 1998; Leung and Giraudat, 1998). Data on gene expression suggest that ABA-dependent and ABA-independent stress signaling pathways interact and converge to activate gene expression (Ishitani et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 1997; Frank et al., 1998). Our studies of PLD activity indicate that ABA is not able to mimic the dehydration-induced stimulation of this enzyme, which suggests either that PLD acts in an ABA-independent pathway or that its activation is upstream of ABA action. The rapid activation of PLD could be triggered by a sensing mechanism in the membrane, which is important for the early response. However, the ABA-induced increase of the CpPLD-2 transcript at a later time may suggest involvement of the encoded PLD protein in an adaptive response. Ritchie and Gilroy (1998), investigating the role of PLD during germination in barley, reported the activation of PLD in aleurone protoplasts by exogenous ABA. They also observed an ABA-like inhibition of α-amylase production and an induction of ABA-upregulated proteins such as amylase subtilisin inhibitor and RAB (for responsive to ABA) when PtdOH was applied to the cells. These results, in contrast with our data, suggest that ABA activates PLD to produce PtdOH, which is involved in triggering the ABA responses of the aleurone cells. Aleurone protoplasts are very specialized cells and may therefore react differently to ABA. In addition, Ritchie and Gilroy used hydrolysis of fluorescent lipids to assay PLD, which may not be as specific as the assay used here.
Subcellular Distribution of PLD Isoforms
Immunoblot experiments showed that the distribution of the PLD protein depended on the water status of the tissue. Although PLD was constitutively present in total protein extracts from leaves and roots, it was detected in the soluble fraction only in the protein samples derived from dried tissues; the PLD isoforms were present after 16 hr of dehydration in roots and after 48 hr of water deficit in leaves. This observation can be explained either by de novo synthesis of PLD during dehydration and subsequent accumulation in the cytosol or by translocation of membrane-bound PLD into the cytosol.
We favor the first assumption, because CpPLD-2 was transcriptionally induced by dehydration at later times. Also, the immunological data correlated well with the enzymatic activity measurements; the PLD protein was constitutively present in total protein extracts and was associated with the membranes in fresh tissues. Membrane localization is required for the catalytic function of PLD, because membranes represent the substrate of this class of enzymes. The membrane-associated enzyme can be easily activated at the onset of dehydration. Studies similar to ours have shown that the distribution of PLD is altered by developmental processes and external stimuli. During leaf senescence, for example, the amount of membrane-associated PLD increased, as would be expected if PLD plays a role in membrane disintegration and plant senescence (Ryu and Wang, 1995). PLD distribution was also altered in rice leaves challenged with Xanthomonas oryzae pv oryzae. In cells undergoing a susceptible response, PLD was uniformly distributed along the plasma membrane after inoculation with the bacteria but was clustered preferentially in membranes adjacent to bacterial cells after bacterial challenge in resistant interactions (Young et al., 1996).
Ca2+ ions are thought to be important in controlling PLD activity and distribution, as concluded from the Ca2+-dependent enzyme activity of most plant PLDs. The presence of calcium binding domains in the predicted products of all the PLD-encoding genes sequenced to date indicates a specific mode of activation. When activated by Ca2+, PLD proteins bind membrane lipids (Ponting and Parker, 1996; Williams and Katan, 1996) and are translocated from the cytosol to membranes, where their substrate is concentrated, thus leading to increased activity. CpPLD-1 and CpPLD-2 from C. plantagineum also contain a calcium binding domain at their N termini; thus, regulation of the proteins is likely to be Ca2+ dependent.
Molecular Analysis of CpPLD-1 and CpPLD-2
When the involvement of PLD in the water stress response was discovered, we isolated two members of the α class of PLDs to analyze the role of this class of enzymes. Whereas all previously characterized α-class PLD proteins have a size of 808 to 812 amino acids (Wang, 1997), the predicted length of the two α class PLD proteins from C. plantagineum is 807 amino acids. The genes encoding PLD are differentially expressed: CpPLD-1 is constitutively expressed and was not affected by dehydration or ABA treatment, whereas CpPLD-2 mRNA was synthesized only in response to dehydration, ABA, or wounding. The constitutive expression of CpPLD is consistent with the increased enzymatic activity of PLD seen during the onset of dehydration. Thus, CpPLD-1 in particular is a potential candidate for encoding the enzyme that is activated upon dehydration. The accumulation of CpPLD-2 transcript, which starts 2 hr after the onset of dehydration, should result in reinforcement of the PLD-dependent signal.
Differential expression of PLD was also found in rice leaves in response to inoculation with a virulent or an avirulent strain of X. o. oryzae (Young et al., 1996). Genes encoding PLD may also play a role in germination, because the transcript was not detected in dry seeds of rice, castor bean, or soybean; it was, however, present during water uptake by the seed after the radicle emerged (Dyer et al., 1994; Ueki et al., 1995; Ryu et al., 1996). This is in contrast to the expression of CpPLD-2, which is induced by dehydration.
The differential expression patterns of the two genes studied, the subcellular distribution of their products, and the data on enzymatic activity suggest specific roles for the two CpPLD forms (summarized in Table 1). The constitutively expressed CpPLD-1 could act in the very early response to dehydration, producing the second messenger PtdOH to amplify the signal after its perception. The dehydration-inducible CpPLD-2 may be involved in phospholipid metabolism and membrane rearrangements, which may be important components of desiccation survival processes in C. plantagineum. Although evidence has been presented here for a role of PLD in dehydration stress signaling, the mechanism is unknown. Putative targets of PtdOH could be protein kinases, Ca2+ fluxes, G proteins, and vesicular trafficking (Munnik et al., 1998a). With the molecular tools available, the production of CpPLD-1 and CpPLD-2 antisense plants will be helpful in determining the functions of these proteins.
Table 1.
Enzymatic and Molecular Analyses of PLD during Dehydration in C. plantagineum
| Response to Dehydration
|
||
|---|---|---|
| Events | Within Minutes | Within Hours |
| PLD protein | ||
| Activity | Activated | Active |
| Localization | Associated with leaf and root membranes |
Shift of leaf and root isoforms to the cytoplasm |
| Transcript accumulation | ||
| CpPLD-1 | Constitutive | Constitutive |
| CpPLD-2 | Not detected | Induced |
METHODS
Plant Material
Plants (Craterostigma plantagineum) originally collected in South Africa were propagated as reported previously (Bartels et al., 1990). For dehydration treatment, whole plants were taken out of their pots and dried in the growth chamber for the times stated above. Detached leaves and leaf discs were desiccated on filter paper. Abscisic acid (ABA) treatments were performed as described by Bartels et al. (1990), if not stated otherwise.
DNA Manipulations and Sequence Analysis
DNA was manipulated according to standard procedures (Sambrook et al., 1989; Ausubel et al., 1996). DNA and protein similarities were determined by using the BLAST network service (Altschul et al., 1990) and analyzed with use of the Genetics Computer Group (Madison, WI) version 9.0 software package. For sequence comparison of CpPLD-1 and CpPLD-2 with other phospholipase D (PLD) proteins of the α class from plants, 11 full-length cDNAs with the following accession numbers were selected from the SwissProt database: Ricinus communis, Q41142, A54850, and U72693; Oryza sativa, Q43007; Zea mays, Q43270; Arabidopsis thaliana, Q38882; Nicotiana tabacum, P93400; Pimpinella brachycarpa, O04883; Vigna unguiculata, O04865; and Brassica oleracea, U85482 and AF090444. Sequence alignment of protein sequences was performed by using the PILEUP program.
Nucleic Acid Extraction and Analysis
Poly(A)+ RNA extraction and RNA gel blot analysis were performed as described previously (Bartels et al., 1990). Genomic DNA was extracted by using the genomic DNA purification kit (Qiagen, Hilden, Germany). DNA digested with restriction enzymes was electrophoretically separated on an 0.8% agarose gel and transferred to a nylon membrane according to Sambrook et al. (1989). Hybridizations of genomic DNA gel blot filters were performed at 65°C in 0.6 M NaCl, 10 mM Pipes, pH 6.8, 1 mM EDTA, 0.1% SDS, 10 × Denhardt's solution (1 × Denhardt's solution is 2% [w/v] BSA, 2% [w/v] Ficoll 400, 2% [w/v] polyvinylpyrrolidone, and 100 μg mL−1 denatured salmon sperm DNA). Washes were performed in 0.5 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C. Hybridizations of filters carrying phage DNA were performed in the same buffer at 65°C (high stringency) and 62°C (low stringency). Washes were performed with 0.2 × SSC and 0.1% SDS at 65°C (high stringency) or with 2 × SSC and 0.1% SDS at 62°C (low stringency). Hybridization signals were removed by incubating the membranes for 30 min at 95°C in 0.1% SDS.
Hybridization Probes
For RNA and DNA gel blot analyses, two fragments of the CpPLD-1 and CpPLD-2 cDNAs were used. A 2239-bp fragment of the CpPLD-1 cDNA was obtained by polymerase chain reaction (PCR) amplification with the following primers: sense primer, 5′-GTGGGTTGAGATACTTGACAAT-3′ (nucleotides 401 to 422); antisense primer, 5′-CCGATAACATAAAACCAAAG-3′ (nucleotides 2639 to 2620). The CpPLD-2 gene-specific probe was the original differential display–reverse transcription–PCR (DD-RT-PCR) fragment subcloned in the pPCR-Script™ cloning vector (Stratagene, La Jolla, CA); this fragment encompasses the coding sequence for the last 44 C-terminal amino acids and the 3′ untranslated region. Dehydration and ABA treatments of the plant material used for RNA extractions were monitored by reprobing the membranes with the drought- and ABA-inducible late embryogenesis abundant (LEA)–type cDNA pcC27-45 from C. plantagineum (Piatkowski et al., 1990). Equal loading of RNA on membranes was monitored by hybridization with an rDNA fragment (Gerlach and Bedbrook, 1979). Random oligonucleotide labeling of DNA fragments with α-32P-dCTP was performed according to Feinberg and Vogelstein (1983).
Isolation of cDNA Clones
The CpPLD-2 cDNA was isolated by DD-RT-PCR, which was performed according to Liang and Pardee (1992) and by using the same cDNAs and primers as described previously (Frank et al., 1998). A cDNA library prepared from poly(A)+ RNA isolated from leaves dried for 1 hr (Bockel et al., 1998) was screened with the DD-RT-PCR fragment of the CpPLD-2 cDNA under high- and low-stringency conditions to obtain full-length cDNA clones of the gene family encoding PLD.
Preparation of the Anti–CpPLD-1 Antiserum
A CpPLD-1–glutathione S-transferase fusion protein was expressed in Escherichia coli by using the pGEX-4T-2 vector (Smith and Johnson, 1988). A 621-bp fragment of the CpPLD-1 cDNA was amplified by using Pfu DNA polymerase (Stratagene) and the following primers: sense primer, 5′-GGAATTCCTTAACTTTCTTCTGCCTCGG-3′ (nucleotides 1870 to 1897); antisense primer, 5′-CCAGAACTCGAGTTAAACAAAGCAC-3′ (nucleotides 2490 to 2466). For directional cloning into the pGEX-4T-2 vector, the primers contained an EcoRI and a XhoI restriction site, respectively (underlined in the sequences above). The fragment was cloned into the EcoRI-XhoI sites of the vector, yielding a translational fusion with glutathione S-transferase. A 56-kD protein was expressed on exposure of plasmid-containing E. coli BL21 to 0.1 mM isopropyl-β-d-thiogalactopyranoside and partially purified by isolation of inclusion bodies (Schmidt et al., 1986). Subsequently, the protein-containing band was excised from a preparative SDS gel and then freeze-dried for direct injection into rabbits (BioGenes, Berlin, Germany).
Protein Extraction
Soluble proteins from leaves and roots were extracted according to a modified protocol of Dyer et al. (1994). Plant material was ground to fine powder under liquid N2. The following steps were performed at 4°C. Proteins from 5 to 10 g of plant material were extracted in 100 mL of buffer A: 50 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM EDTA, 0.5 M sucrose, 0.5 mM phenylmethylsulfonyl fluoride, and 2 mM DTT. The homogenate was filtered through three layers of cheesecloth and centrifuged at 10,000g for 20 min. The supernatant was centrifuged at 110,000g for 1 hr. Proteins in the supernatant were precipitated by adding 0.313 g mL−1 (NH)4SO4 (50% saturation). The precipitated proteins were pelleted at 12,000g for 20 min and dissolved in 0.5 to 1 mL of buffer B (the same as buffer A, except that sucrose was omitted and 14% glycerol was added). Insoluble material was removed by centrifugation at 10,000g for 5 min. Soluble proteins were dialyzed against buffer B and stored in aliquots at −70°C. The protein concentration was determined according to the method of Bradford (1976). Total proteins from leaves and roots were extracted by boiling 400 mg of ground material in 1 mL of SDS loading buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.1% bromophenol blue) for 10 min. After centrifugation for 10 min at 14,000g, the supernatant was collected and stored at −20°C. Before the determination of protein concentration according to Bradford (1976), SDS was precipitated by the addition of 4 volumes 0.1 M KH2PO4/K2HPO4, pH 7.2, to a 1:5 dilution of the samples.
Electrophoresis and Immunoblotting
Electrophoresis under denaturing conditions was performed as described by Laemmli (1970) using 10% SDS–polyacrylamide gels. Native isoelectric focusing was performed according to Robertson et al. (1987), with the following modifications. The gels contained 3.5% (v/v), pH 5.0 to 8.0, ampholyte and 1.5% (v/v), pH 3.5 to 9.5, ampholyte. Proteins were mixed with an equal volume of 50% (v/v) glycerol and 4% (v/v) ampholytes in the pH range used to prepare the gel. Before blotting, native gels were equilibrated in blotting buffer (39 mM glycine, 48 mM Tris, 0.0375% SDS, and 20% methanol) for 10 min, whereas denaturing SDS gels were blotted directly after electrophoresis. The proteins were blotted onto polyvinyl difluoride membrane at 2 mA cm−2 for 1 hr by using a semidry electroblotter (Sartorius, Göttingen, Germany). Protein gel blots were probed with the anti–CpPLD-1 antiserum diluted 1:5000 before incubation with anti–rabbit IgG (Sigma, Deisenhofen, Germany; 1:10,000 dilution). The protein–antibody complex was detected with a chemiluminescence protein gel blotting detection system (ECL; Amersham Buchler, Braunschweig, Germany).
Phospholipid Labeling and PLD Assay
The in vivo production of 32P-labeled phosphatidylbutanol (PtdButOH) was the basis of PLD activity measurements, as described by Munnik et al. (1995) and de Vrije and Munnik (1997). Leaf discs (4 mm diameter) were excised from C. plantagineum leaves, and phospholipids were metabolically labeled by incubating for 16 hr a single disc with 370 kBq of carrier-free 32PO43− (32Pi) (Amersham) in 90 μL of 10 mM Mes, pH 5.5 (KOH). For the dehydration treatment, the prelabeled leaf discs were removed from the labeling buffer and dried on filter paper under the hood for the times indicated in Figure 1A. Mastoparan or mas17 (Peninsula Laboratories, Belmont, CA) and ABA treatments were performed by adding fresh stock solution for the times and concentrations indicated in Figure 3A. To assay PLD activity, we vacuum-infiltrated the prelabeled leaf discs in 180 μL of 0.75% (v/v) n-butanol and 10 mM Mes, pH 5.5. After vacuum infiltration (0.95 bar) for 5 min, the vacuum was released, and the leaf discs were incubated for another 25 min at room temperature. PLD activity was measured by adding 40 μL of n-butanol and then vacuum-infiltrating and incubating the discs as described above. The reactions were stopped by adding 20 μL of ice-cold 50% (v/v) perchloric acid, vortex-mixing for 5 sec, and freezing the samples in liquid N2.
Samples were extracted by adding 750 μL of CHCl3/MeOH/HCl (50:100:1 [v/v]). The samples were vortex-mixed for 5 sec and again frozen in N2 for 5 min. After thawing, the samples were shaken on an Eppendorf mixer (model 5432; Eppendorf, Hamburg, Germany) for 1 hr at room temperature. Two liquid phases were induced by the addition of 200 μL 0.9% (w/v) NaCl and 750 μL CHCl3. The lipids were subsequently extracted as described by Munnik et al. (1996).
32P-labeled PtdButOH was separated from phosphatidic acid (PtdOH) and the rest of the phospholipids by a modified ethyl acetate thin-layer chromatography (TLC) system (the organic upper phase consisted of a mixture of ethyl acetate/iso-octane/HCOOH/H2O (13:2:3:10 [v/v], as described by Munnik et al. [1998b]). Radiolabeled phospholipids were detected by autoradiography and by using a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Radioactivity on TLC plates was quantified by using ImageQuant software (Molecular Dynamics). After subtraction of the background, the quantities of PtdButOH and PtdOH are given as percentages of the total counts per minute per lane.
Materials
ABA (mixed isomers; Sigma) was dissolved in ethanol at a concentration of 100 mM, and aliquots were stored in a light-proof container at −20°C. Aliquots of mastoparan were dissolved in water at a concentration of 1 mM and stored at −20°C. Every experiment was performed with a fresh aliquot of ABA or mastoparan. Reagents for lipid extraction and subsequent analyses as well as the Silica 60 TLC plates (20 × 20 cm) were from Merck (Darmstadt, Germany).
Acknowledgments
This article is dedicated to Prof. J. Schell with thanks for his support.
We thank Barbara Eilts for technical assistance, Dr. Alan Musgrave for his support and interest, and the European Union biotechnology program for financial support (Contract No. BIO4-CT.960062).
References
- Altschul, S.F., Gish, W., Miller, W., Meyers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1996). Current Protocols in Molecular Biology. (New York: Wiley Interscience).
- Bartels, D., Schneider, K., Terstappen, G., Piatkowski, D., and Salamini, F. (1990). Molecular cloning of abscisic acid–modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta 181 27–34. [DOI] [PubMed] [Google Scholar]
- Bockel, C., Salamini, F., and Bartels, D. (1998). Isolation and characterization of genes expressed during early events of the dehydration process in the resurrection plant Craterostigma plantagineum. J. Plant Physiol. 152 158–168. [Google Scholar]
- Bonitta, D., and McCourt, P. (1998). Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 3 231–235. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2 48–54. [Google Scholar]
- Chapman, D. (1998). Phospholipase activity during plant growth and development and in response to environmental stress. Trends Plant Sci. 3 419–426. [Google Scholar]
- de Vrije, T., and Munnik, T. (1997). Activation of phospholipase D by calmodulin antagonists and mastoparan in carnation petals. J. Exp. Bot. 48 1631–1637. [Google Scholar]
- Dyer, J.H., Ryu, S.B., and Wang, X. (1994). Multiple forms of phospholipase D following germination and during leaf development of castor bean. Plant Physiol. 105 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, D. (1996). Phosphatidic acid: A lipid messenger involved in intracellular and extracellular signaling. Cell. Signaling 8 341–347. [DOI] [PubMed] [Google Scholar]
- English, D., Cui, Y., and Siddiqui, R.A. (1996). Messenger functions of phosphatidic acid. Chem. Phys. Lipids 80 117–132. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132 6–13. [DOI] [PubMed] [Google Scholar]
- Frank, W., Phillips, J., Salamini, F., and Bartels, D. (1998). Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain–leucine zipper proteins. Plant J. 15 413–421. [DOI] [PubMed] [Google Scholar]
- Furini, A., Koncz, C., Salamini, F., and Bartels, D. (1997). High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. EMBO J. 16 3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaff, D.F. (1971). Desiccation tolerant flowering plants in southern Africa. Science 174 1033–1034. [DOI] [PubMed] [Google Scholar]
- Gerlach, W.L., and Bedbrook, J.R. (1979). Cloning and characterisation of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T., Ohto, C., Mizoguchi, T., and Shinozaki, K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt-stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92 3903–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid–dependent and abscisic acid–independent pathways. Plant Cell 9 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, S., Suh, S., Seju, K., Crain, R.C., Kwak, J.M., Nam, H.G., and Lee, Y. (1997). Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 12 547–556. [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 199–222. [DOI] [PubMed] [Google Scholar]
- Liang, P., and Pardee, A.B. (1992). Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257 967–971. [DOI] [PubMed] [Google Scholar]
- Liscovitch, M., and Cantley, L.C. (1994). Lipid second messengers. Cell 77 329–334. [DOI] [PubMed] [Google Scholar]
- Liscovitch, M., and Cantley, L.C. (1995). Signal transduction and membrane traffic: The PITP/phosphoinositide connection. Cell 81 659–662. [DOI] [PubMed] [Google Scholar]
- Mikami, K., Katagiri, T., Iuchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 15 563–568. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Arisz, S.A., de Vrije, T., and Musgrave, A. (1995). G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7 2197–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik, T., de Vrije, T., Irvine, R.F., and Musgrave, A. (1996). Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271 15708–15715. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Irvine, R.F., and Musgrave, A. (1998. a). Phospholipid signaling in plants. Biochim. Biophys. Acta 1389 222–272. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Vanhimbergen, J.A.J., Terriet, B., Braun, F.J., Irvine, R.F., Vandenende, H., and Musgrave, A. (1998. b). Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipases C and D in Chlamydomonas cells treated with non-permeabilizing concentrations of mastoparan. Planta 207 133–145. [Google Scholar]
- Paliyath, G., and Droillard, M.J. (1992). The mechanisms of membrane deterioration and disassembly during senescence. Plant Biochem. Physiol. 30 789–812. [Google Scholar]
- Park, C., Park, M.H., and Chae, Q. (1996). Identification and characterization of phytochrome-regulated phospholipase D in oat cells (Avena sativa L.). J. Biochem. Mol. Biol. 29 535–539. [Google Scholar]
- Piatkowski, D., Schneider, K., Bartels, D., and Salamini, F. (1990). Characterization of five abscisic acid–responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol. 94 1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P., and Parker, P.J. (1996). Extending the C2 domain family: C2s in PKCs δ, ε, η, ϑ, phospholipases, GAPs and perforin. Protein Sci. 5 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, S., and Gilroy, S. (1998). Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 95 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, E.F., Dannelly, H.K., Malloy, P.J., and Reeves, H.C. (1987). Rapid isoelectric focussing in a vertical polyacrylamide minigel system. Anal. Biochem. 167 290–294. [DOI] [PubMed] [Google Scholar]
- Ryu, S.B., and Wang, X. (1995). Expression of phospholipase D during castor bean leaf senescence. Plant Physiol. 108 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, S.B., and Wang, X. (1996). Activation of phospholipase D and the possible mechanisms of activation in wound-induced lipid hydrolysis in castor leaves. Biochim. Biophys. Acta 1303 243–250. [DOI] [PubMed] [Google Scholar]
- Ryu, S.B., Zheng, L., and Wang, X. (1996). Changes in phospholipase D expression in soybeans during seed development and germination. J. Am. Oil Chem. Soc. 73 1171–1176. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schmidt, J., John, M., Wieneke, U., Krüssmann, H.-D., and Schell, J. (1986). Expression of the nodulation gene nodA in Rhizobium meliloti and localization of the gene product in the cytosol. Proc. Natl. Acad. Sci. USA 83 9581–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, K., Wells, B., Schmelzer, E., Salamini, F., and Bartels, D. (1993). Desiccation leads to the rapid accumulation of both cytosolic and chloroplastic proteins in the resurrection plant Craterostigma plantagineum Hochst. Planta 189 120–131. [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R.B., Davletov, B.A., Berghuis, A.M., Sudhof, T.C., and Sprang, S.R. (1995). Structure of the first C2 domain of synaptotagmin: A novel Ca2+/phospholipid-binding fold. Cell 80 929–938. [DOI] [PubMed] [Google Scholar]
- Smith, D.B., and Johnson, K.S. (1988). Single step purification of polypeptides expressed in Escherichia coli as fusion with glutathione S-transferase. Gene 67 31–40. [DOI] [PubMed] [Google Scholar]
- Ueki, J., Morioka, S., Komari, T., and Kumashiro, T. (1995). Purification and characterization of phospholipase D (PLD) from rice (Oryza sativa L.) and cloning of cDNA for PLD from rice and maize (Zea mays L.). Plant Cell Physiol. 36 903–914. [DOI] [PubMed] [Google Scholar]
- Wang, X. (1997). Molecular analysis of phospholipase D. Trends Plant Sci. 2 261–266. [Google Scholar]
- Williams, R.L., and Katan, M. (1996). Structural views of phosphoinositide-specific phospholipase C: Signalling the way ahead. Structure 4 1387–1394. [DOI] [PubMed] [Google Scholar]
- Young, S., Wang, X., and Leach, J.E. (1996). Changes in the plasma membrane distribution of rice phospholipase D during resistant interactions with Xanthomonas oryzae pv oryzae. Plant Cell 8 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]