Abstract
In the green alga Chlamydomonas reinhardtii, the ClpP protease is encoded by an essential chloroplast gene. Mutating its AUG translation initiation codon to AUU reduced ClpP accumulation to 25 to 45% of that of the wild type. Both the mature protein and the putative precursor containing its insertion sequence were present in reduced amounts. Attenuation of ClpP did not affect growth rates under normal conditions but restricted the ability of the cells to adapt to elevated CO2 levels. It also affected the rate of degradation of the cytochrome b6f complex of the thylakoid membrane in two experimental situations: (1) during nitrogen starvation, and (2) in mutants deficient in the Rieske iron–sulfur protein. The ClpP level also controls the steady state accumulation of a mutated version of the Rieske protein. In contrast, attenuation of ClpP did not rescue the fully unassembled subunits in other cytochrome b6f mutants. We conclude that proteolytic disposal of fully or partially assembled cytochrome b6f is controlled by the Clp protease.
INTRODUCTION
Most intracellular proteolysis is performed by processive ATP-dependent proteases. These enzymes, which play a critical role in the removal of damaged proteins and in the fine control of some key cellular components, combine a peptidase and a chaperone activity (Schirmer et al., 1996; Suzuki et al., 1997). In Escherichia coli, five enzymes account for the bulk of the ATP-dependent proteolytic activity within the cell: the cytoplasmic ClpAP, ClpXP, Lon, and HslUV proteases and membrane-bound FtsH (reviewed in Gottesman, 1996). Although very diverse in sequence and enzymatic mechanism, they share a unique property that explains both their restricted specificity and their processive character: the recognition of protein substrates and the presentation of these substrates to the proteolytic active site are performed by an ATP-dependent molecular chaperone that is associated with the protease. The chaperone and protease activities can be carried by two distinct domains of the same protein (e.g., with Lon and FtsH) or by different subunits (e.g., with HslUV, ClpAP, and ClpXP). The proteolytic components HslV and ClpP are, respectively, threonine and serine proteases of completely different origin, but their associated regulatory subunits (HslU and ClpA or ClpX) are related to each other and to eukaryotic Hsp100 chaperones (Schirmer et al., 1996). The chaperone specifically recognizes the protein substrates and either remodels/refolds them or unfolds and presents them to the protease (Schirmer et al., 1996; Gottesman et al., 1997). The eukaryotic proteasome functions in a similar manner, with a proteolytic 20S core and a 19S particle involved in substrate recognition and presentation.
The elucidation of the structure of several ATP-dependent proteases has unraveled interesting common traits in their structure and mechanism (Lowe et al., 1995; Baumeister and Lupas, 1997; Bochtler et al., 1997; Groll et al., 1997; Wang et al., 1997), and the term self-compartmentalizing proteases has been coined for these barrel-shaped assemblies (Lupas et al., 1997). The central proteolytic core is made of two to four stacked rings of either six (HslV) or seven subunits (the proteasome, ClpP). This core is capped on each side by six-membered rings of chaperone subunits (Kessel et al., 1995; Grimaud et al., 1998). In all cases, the proteolytic subunits are arranged to form a proteolytic chamber separated from the cytoplasm, with the active sites oriented inward. Access to this chamber is through a narrow axial pore wide enough to give entry only to small peptides or polypeptides in an extended conformation. The chaperones restrict access to the entry pore and are believed to feed the unfolded polypeptide substrates into the chamber.
In view of the endosymbiontic origin of chloroplasts and mitochondria, it is not surprising that their proteolytic machineries are related in part to that of eubacteria (reviewed in Adam, 1996). In the chloroplast, ATP-dependent proteolysis is the major route for degradation of nonassembled polypeptides (Liu and Jagendorf, 1984, 1985), and homologs of ClpP and FtsH have been localized to the stroma and thylakoid membrane, respectively (Shanklin et al., 1995; Lindahl et al., 1996). A clpP gene has been found in all chloroplastic genomes sequenced, with the exception of those of Euglena and Odontella (Gray et al., 1990; Hallick et al., 1993; Kowallik et al., 1995). In addition, several chloroplast-targeted members of the Hsp100 family, belonging to the ClpX, ClpC, or ClpD subfamilies (Schirmer et al., 1996), are present in the nuclear genome of plants (Gottesman et al., 1990; Kiyosue et al., 1993). ClpC can substitute for ClpA in a protease assay (Shanklin et al., 1995) and associates with ClpP in in organello import experiments (Sokolenko et al., 1998). However, its in vivo function is unclear, because it has also been found as a component of the chloroplastic import machinery (Nielsen et al., 1997). In the absence of a clear identification of the chaperone partner of ClpP in chloroplasts, we refer to a protease formed around ClpP as a Clp protease.
Despite our growing knowledge of ATP-dependent chloroplast proteases, none of them has been definitely implicated in any of the protein degradation processes that have been identified in vivo in the chloroplast compartment. The goal of our work is to understand how proteases contribute to the biogenesis and degradation of thylakoid membrane proteins. Most such proteins are large oligomeric complexes in which polypeptide subunits synthesized in the chloroplast and in the cytoplasm are associated with cofactors and pigments. Proteolysis plays a major role in their biogenesis by removing those subunits that are incorrectly folded or made in excess of their assembly partners (reviewed in Wollman et al., 1999). This process is best studied in photosynthesis-deficient mutants affected in the biogenesis of a single subunit in a protein complex. These studies have been conducted mostly in the unicellular green alga Chlamydomonas reinhardtii (reviewed in Rochaix et al., 1998; de Vitry and Vallon, 1999) and more recently in higher plants as well (Meurer et al., 1998; Monde et al., 2000). The general picture that emerges is that subunits synthesized in the absence of their cofactors or assembly partners are rapidly degraded and fail to accumulate.
Changes in the environment also can induce the cell to specifically downregulate the amount of a protein complex in the membrane. For instance, transfer of plants from low to high light is accompanied by a large reduction in light-harvesting complex II content due to induction of an acclimative proteolytic system (Yang et al., 1998). Similarly, transfer of Chlamydomonas cells to culture medium that lacks nitrogen or sulfur leads to rapid loss of cytochrome b6f or photosystem II, respectively (Bulté and Wollman, 1992; Davies et al., 1996). During photoinhibition, probably the best-studied situation of stress-induced proteolysis, the photosystem II reaction center and principally its D1 subunit are subject to intense proteolytic degradation (Ohad et al., 1994).
In this study, we demonstrate that the rate of degradation of the cytochrome b6f complex in Chlamydomonas is controlled in vivo by the level of expression of the chloroplast clpP gene. The Chlamydomonas clpP gene lies downstream of the petB gene that encodes cytochrome b6. Transformants with an inactivated clpP gene can be recovered only in a heteroplasmic state, suggesting that the gene is essential for cell viability (Huang et al., 1994). For this reason, we have sought to reduce but not abolish its expression by a strategy that we hereafter refer to as translational attenuation. Based on the study by Stern and colleagues (Chen et al., 1993, 1995) of the initiation of translation in the Chlamydomonas petA and petD genes, coding for cytochrome f and subunit IV of the cytochrome b6f complex, respectively, we have mutated the AUG initiation codon of clpP to AUU. We found that the ensuing reduction in ClpP amount retards proteolytic removal of the cytochrome b6f complex in situations in which it is known to be actively degraded. Thus, we provide definitive evidence for a physiological role of a chloroplast-located protease in the degradation of a thylakoid membrane protein.
RESULTS
Generation of clpP Initiation Codon Mutants Using Chloroplast Transformation
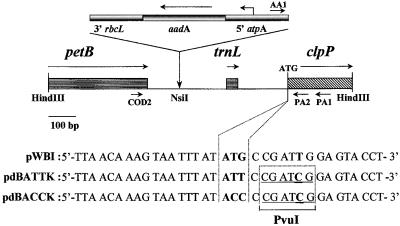
A map of the petB–trnL–clpP region of the chloroplast genome is shown in Figure 1. Chlamydomonas strains carrying the mutations AUU and ACC at the initiation codon position of the clpP gene were created by particle bombardment. For screening purposes, we also introduced a restriction fragment length polymorphism marker (a PvuI restriction site) by a silent T→C change at the last position of the third codon. For selection of transformants, the aadA spectinomycin and streptomycin resistance cassette was introduced upstream of clpP. As a control, we also constructed strain BI carrying the resistance cassette linked to a wild-type clpP gene. Putative transformants were analyzed by polymerase chain reaction (PCR) and restriction analysis. Two pairs of primers were used (Figure 1) that amplify the wild-type and mutant copies, respectively (see Methods). Transformants obtained with the plasmid carrying the AUG→AUU mutation initially presented both types of fragments, and four rounds of single-colony isolation were necessary to obtain homoplasmic strains containing the aadA cassette and the PvuI restriction site. In one of these strains, the PCR product amplified using the AA1-PA1 primer pair was sequenced to verify the presence of the AUU mutation. This strain was called clpP-AUU.
Figure 1.
Map of the Inserts in the Plasmids Used for Mutagenesis.
The positions of the genes and their translation direction are indicated, together with those of the primers used for PCR. The sequence surrounding the translation initiation site is indicated for the pWBI (control) and pdBATTK and pdBACCK (mutant) plasmids. Mutations introduced are indicated in bold, and the PvuI site is underlined.
In contrast, we were unable to recover a homoplasmic strain carrying the clpP-ACC mutation. After nine or ten rounds of single-colony replication on selective medium (containing Tris-acetate [TAP] and spectinomycin), the transformants became homoplasmic for the aadA cassette. However, the PvuI restriction site was no longer present in those strains (five independent transformants), and we assume that the clpP-ACC mutation had been lost as well by recombination with the wild-type copy. We interpret this failure to isolate homoplasmic clpP-ACC transformants as an indication that the level of translational attenuation caused by the clpP-ACC mutation was too extensive to preserve cell viability. Indeed, Chen et al. (1993)(1995) reported that the petA-ACC mutant expressed only 1 to 2% of the wild-type content in cytochrome f. Thus, our results are in agreement with the earlier conclusion of Huang et al. (1994) that the chloroplast clpP gene is an essential gene in Chlamydomonas.
The Amount of Two Forms of ClpP Is Reduced in the clpP-AUU Strain
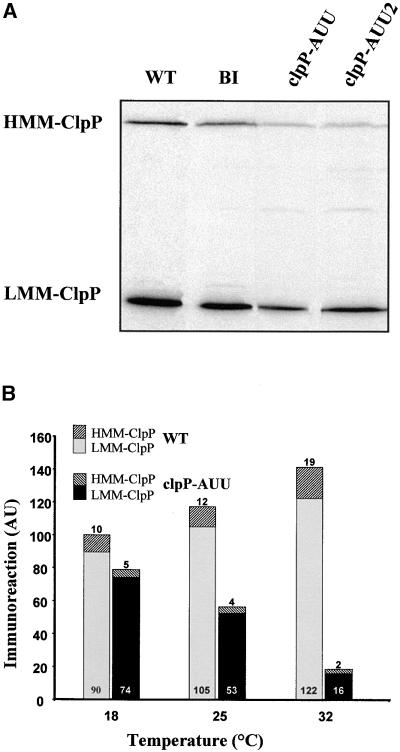
The protein sequence deduced from the C. reinhardtii clpP gene (Huang et al., 1994) is considerably longer than are ClpP sequences found in other organisms (524 residues instead of the usual 200 or so). This is due to the presence of an insertion sequence (IS1) that is not spliced out from the mRNA. The insertion conserves the correct reading frame for ClpP, and it has been proposed that IS1 is translated together with the ClpP sequence, then eliminated by a mechanism similar to the splicing of inteins (Huang et al., 1994). We raised polyclonal antibodies against two overexpressed versions of full-length ClpP either alone or fused with thioredoxin. Both antibodies recognized two bands in Chlamydomonas (Figure 2A), a high molecular mass band (HMM-ClpP), presumably corresponding to the full-size translation product, which has a predicted molecular mass of 59.5 kD, and a low molecular mass band of 25 kD (LMM-ClpP), which we identify as the mature ClpP protein (26.8 kD calculated molecular mass, assuming the sequence predicted in Huang et al., 1994). In the wild-type and BI strains, HMM-ClpP represented 5 to 10% of the total immunoreactivity.
Figure 2.
Attenuation of ClpP by the clpP-AUU Mutation.
(A) Immunoblot of total cell proteins taken from exponential-phase TAP cultures of the wild-type (WT), BI, clpP-AUU, and clpP-AUU2 (an independent transformant carrying the clpP-AUU mutation) strains. (B) Temperature-dependent attenuation of ClpP. Samples were taken from cultures of the wild type (WT) or clpP-AUU in exponential phase and grown at 18, 25, or 32°C. The amount of LMM-ClpP and HMM-ClpP was determined by immunoblotting with a ClpP antibody and PhosphorImager analysis. The value 100 represents the total immunoreactivity in the wild type at 18°C. AU, arbitrary units.
The clpP-AUU mutation reduced accumulation of both forms of ClpP (Figure 2A). In two independent clpP-AUU–expressing transformants grown in mixotrophic or phototrophic conditions at 25°C, the steady state levels of the HMM-ClpP and LMM-ClpP ranged between 20 and 25% and 25 and 45% of those in the BI strain, respectively. The ClpP levels in the BI strain were similar to those in the wild type, indicating that introduction of the aadA cassette itself had no deleterious effect on the expression of clpP. In sexual crosses involving clpP-AUU strains, all the progeny clones that we have analyzed carried the restriction fragment length polymorphism in clpP and the aadA cassette, as expected from the uniparental inheritance of chloroplast genes. In all cases, we observed attenuation of both forms of ClpP. These observations strongly argue that both the immunoreactive bands correspond to genuine products of the single chloroplast clpP gene that we have mutated.
In agreement with the reports by Chen et al. (1993)(1995) that translational attenuation in petA-AUU– and petD-AUU–expressing strains was strongly temperature dependent, we observed that attenuation of ClpP in the clpP-AUU strain was more pronounced at higher temperatures. In the typical experiment shown in Figure 2B, accumulation of LMM-ClpP in the clpP-AUU strain was 82% of that in the wild type at 18°C, 50% at 25°C, and 13% at 32°C. The same temperature effect was observed for HMM-ClpP, being 50% of that in the wild type at 18°C, 33% at 25°C, and 11% at 32°C. We note that HMM-ClpP amounts were consistently more attenuated than were those of LMM-ClpP in the mutant.
Growth Phenotype of the clpP-AUU Strain
Because the chloroplast clpP gene is essential for cell viability (Huang et al., 1994; this manuscript), we first looked for possible growth defects in the clpP-AUU strain. We observed no difference in growth rate among the clpP-AUU, the wild-type, or the BI strains in standard conditions (TAP or minimal medium, light ⩽ 60 μE m−2 sec−1) at 18, 25, or 32°C. However, increasing light intensity had a marked inhibitory effect on the clpP-AUU strain when it was combined with a change in culture conditions. In one experiment, cells grown in liquid TAP medium at 60 μE m−2 sec−1 were plated to single colonies onto solid TAP medium at 200 μE m−2 sec−1. Compared with the control wild-type or BI strains, the clpP-AUU mutants (two independent transformants and two clpP-AUU segregants from genetic crosses) showed a marked lag (∼3 to 5 days) in colony formation, after which colonies started to grow normally. This lag was not observed at fluence rates 40 μE m−2 sec−1 or lower or when cells were plated on minimal medium.
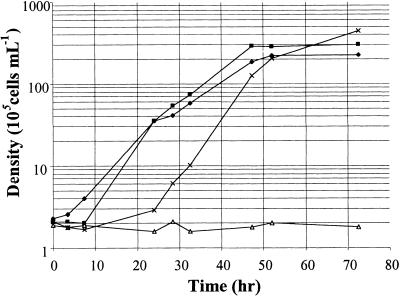
We also observed a pronounced growth defect in CO2-enriched medium. The clpP-AUU strain grew well in TAP medium at 200 μE m−2 sec−1, unless CO2 was bubbled into the culture (Figure 3). The growth defect was more pronounced in TAP than in minimal medium and was not observed in low light (30 μE m−2 sec−1). We note that a pronounced lag was observed in the control strain upon transfer to high CO2 (Figure 3). This observation suggests that a sharp increase in CO2 concentration normally causes a shock to the cells, from which they recover in a ClpP-dependent manner. In conclusion, ClpP attenuation appears to thwart the ability of the cell to adapt to high-light conditions when combined with other major changes in their environmental conditions, such as elevation of CO2 concentration and transition from liquid to plate culture.
Figure 3.
Growth Curves of the BI and clpP-AUU Strains.
Cells grown in TAP medium at 60 μE m−2 sec−1 were diluted at 2 × 105 cells per mL in fresh medium and placed at 320 μE m−2 sec−1 with (clpP-AUU, open triangles; BI, X's) or without (clpP-AUU, filled diamonds; BI, filled squares) CO2 bubbling (5% in air).
Effect of ClpP Attenuation in Mutants Fully Defective in Cytochrome b6f Assembly
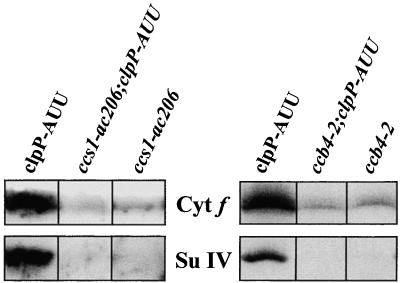
Earlier comparison of various mutants defective in cytochrome b6f function has shown that cytochrome b6, subunit IV, and the Rieske protein are rapidly degraded when they cannot assemble with cytochrome f (Kuras and Wollman, 1994). To analyze a possible role of ClpP in this degradation process, we crossed the clpP-AUU chloroplast mutation into nuclear contexts that impair synthesis of holocytochrome f or holocytochrome b6. The ccs1-ac206 mutation prevents heme binding to cytochrome f (Xie et al., 1998), whereas the ccb4-2 mutation (Kuras et al., 1997) blocks the conversion of apocytochrome to holocytochrome b6. Both mutations cause an almost complete absence of all cytochrome b6f subunits by preventing assembly at an early stage. As determined from immunoblotting analysis (Figure 4) and 3,3′,5,5′-tetramethyl benzidin (TMBZ) staining (not shown), the clpP-AUU mutation did not lead to an increase in the level of the unstable subunits of cytochrome b6f either in a ccs1-ac206 or in a ccb4-2 nuclear background. This was true during the exponential phase of growth as well as during the stationary phase. We conclude that the degradation rate of the highly unstable nonassembled cytochrome b6f subunits in these mutants is not limited by ClpP, at least at the level of ClpP attenuation attained in the present study.
Figure 4.
Immunoblot Analysis of ccs1-ac206 clpP-AUU and ccb4-2 clpP-AUU Double Mutants.
Cells were harvested during the exponential phase of growth (0.5 to 1 × 106 cells per mL). Blots were reacted with antibodies to cytochrome (Cyt) f and subunit (Su) IV.
ClpP Is Involved in the Degradation of the Cytochrome b6f Complex in Mutants of the Rieske Protein
In contrast to the above-described situations, in which mutants affected in one subunit of the cytochrome b6f complex show an almost complete deficiency in all the other subunits, mutants unable to assemble the Rieske iron–sulfur protein still accumulate the cytochrome moiety of the protein complex (Lemaire et al., 1986). This subcomplex sediments in a gradient as a dimer, only slightly lighter than the wild-type complex (de Vitry et al., 1999). The level of accumulation of this subcomplex is largely dependent on growth phase, ranging from 40 to 60% of the wild type during early exponential growth (up to 5 × 105 cells per mL) to a complete loss in stationary phase. To investigate a possible role of ClpP in the turnover of this Rieske-less subcomplex, we have analyzed its accumulation level and degradation rate in petC mutants carrying the clpP-AUU mutation.
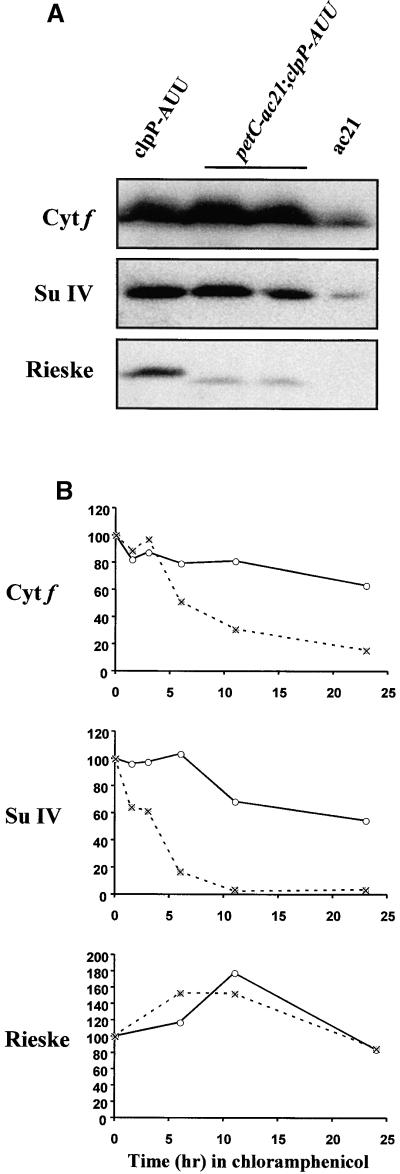
Figure 5A shows an immunoblot analysis of the ac21 strain, harboring a mutation in the nuclear PETC gene that causes replacement of Arg 163 by Trp (de Vitry et al., 1998). In accordance with previous studies (Lemaire et al., 1986), we found that ac21 contained significant although reduced amounts of the chloroplast-encoded cytochrome b6f subunits (cytochromes f and b6, subunit IV). The Rieske protein was not detected in ac21 in this experiment, but we and others (de Vitry et al., 1998) have detected it as a faint substochiometric band migrating slightly ahead of the wild-type protein. In double mutants carrying petC-ac21 together with the clpP-AUU mutation, the level of accumulation of all cytochrome b6f subunits was markedly increased (Figure 5A). This was true not only for the cytochrome subcomplex but also for the mutant Rieske protein. It is worth noting that the mutant Rieske protein still remained substoichiometric with respect to the cytochrome moiety of the protein complex (in exponential phase 12 to 20% of the wild type versus 45 to 115% for cytochrome f and subunit IV).
Figure 5.
Analysis of petC-ac21 clpP-AUU Double Mutants.
(A) Immunoblot of total cell proteins from parental strains ac21 and clpP-AUU, and two double mutant progeny from the cross, harvested during exponential phase. (B) Stability of cytochrome f, subunit IV, and the Rieske protein in the ac21 (X's) and petC-ac21 clpP-AUU mutant (open circles) strains. Chloramphenicol was added at time 0 in early exponential- phase cultures. Immunoreactivity is plotted as the percentage of the initial level in each culture. Cyt f, cytochrome f; Su IV, subunit IV.
To measure directly the degradation rate of chloroplast-encoded cytochrome b6f subunits in the ac21 strain and the petC-ac21 clpP-AUU double mutant without interference from de novo synthesis, a protein chase experiment was performed in which chloramphenicol (100 μg/mL) was used to block chloroplast translation. Under these conditions, degradation of preexisting proteins can be followed directly by immunoblotting (Figure 5B). The approximate half-life of cytochrome f and subunit IV increased from 7 and 4 hr, respectively, in ac21 to >25 hr in the ClpP-attenuated strain. Therefore, we conclude that the increased accumulation of the chloroplast-encoded cytochrome b6f subunits in the petC-ac21 clpP-AUU double mutant is indeed due to a decreased rate of degradation. Note that the addition of chloramphenicol did not markedly affect the amount of mutant Rieske protein in either strain. This is as expected for a nuclear-encoded protein, the synthesis of which is not prevented by chloramphenicol.
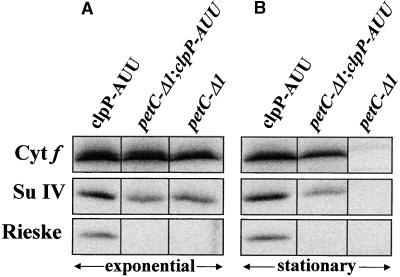
Because de Vitry et al. (1999) have shown that at least a portion of the mutant Rieske protein in ac21 is associated with the cytochrome moiety, we wondered whether the effect of the clpP-AUU mutation was dependent on the presence of a mutant Rieske protein. Therefore, we analyzed another petC mutant, petC-Δ1, which harbors a frameshift in the coding region leading to a premature stop codon (de Vitry et al., 1998). This mutant fails to accumulate any immunoreactive form of Rieske protein but still accumulates in the exponential phase of growth a cytochrome moiety with characteristics similar to that of ac21 (Figure 6A). As in ac21, the subcomplex was lost when the cells entered stationary phase (Figure 6B). When the clpP-AUU mutation was combined with petC-Δ1, we observed an increased accumulation of the partially assembled complex (Figure 6). In this particular experiment, because of the high level of accumulation of the cytochrome moiety in the petC-Δ1 mutant in exponential phase, the protection effect of the clpP-AUU mutation is best seen in stationary phase. Chloramphenicol chase experiments with these strains confirmed the decreased turnover of the cytochrome b6f subunits in the petC-Δ1 clpP-AUU double mutant (data not shown).
Figure 6.
Analysis of the petC-Δ1 clpP-AUU Double Mutant.
Cells were harvested during the exponential phase of growth (A) or in stationary phase, 3 days later (B). Blots were reacted with antibodies to cytochrome f (Cyt f), subunit IV (Su IV), and the Rieske protein.
Therefore, we conclude that attenuation of ClpP has a marked protection effect against degradation on the cytochrome moiety of cytochrome b6f in petC mutants. In the ac21 strain, this effect is distinct from the protective effect on the mutant Rieske protein itself.
ClpP Is Involved in the Degradation of the Assembled Cytochrome b6f Complex during Nitrogen Starvation
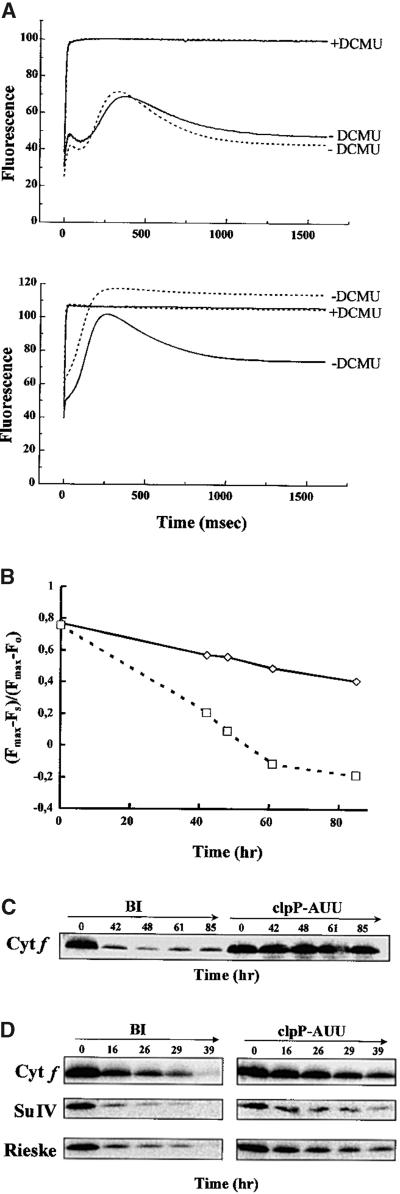
Because the stabilization effect of ClpP attenuation appeared to be restricted to those cytochrome b6f mutants in which the protein complex was partially assembled, we wondered whether ClpP also would control the degradation of fully assembled cytochrome b6f. We have reported previously that a selective degradation of the cytochrome b6f complex, leading to photosynthesis inactivation, takes place when Chlamydomonas is starved for nitrogen (Bulté and Wollman, 1992). Thus, we placed the clpP-AUU and the BI control strains in nitrogen-free medium and followed inactivation of photosynthetic electron transport by in vivo measurement of the chlorophyll fluorescence induction kinetics (Figure 7A). The fluorescence yield of photosystem II increases when the primary quinone acceptor QA is placed in a reduced state. In the wild type, after an initial rise from the dark-adapted fluorescence level (F0), fluorescence declines to a stationary level (Fs) due to plastoquinol reoxidation by the cytochrome b6f complex. In contrast, any block in electron flow between photosystem II and photosystem I will lead to a rise in the fluorescence level as cells are illuminated, eventually reaching the maximal fluorescence (Fmax) level when the acceptor side of photosystem II is fully reduced. Experimentally, the value of Fmax is obtained by illuminating cells in the presence of DCMU, an inhibitor of the photosystem II acceptor side. The difference between Fs and Fmax gives a relative measure of the electron transfer capacity beyond the plastoquinone pool. In TAP-grown cells, the fluorescence parameters in the BI and clpP-AUU strains were similar to those in the wild type. After 61 hr in nitrogen-free medium, the BI strain had lost photosynthetic electron transfer: the parameter (Fmax − Fs)/(Fmax − F0) even became negative (Figure 7B) due to the dequenching effect of plastoquinol (Vernotte et al., 1979). The loss of cytochrome b6f was confirmed by immunoblotting with specific antibodies (Figure 7C). In contrast, the clpP-AUU strain, while still showing some changes in the shape of the fluorescence induction curve, maintained a fair level of electron transfer activity during the 85 hr of the experiment (Figures 7A and 7B). Accordingly, the levels of cytochrome b6f subunits, determined by immunoblotting and TMBZ staining, decreased much less rapidly in clpP-AUU compared with BI (Figure 7C).
Figure 7.
The clpP-AUU Mutation Retards Loss of Cytochrome b6f in Nitrogen Starvation.
(A) Chlorophyll fluorescence induction kinetics, in the presence (+) or absence (−) of DCMU, of TAP-grown cells (top) and cells starved for nitrogen for 61 hr (bottom). Solid lines, clpP-AUU; dotted lines, BI. (B) Evolution of the (Fmax − Fs)/(Fmax − F0) parameter during nitrogen starvation. Solid line, clpP-AUU; dotted line, BI. (C) Immunoblot of total cell proteins as given for (A) and (B), reacted with an antibody to cytochrome f (Cyt f). (D) Protein chase experiment under nitrogen starvation conditions with antibodies to cytochrome f (Cyt f), subunit IV (Su IV), and the Rieske protein. Chloramphenicol was added at time 0.
Because the loss of cytochrome b6f in these conditions is not due to a specific decrease in translation but rather to an increased proteolysis rate (Bulté and Wollman, 1992), our observations suggested that attenuation of ClpP was delaying degradation of cytochrome b6f. This was further confirmed by a chloramphenicol chase experiment (Figure 7D). As noted before (Bulté and Wollman, 1992), addition of chloramphenicol slightly accelerated disappearance of cytochrome b6f, and this was observed in both strains. The half-life of cytochrome b6f subunits almost doubled in the clpP-AUU strain compared with BI (21 versus 12 hr for cytochrome f, 14 versus 8 hr for subunit IV, and 14 versus 9 hr for the Rieske protein). Moreover, the amount of nuclear-encoded Rieske protein decreased at the same rate as the chloroplast-encoded subunits, even though its synthesis was not altered by the treatment. Therefore, the proteolytic susceptibility of the complex appears to be controlled as a whole, with little chance for unassembled subunits to accumulate to any significant degree. These experiments show that ClpP is involved, either directly or indirectly, in the degradation of cytochrome b6f complex during nitrogen starvation. The amount of ClpP did not change significantly in either strain during the starvation period, even in the presence of chloramphenicol (data not shown).
DISCUSSION
Previous studies from our laboratory (Lemaire et al., 1986; Kuras and Wollman, 1994; Wollman, 1998) have led to the description of cytochrome b6f assembly as a multistep process. As is the case for photosystem II (de Vitry et al., 1989), the intermediate states of assembly are revealed only when frozen by mutational blocks, but the model is believed to apply to the wild type as well. The initial stage of stabilization involves post-translational modifications of the subunits: apocytochromes f and b6 will remain in a highly protease-sensitive state until heme binding is achieved (Kuras et al., 1995, 1997). Whereas holocytochrome f can accumulate to some extent in the absence of its assembly partners, stabilization of cytochrome b6 requires interaction first with subunit IV and then with cytochrome f. The state of assembly of the complex can be assessed indirectly by monitoring the rate of translation of cytochrome f, which is downregulated whenever assembly is prevented (Choquet et al., 1998). Once formed, the cytochrome moiety is further stabilized by binding of the Rieske protein (Lemaire et al., 1986). Both the Rieske protein and the PetL subunit contribute to the stability of the functional dimer (Breyton et al., 1997). The fully assembled complex is extremely long-lived, possessing a half-life in the order of days. This stability can be compromised when the cells are placed in adverse physiological conditions, such as nitrogen starvation (Bulté and Wollman, 1992).
The goal of the present study was to examine whether a Clp protease plays a role in the biogenesis of cytochrome b6f at any of the stages described above. Due to the essential nature of the clpP gene in Chlamydomonas, we had to devise a method for reducing its expression level while still retaining enough protein for cell viability. Following the strategy of Chen et al. (1993)(1995), we have designed mutations in the translation initiation codon of the clpP gene, which proved efficient in reducing ClpP protein accumulation. The attenuated clpP-AUU mutant was viable and appeared normal, except for a reduced ability to recover from abrupt changes in growth condition in mixotrophic cultures. We have used it to investigate a possible role of ClpP in the degradation of the various intermediate forms found along the cytochrome b6f assembly pathway.
When analyzing mutations that block assembly at an early stage (heme binding to cytochromes b6 or f ), we have found no evidence for a role of ClpP in the degradation of the unassembled subunits. In these genetic contexts, neither the defective subunits nor their assembly partners were stabilized by the clpP-AUU mutation. This can be taken as evidence that ClpP is not involved in their proteolytic removal and that other proteases actually fulfill this role. The FtsH and DegP chloroplast homologs (Lindahl et al., 1996; Itzhaki et al., 1998) are obvious candidates, but any of the numerous unidentified SDS-stable proteases in the thylakoid membrane (Sokolenko et al., 1997) could be involved as well. It should be noted, however, that 25 to 40% of chloroplast ClpP was still present in our attenuated strains so that a role for ClpP in the proteolytic disposal of these unstable subunits cannot be ruled out. The proteolytic system also may be partly redundant, with several proteases being able to replace each other.
In contrast, our experiments with petC mutants and nitrogen-starved cells indicate that the stability of the partially or fully assembled forms of cytochrome b6f is strongly dependent on the level of ClpP in the chloroplast. ClpP may participate directly in the degradation mechanism of assembled cytochrome b6f complexes but it may also have an indirect effect: a general increase in misfolded proteins in the chloroplast upon ClpP attenuation may saturate the proteolytic pathways that normally carry out degradation of cytochrome b6f. Whatever the actual mechanism, the present study clearly shows that a Clp protease controls the rate of degradation of a transmembrane protein in the chloroplast.
As in other systems, ClpP in Chlamydomonas fractionates almost exclusively as a soluble protein (data not shown). It is somewhat unexpected that a soluble Clp protease would control the lifetime of a transmembrane thylakoid protein. In mitochondria, proteolytic disposal of the nonassembled subunits of the respiratory complexes of the inner membrane has been attributed to two membrane-bound proteases homologous to FtsH, Yme1p, and the Afg3p-Rca1p complex (Arlt et al., 1996; Weber et al., 1996). FtsH also degrades the polytopic membrane protein SecY in E. coli (Akiyama et al., 1996). However, the involvement of soluble proteases in the degradation of membrane proteins is not unprecedented. Whereas most of the known ClpAP or ClpXP substrates in E. coli are soluble proteins (reviewed in Gottesman, 1996), ClpP has been implicated in the degradation of a set of membrane proteins induced by carbon starvation (Damerau and St. John, 1993). More recently, the 26S proteasome has been shown to degrade membrane proteins of the endoplasmic reticulum (Wiertz et al., 1996; Pilon et al., 1997; Yu et al., 1997; Mayer et al., 1998). The process involves their dislocation into the cytosol through the Sec61p channel, with the AAA chaperones of the proteasome providing the driving force for retrotransfer. Interestingly, a similar mechanism appears to be at play in the FtsH-mediated proteolysis of membrane proteins in E. coli (Kihara et al., 1999). In view of the functional and sequence similarity between the substrate binding domains of Hsp100 and AAA proteins (Smith et al., 1999), we speculate that Clp and other ATP-dependent proteases can use a mechanism of this type to degrade their membrane protein substrates.
For a multisubunit complex such as cytochrome b6f, numerous questions arise as to the molecular basis of protease–substrate interaction. Which particular subunit, if any, constitutes the primary site of attack by the protease? If degradation indeed proceeds in a processive manner, which end is degraded first: N-terminal, as in λO and RepA (Paetzel et al., 1998; Gonciarz-Swiatek et al., 1999), or C-terminal, as in the Mu transposase or SsrA-tagged proteins (Levchenko et al., 1997; Gottesman et al., 1998)? Exposure of hydrophobic sequences that are normally masked by folding or by interaction with assembly partners has often been invoked as a signal for proteolytic degradation (e.g., Levchenko et al., 1997). The N-terminal portion of the Rieske protein is normally exposed to the stromal side of the membrane: its absence in petC mutants might expose target sites for proteases on the other subunits either directly or by preventing dimerization. In the case of nitrogen starvation, a different mechanism must be at play; because all subunits are present under these conditions, we have to consider that a more subtle chemical or conformational change occurs in the complex due to nitrogen starvation that renders it a better substrate for the protease. Alternatively, because the ClpP level does not increase, a change in the specificity of the protease may be brought about by a modification of its chaperone partner(s).
In the petC mutants, we have observed that the clpP-AUU mutation, besides protecting from degradation the cytochrome moiety of the protein complex, also increased the accumulation of the mutant Rieske protein. This raises the possibility that the mutant protein is by itself a substrate for ClpP, independent of a stable association with the cytochrome moiety. Experiments are under way to test this hypothesis. Note that in isolated plant chloroplasts, most of the newly imported Rieske protein remains accessible on the stromal surface of the membrane, where it can be substrate for a metalloprotease, possibly FtsH (Ostersetzer and Adam, 1997).
Our understanding of the role of ClpP in Chlamydomonas will have to include a better description of its biogenesis. Most conspicuous is the presence in the ClpP coding sequence of the long IS1 (Huang et al., 1994). Our immunoblotting experiments revealed the presence of an immunoreactive polypeptide of exactly the predicted size for the complete reading frame. Furthermore, both this band and the 25-kD polypeptide that we identify as mature ClpP were attenuated in the clpP-AUU mutant. Because chloroplast genomes, including that of Chlamydomonas, contain a single clpP gene, we propose that the two immunoreactive forms are two different products of the same gene. Nuclear clpP genes have been described in higher plants (Schaller and Ryan, 1995; Sokolenko et al., 1998) and may exist in Chlamydomonas as well, but no cross-reaction has been observed with antibodies to the chloroplast-encoded ClpP (Sokolenko et al., 1998).
How the chloroplast clpP gene can generate the low molecular weight form is at present unknown. Huang et al. (1994) detected only one clpP transcript, containing the entire reading frame. As they originally pointed out, generation of mature ClpP could involve a protein splicing event whereby the intervening IS1 sequence would be removed while the N- and C-terminal sequences would be religated to form a single polypeptide chain. Protein splicing has been demonstrated for a host of intein-containing proteins found in all kingdoms of life (Perler et al., 1992; Perler, 1999). However, the C. reinhardtii IS1 does not appear to be a classical intein by several criteria. IS1 sequences are published for C. reinhardtii and C. eugametos, and similar sequences have been reported in the clpP genes of eight Chlamydomonas spp examined (Huang et al., 1994). Yet, our database searches using BLASTP and TBLASTN failed to identify similar sequences outside of the genus. In particular, the consensus motifs of classical inteins (LAGLI/DADG, C-terminal HN dipeptide, etc.; see Pietrokovski, 1994; Shao et al., 1995) were missing. The only shared sequence feature of IS1s and inteins is the presence of a nucleophilic side chain at the N-terminal positions of the spacer and C-terminal domains (S in IS1s, and C or S in inteins), thought to be required for the essential acyl rearrangements (Shao et al., 1996). In addition, splicing of inteins is usually a very efficient process that can occur in heterologous organisms, in heterologous genes, or even in vitro (Chong et al., 1998). In contrast, we have seen no conversion of HMM-ClpP into the LMM form in our chloramphenicol chase experiments (data not shown). This, together with the high abundance of the HMM form of ClpP (Figure 2), indicates that this splicing must either be very slow or be restricted to a narrow window of time after translation. In other words, if the IS1 sequence of C. reinhardtii is indeed spliced out post-translationally, this represents a completely novel category of inteins, probably exhibiting a unique splicing mechanism.
METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii wild type (WT11 mt+, derived from strain 137c) and photosynthetic transformants were grown on Tris-acetate (TAP) medium (Harris, 1989) at 25°C under 30 μE m−2 sec−1 continuous illumination, unless otherwise noted. Nonphotosynthetic strains, carrying mutations tbc1-F34 (Chua and Bennoun, 1975), ccs1-ac206 (Xie et al., 1998), ccb4-2 (Kuras et al., 1997), petC-ac21 (Levine and Smillie, 1962; de Vitry et al., 1998), or petC-Δ1 (de Vitry et al., 1998), were grown at 6 μE m−2 sec−1. The clpP-AUU mutation was combined with the photosynthesis mutations by sexual crossing (Harris, 1989). In each case, two independent double mutant segregants were analyzed. The presence of the mutated clpP gene and the reduced ClpP protein level was checked by polymerase chain reaction (PCR) and immunoblotting, respectively.
Oligonucleotides, Mutagenesis, and Plasmids
Plasmids that carry mutations of the clpP AUG initiation codon to AUU or ACC were constructed. In plasmid pdB1.7 (Buschlen et al., 1991), a HindIII fragment of C. reinhardtii chloroplast DNA encompassing the C-terminal coding region of petB, the trnL gene, and the clpP N-terminal coding region is cloned into the pBluescript KS− vector (Stratagene, LaJolla, CA). The ATT and ACC mutations in the clpP translation initiation codon were introduced in this plasmid by site-directed mutagenesis (Kunkel, 1985), using the following synthetic nucleotides: P-ATT, 5′-ATACGAGGTACTCCGATCGGAATATAAATTACTTTGTTAATAAA-3′; and P-ACC, 5′-ATACGAGGTACT-CCGATCGGGGTATAAATTACTTTGTTAATAAA-3′. Nucleotides differing from the wild-type sequence are written in boldface. The resulting plasmids, pdBATT and pdBACC, contain a clpP initiation codon mutation and a PvuI restriction site (underlined) to facilitate screening of transformants by restriction fragment length polymorphism analysis.
In a second step, plasmids pdBAUUK and pdBACCK (Figure 1) were created by insertion of the 1.9-kb EcoRV-SmaI fragment from the plasmid pUC-atpA-aadA (Goldschmidt-Clermont, 1991), containing the aadA cassette, into the unique NsiI site upstream of clpP, previously blunt-ended with T4 polymerase. In those plasmids, the aadA cassette, flanked by expression signals from atpA and rbcL, is located 376 bp from the clpP initiation codon and is transcribed in the opposite direction to clpP, as illustrated in Figure 1. The pWBI control plasmid was created by Y. Choquet in our laboratory by introducing the aadA cassette into the NsiI site of pWB (Kuras and Wollman, 1994) in the same orientation as in pdBAUUK and pdBACCK.
Overexpression of ClpP in Escherichia coli and Antibody Production
The entire clpP coding sequence was amplified by PCR from a Chlamydomonas DNA preparation, with primers CLPP1 (5′-CCGGATCCCTAACTATTACTTTCTTGC-3′) and CLPP2 (5′-GCCATA-TGCCGATTGGAGTAC-3′). These primers introduce BamHI and NdeI restriction sites (underlined) that were used to clone this fragment between the corresponding sites of the pet15 and pet32a vectors (Novagen, Madison, WI). The resulting plasmids, containing the entire clpP sequence either alone (pet15-clpP) or fused downstream to thioredoxin-A and a histidine tag (pet32-clpP), were transformed into the BL21 E. coli strain. After isopropylthio-β-galactoside induction, the ClpP and Trx-ClpP proteins were recovered in inclusion bodies. This fraction was separated by polyacrylamide gel electrophoresis, and the proteins were recovered by electroelution. The antibodies were prepared in rabbits by Neosystem (Strasbourg, France).
Chloroplast Transformation in Chlamydomonas
The wild-type strain was transformed by tungsten particle bombardment (Boynton et al., 1988), with a compressed-air shotgun device built in the laboratory by D. Béal and P. Bennoun. Cells were grown in liquid TAP medium to a density of 5 × 106 cells per mL. Cells (108) were plated on TAP medium and bombarded with 1.2-μm tungsten particles coated with the appropriate DNA. After overnight incubation in dim light at 25°C, cells were replated on TAP medium containing 100 μg/mL spectinomycin. Colonies were picked after 2 weeks of growth. Transformants were cultured on TAP plates containing spectinomycin and streptomycin, as previously described (Kuras and Wollman, 1994).
The presence of the transforming DNA was followed by PCR. Primers PA1 (5′-GCAGAATCTTTGTCTTGATTAGGTG-3′) and AA1 (5′-CACTGCCTCTAATAAAGTCATCG-3′) (Figure 1) were used to amplify the sequence between clpP and the aadA cassette (0.91 kb). Amplification products were digested by PvuI to check for introduction of the mutation. The wild-type copy was amplified with primers PA2 (5′-CCACAGATTGACGTTTTGC-3′) and COD2 (5′-ACAGCTGTTTTCATGTTAATGCAC-3′), permitting amplification between petB and clpP on the wild-type copy (1.16 kb) but not on copies containing the aadA cassette. PCR was performed on crude DNA preparations (Roffey et al., 1991) or on lysates obtained from plate-grown cells. Lysates were prepared by resuspending ∼5 × 104 cells in 20 μL lysis buffer (0.1 M Tris-HCl, pH 8.0, 0.1 M EDTA, 0.25 M NaCl, and 0.1% SDS), freezing at −80°C, and thawing for 10 min at 70°C.
Biochemical Analysis
SDS-PAGE was performed on 12 to 18% urea-containing gels (Piccioni et al., 1981); samples were loaded on an equal chlorophyll basis. Proteins were electroblotted onto nitrocellulose (Towbin et al., 1979), immunodetection was performed using 125I-Protein A, and the reactions were quantified with a PhosphorImager. The antisera to thy-lakoid membrane proteins have been described previously (Pierre and Popot, 1993; Breyton et al., 1994).
Acknowledgments
The authors thank Catherine de Vitry for sharing unpublished data on petC mutants. W.M. is a recipient of a fellowship from the Ministère de l'Education Nationale. This work was supported by the Centre National de la Recherche Scientifique (UPR 1261).
References
- Adam, Z. (1996). Protein stability and degradation in chloroplasts. Plant Mol. Biol. 32 773–783. [DOI] [PubMed] [Google Scholar]
- Akiyama, Y., Kihara, A., Tokuda, H., and Ito, K. (1996). FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271 31196–31201. [DOI] [PubMed] [Google Scholar]
- Arlt, H., Tauer, R., Feldmann, H., Neupert, W., and Langer, T. (1996). The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85 875–885. [DOI] [PubMed] [Google Scholar]
- Baumeister, W., and Lupas, A. (1997). The proteasome. Curr. Opin. Struct. Biol. 7 273–278. [DOI] [PubMed] [Google Scholar]
- Bochtler, M., Ditzel, L., Groll, M., and Huber, R. (1997). Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA 94 6070–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton, J.E., et al. (1988). Chloroplast transformation in Chlamy-domonas with high velocity microprojectiles. Science 240 1534–1538. [DOI] [PubMed] [Google Scholar]
- Breyton, C., de Vitry, C., and Popot, J.-L. (1994). Membrane association of cytochrome b6f subunits: The Rieske protein of Chlamydomonas reinhardtii is an extrinsic protein. J. Biol. Chem. 269 7597–7602. [PubMed] [Google Scholar]
- Breyton, C., Tribet, C., Olive, J., Dubacq, J.P., and Popot, J.L. (1997). Dimer to monomer conversion of the cytochrome b6f complex: Causes and consequences. J. Biol. Chem. 272 21892–21900. [DOI] [PubMed] [Google Scholar]
- Bulté, L., and Wollman, F.A. (1992). Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 204 327–336. [DOI] [PubMed] [Google Scholar]
- Buschlen, S., Choquet, Y., Kuras, R., and Wollman, F.A. (1991). Nucleotide sequences of the continuous and separated petA, petB and petD chloroplast genes in Chlamydomonas reinhardtii. FEBS Lett. 284 257–262. [DOI] [PubMed] [Google Scholar]
- Chen, X., Kindle, K., and Stern, D. (1993). Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 12 3627–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Kindle, K.L., and Stern, D.B. (1995). The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell 7 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, S., Montello, G.E., Zhang, A., Cantor, E.J., Liao, W., Xu, M.Q., and Benner, J. (1998). Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acids Res. 26 5109–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, N.H., and Bennoun, P. (1975). Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. USA 72 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerau, K., and St. John, A.C. (1993). Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J. Bacteriol. 175 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J.P., Yildiz, F.H., and Grossman, A. (1996). Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15 2150–2159. [PMC free article] [PubMed] [Google Scholar]
- de Vitry, C., and Vallon, O. (1999). Mutants of Chlamydomonas: Tools to study thylakoid membrane structure, function and biogenesis. Biochimie 81 1–13. [DOI] [PubMed] [Google Scholar]
- de Vitry, C., Olive, J., Drapier, D., Recouvreur, M., and Wollman, F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry, C., Choquet, Y., Baymann, F., Finazzi, G., and Kallas, T. (1998). Deficient mutants and site-directed mutagenesis of the nuclear-encoded chloroplast Rieske 2Fe-2S protein. In Photosynthesis: Mechanisms and Effects, G. Garab, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 3151–3154.
- de Vitry, C., Finazzi, G., Baymann, F., and Kallas, T. (1999). Analysis of the nucleus-encoded and chloroplast-targeted Rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell 11 2031–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonciarz-Swiatek, M., Wawrzynow, A., Um, S.J., Learn, B.A., McMacken, R., Kelley, W.L., Georgopoulos, C., Sliekers, O., and Zylicz, M. (1999). Recognition, targeting, and hydrolysis of the λO replication protein by the ClpP/ClpX protease. J. Biol. Chem. 274 13999–14005. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. (1996). Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30 465–506. [DOI] [PubMed] [Google Scholar]
- Gottesman, S., Squires, C., Pichersky, E., Carrington, M., Hobbs, M., Mattick, J.S., Dalrymple, B., Kuramitsu, H., Shiroza, T., and Foster, T. (1990). Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. USA 87 3513–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, S., Wickner, S., and Maurizi, M.R. (1997). Protein quality control: Triage by chaperones and proteases. Genes Dev. 11 815–823. [DOI] [PubMed] [Google Scholar]
- Gottesman, S., Roche, E., Zhou, Y., and Sauer, R.T. (1998). The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J.C., Hird, S.M., and Dyer, T.A. (1990). Nucleotide sequence of a wheat chloroplast gene encoding the proteolytic subunit of an ATP-dependent protease. Plant Mol. Biol. 15 947–950. [DOI] [PubMed] [Google Scholar]
- Grimaud, R., Kessel, M., Beuron, F., Steven, A.C., and Maurizi, M.R. (1998). Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273 12476–12481. [DOI] [PubMed] [Google Scholar]
- Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H.D., and Huber, R. (1997). Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386 463–471. [DOI] [PubMed] [Google Scholar]
- Hallick, R.B., Hong, L., Drager, R.G., Favreau, M.R., Monfort, A., Orsat, B., Spielmann, A., and Stutz, E. (1993). Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 21 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H., ed (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Huang, C., Wang, S., Chen, L., Lemieux, C., Otis, C., Turmel, M., and Liu, X.Q. (1994). The Chlamydomonas chloroplast clpP gene contains translated large insertion sequences and is essential for cell growth. Mol. Gen. Genet. 244 151–159. [DOI] [PubMed] [Google Scholar]
- Itzhaki, H., Naveh, L., Lindahl, M., Cook, M., and Adam, Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273 7094–7098. [DOI] [PubMed] [Google Scholar]
- Kessel, M., Maurizi, M.R., Kim, B., Kocsis, E., Trus, B.L., Singh, S.K., and Steven, A.C. (1995). Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J. Mol. Biol. 250 587–594. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. (1999). Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18 2970–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A, B-like protein in Arabidopsis thaliana L. Biochem. Biophys. Res. Commun. 196 1214–1220. [DOI] [PubMed] [Google Scholar]
- Kowallik, K.V., Stoebe, B., Schaffran, I., Kroth-Pancic, P., and Freier, U. (1995). The chloroplast genome of a chlorophyll a+c–containing alga, Odontella sinensis. Plant Mol. Biol. Rep. 13 336–342. [Google Scholar]
- Kunkel, T.A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., and Wollman, F.-A. (1994). The assembly of cytochrome b6f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., Buschlen, S., and Wollman, F.A. (1995). Maturation of pre-apocytochrome f in vivo: A site-directed mutagenesis study in Chlamydomonas reinhardtii. J. Biol. Chem. 270 27797–27803. [DOI] [PubMed] [Google Scholar]
- Kuras, R., de Vitry, C., Choquet, Y., Girard-Bascou, J., Culler, D., Buschlen, S., Merchant, S., and Wollman, F.A. (1997). Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272 32427–32435. [DOI] [PubMed] [Google Scholar]
- Lemaire, C., Girard-Bascou, J., Wollman, F.-A., and Bennoun, P. (1986). Studies on the cytochrome b6f complex. I. Characterization of the complex subunits in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 851 229–238. [Google Scholar]
- Levchenko, I., Yamauchi, M., and Baker, T.A. (1997). ClpX and MuB interact with overlapping regions of Mu transposase: Implications for control of the transposition pathway. Genes Dev. 11 1561–1572. [DOI] [PubMed] [Google Scholar]
- Levine, R.P., and Smillie, R.M. (1962). The pathway of triphosphopyridine nucleotide photoreduction in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 48 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, M., Tabak, S., Cseke, L., Pichersky, E., Andersson, B., and Adam, Z. (1996). Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 271 29329–29334. [DOI] [PubMed] [Google Scholar]
- Liu, X.-Q., and Jagendorf, A.T. (1984). ATP-dependent proteolysis in pea chloroplasts. FEBS Lett. 166 248–252. [Google Scholar]
- Liu, X.-Q., and Jagendorf, A.T. (1985). Role of ATP-dependent and ATP-independent proteases of pea chloroplast in regulation of the plastid translation products. Physiol. Vég. 23 749–755. [Google Scholar]
- Lowe, J., Stock, D., Jap, B., Zwickl, P., Baumeister, W., and Huber, R. (1995). Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268 533–539. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Flanagan, J.M., Tamura, T., and Baumeister, W. (1997). Self-compartmentalizing proteases. Trends Biochem. Sci. 22 399–404. [DOI] [PubMed] [Google Scholar]
- Mayer, T.U., Braun, T., and Jentsch, S. (1998). Role of the proteasome in membrane extraction of a short-lived ER transmembrane protein. EMBO J. 17 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Grevelding, C., Westhoff, P., and Reiss, B. (1998). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258 342–351. [DOI] [PubMed] [Google Scholar]
- Monde, R.A., Zito, F., Olive, J., Wollman, F.-A., and Stern, D.B. (2000). Post-translational defects in tobacco chloroplast mutants lacking the cytochrome b6f complex. Plant J., in press. [DOI] [PubMed]
- Nielsen, E., Akita, M., Davila-Aponte, J., and Keegstra, K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, I., Keren, N., Zer, H., Gong, H., Mor, T.S., Gal, A., Tal, S., and Domovich, Y. (1994). Light-induced degradation of the photosystem II reaction centre D1 protein in vivo: An integrative approach. In Photoinhibition from the Molecule to the Field, N. Baker, ed (Oxford, UK: Bios Scientific Publishers), pp. 161–177.
- Ostersetzer, O., and Adam, Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: The possible role of the FtsH protease. Plant Cell 9 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel, M., Dalbey, R.E., and Strynadka, N.C. (1998). Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature 396 186–190. Erratum. Nature 396, 707. [DOI] [PubMed] [Google Scholar]
- Perler, F.B. (1999). InBase, the New England Biolabs Intein Database. Nucleic Acids Res. 27 346–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler, F.B., Comb, D.G., Jack, W.E., Moran, L.S., Qiang, B., Kucera, R.B., Benner, J., Slatko, B.E., Nwankwo, D.O., and Hempstead, S.K. (1992). Intervening sequences in an Archaea DNA polymerase gene. Proc. Natl. Acad. Sci. USA 89 5577–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccioni, R.G., Bennoun, P., and Chua, N.H. (1981). A nuclear mutant of Chlamydomonas reinhardtii defective in photosynthetic photophosphorylation: Characterization of the algal coupling factor ATPase. Eur. J. Biochem. 117 93–102. [DOI] [PubMed] [Google Scholar]
- Pierre, Y., and Popot, J.-L. (1993). Identification of two 4kDa miniproteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C. R. Acad. Sci. Ser. III Sci. Vie 316 1404–1409. [PubMed] [Google Scholar]
- Pietrokovski, S. (1994). Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 3 2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon, M., Schekman, R., and Romisch, K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.-D., Goldschmidt-Clermont, M., and Merchant, S., eds (1998). The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Roffey, R.A., Golbeck, J.H., Hille, C.R., and Sayre, R.T. (1991). Photosynthetic electron transport in genetically altered photosystem II reaction centers of chloroplasts. Proc. Natl. Acad. Sci. USA 88 9122–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A., and Ryan, C.A. (1995). Cloning of a tomato cDNA (Genbank L38581) encoding the proteolytic subunit of a Clp-like energy dependent protease (PGR95-001). Plant Physiol. 108 1341.12228545 [Google Scholar]
- Schirmer, E.C., Glover, J.R., Singer, M.A., and Lindquist, S. (1996). HSP100/Clp proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 21 289–296. [PubMed] [Google Scholar]
- Shanklin, J., DeWitt, N.D., and Flanagan, J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y., Xu, M.Q., and Paulus, H. (1995). Protein splicing: Characterization of the aminosuccinimide residue at the carboxyl terminus of the excised intervening sequence. Biochemistry 34 10844–10850. [DOI] [PubMed] [Google Scholar]
- Shao, Y., Xu, M.Q., and Paulus, H. (1996). Protein splicing: Evidence for an N-O acyl rearrangement as the initial step in the splicing process. Biochemistry 35 3810–3815. [DOI] [PubMed] [Google Scholar]
- Smith, C.K., Baker, T.A., and Sauer, R.T. (1999). Lon and clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. USA 96 6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko, A., Altschmied, L., and Herrmann, R.G. (1997). Sodium dodecyl sulfate-stable proteases in chloroplasts. Plant Physiol. 115 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko, A., Lerbs-Mache, S., Altschmied, L., and Herrmann, R.G. (1998). Clp protease complexes and their diversity in chloroplasts. Planta 207 286–295. [DOI] [PubMed] [Google Scholar]
- Suzuki, C.K., Rep, M., Maarten van Dijl, J., Suda, K., Grivell, L.A., and Schatz, G. (1997). ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22 118–123. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernotte, C., Etienne, A.-L., and Briantais, J.-M. (1979). Quenching of the system II chlorophyll fluorescence by the plastoqinone pool. Biochim. Biophys. Acta 545 519–527. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hartling, J.A., and Flanagan, J.M. (1997). The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91 447–456. [DOI] [PubMed] [Google Scholar]
- Weber, E.R., Hanekamp, T., and Thorsness, P.E. (1996). Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol. Biol. Cell 7 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz, E.J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T.R., Rapoport, T.A., and Ploegh, H.L. (1996). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384 432–438. [DOI] [PubMed] [Google Scholar]
- Wollman, F.-A. (1998). The structure, function and biogenesis of cytochrome b6f complexes. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 459–476.
- Wollman, F.A., Minai, L., and Nechushtai, R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411 21–85. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Culler, D., Dreyfuss, B.W., Kuras, R., Wollman, F.A., Girard-Bascou, J., and Merchant, S. (1998). Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: One chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.H., Webster, J., Adam, Z., Lindahl, M., and Andersson, B. (1998). Induction of acclimative proteolysis of the light-harvesting chlorophyll a/b protein of photosystem II in response to elevated light intensities. Plant Physiol. 118 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Kaung, G., Kobayashi, S., and Kopito, R.R. (1997). Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J. Biol. Chem. 272 20800–20804. [DOI] [PubMed] [Google Scholar]