Abstract
A diverse cadre of metazoan transcription factors mediate repression by recruiting protein complexes containing the SMRT (silencing mediator of retinoid and thyroid hormone receptor) or N-CoR (nuclear receptor corepressor) corepressors. SMRT and N-CoR nucleate the assembly of still larger corepressor complexes that perform the specific molecular incantations necessary to confer transcriptional repression. Although SMRT and N-CoR are paralogs and possess similar molecular architectures and mechanistic strategies, they nonetheless exhibit distinct molecular and biological properties. It is now clear that the functions of both SMRT and N-CoR are further diversified through alternative mRNA splicing, yielding a series of corepressor protein variants that participate in distinctive transcription factor partnerships and display distinguishable repression properties. This review will discuss what is known about the structure and actions of SMRT, N-CoR, and their splicing variants, and how alternative splicing may allow the functions of these corepressors to be adapted and tailored to different cells and to different developmental stages.
Worms, humans, and gene numerology
Genome-level sequence analysis has led to a recent and surprising realization: relatively few genes are necessary to produce multicellular organisms, and the difference in gene number between nematodes and New Yorkers (or Nebraskans for that matter) is quite modest [ Pennisi, 2003 ]. How can only 30,000 genes construct entities as complex as human beings? One explanation originates in a prior, equally unexpected discovery: multiple distinct mRNAs can be generated from a single genetic locus through alternative mRNA splicing. Alternative mRNA splicing diversifies the functions of a single gene by creating multiple protein variants, each possessing distinct domains and distinct properties [ Stamm et al., 2005 ]. In this manner a limited number of genetic loci have been elaborated by evolution to perform a multitude of roles. In this review we will describe recent revelations as to how important transcriptional coregulators, the SMRT and N-CoR corepressors, are customized, tailored, and adapted through alternative mRNA splicing, and the implications of this phenomenon for the regulation of metazoan gene expression.
Bipolar transcription: manic repression
The corepressor story begins with the recognition that many eukaryotic transcription factors can function both as activators and as repressors. This ambidextrous transcriptional control is most readily observed for the nuclear receptors: transcription factors that bind to specific hormone ligands, bind to specific DNA sequences and regulate the expression of adjacent target genes. Many members of the nuclear receptor family, including the thyroid hormone receptors (TRs) and retinoic acid receptors (RARs), repress target gene expression in the absence of hormone ligand, yet activate target gene expression in the presence of a hormone agonist [ Mangelsdorf et al., 1995 ]. Still additional nuclear receptors, such as the estrogen and androgen receptors, are neutral in the absence of hormone yet can be toggled between repressive and activating states by the alternative binding of antagonist or agonist hormone ligands [ Liu et al., 2002; Schulz et al., 2002; Shang and Brown, 2002; Wagner et al., 1998; Zhang et al., 1998 ]. Indeed, this type of pharmacologically-driven receptor modulation is the molecular basis behind clinical interventions in important diseases, such as the use of the anti-estrogen tamoxifen to treat mammary cancers (e.g. [ Keeton and Brown, 2005; Takimoto et al., 1999 ]). Many other transcription factors, even those not regulated by hormone, display a similar transcriptional dualism, either repressing or activating gene expression depending on DNA binding site, promoter context, cell type, and the influence of various cell signaling pathways [ Privalsky, 2001; Privalsky, 2004 ].
A single transcription factor can display these bipolar gene regulatory properties through its ability to alternatively recruit auxiliary proteins denoted corepressors and coactivators [ Glass and Rosenfeld, 2000; Privalsky, 2001 ]. Coactivators include a wide assortment of acetyl transferases, methyl transferases, ATP-dependent chromatin remodeling complexes, and Mediator subunits [ Lonard and O'Malley, 2005; Roeder, 2005 ]. Coactivators enhance transcription by covalently modifying chromatin (i.e. changing the "histone code"), by rendering the DNA template more accessible, and by recruiting components of the general transcriptional machinery to a target promoter [ Freiman and Tjian, 2003; Jenuwein and Allis, 2001; Tsai and Fondell, 2004 ]. Corepressor complexes, conversely, contain a variety of histone deacetylase activities, and thus can strip the acetate activation marks from chromatin; some corepressors also make inhibitory contacts with the general transcriptional machinery and interfere with assembly of the preinitiation complex [ Hu and Lazar, 2000; Jepsen and Rosenfeld, 2002; Privalsky, 2004; Urnov et al., 2001 ]. A given transcription factor, by physically exchanging corepressor complexes for coactivator complexes, can convert from a repressor to an activator of gene expression [ Glass and Rosenfeld, 2000; Perissi et al., 2004; Privalsky, 2004 ].
Corepressor paralogs and functional parallels: SMRT and N-CoR
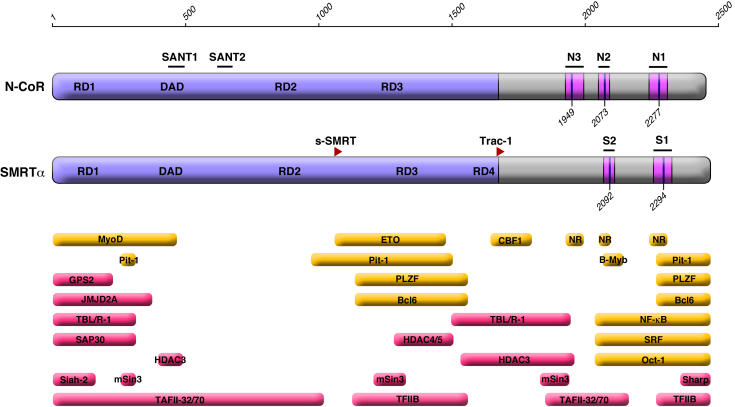
The extensive role of corepressors and transcriptional repression in metazoan gene regulation has only recently become appreciated. Two proteins, Nuclear Receptor Corepressor (N-CoR) and Silencing Mediator of Retinoid and Thyroid hormone Receptors (SMRT), serve as key corepressors for an large assortment of different vertebrate transcription factors, including the nuclear hormone receptors, PLZF, c-Jun, SMADs, CBF-1, B-Myb, Pit-1, ETO-1/2, MyoD, NF-κB, and BCL-6, among others (Figure 1) [ Ordentlich et al., 2001; Privalsky, 2004 ]. N-CoR and SMRT are encoded by distinct genetic loci, but are paralogs of one another and share a common molecular architecture, form similar complexes with other corepressor proteins, and exert overlapping biological functions [ Ordentlich et al., 2001; Privalsky, 2004 ]. Both N-CoR and SMRT operate by tethering to their transcription factor partners and nucleating the assembly of a larger, functional corepressor complex [ Ordentlich et al., 2001; Privalsky, 2004 ]. SMRT and N-CoR tether to nuclear receptors, for example, through a set of C-terminal "CoRNR box" motifs (Figure 1) [ Hu and Lazar, 1999; Nagy et al., 1999; Perissi et al., 1999; Xu et al., 2002 ]. A secondary hierarchy of corepressor subunits, including histone deacetylases, TBL-1, TBLR-1, GPS-2, and a number of other modulatory and effector proteins are recruited through docking surfaces (repression domains, RDs) located principally in the N-terminal and central regions of SMRT and N-CoR (Figure 1) [ Ordentlich et al., 2001; Privalsky, 2004 ]. Significantly, SMRT and N-CoR not only physically recruit, but can also kindle the enzymatic activity of their associated histone deacetylases [ Codina et al., 2005; Guenther et al., 2001 ].
Figure 1. Schematic of the N-CoR and SMRT corepressors.
The primary structure of the human N-CoR and murine SMRTα are sketched from N- to C-terminus [Horlein et al., 1995; Ordentlich et al., 1995]. Codon numbering is indicated on top. The locations of the repression domains (RD1 to RD4), the deacetylase activating domain (DAD), the conserved SANT motifs that include sites of histone interaction, and of the CoRNR box/nuclear receptor interaction sites (N1, N2, and N3 in N-CoR verses S1 and S2 in SMRT) are indicated within each corepressor schematic [Ordentlich et al., 2001]. The initiation codons for the N-terminally truncated s-SMRT and Trac-1 polypeptides are shown as red arrows. Interaction sites for transcription factors that utilize SMRT and/or N-CoR for repression are indicated in yellow, whereas interaction sites for additional components of the corepressor complex or the general transcriptional machinery are shown in red. The actual sites of contact may be smaller than the experimentally mapped domains shown, and not all interacting proteins have been proven to interact with both N-CoR and SMRT.
The generation of SMRT and N-CoR, and their maintenance as distinct paralogs throughout the evolution of the vertebrate lineage, implies that these two superficially similar corepressors must mediate distinct biological functions. Indeed, one important function, the ability to interact with different transcription factor partners, clearly differs. For example, TRs can interact more strongly with N-CoR than with SMRT, whereas the reciprocal appears true for RARs [ Zamir et al., 1997 ]. Differences in amino acid sequence within and flanking the CoRNR box motifs help define these distinct interactions [ Cohen et al., 2001; Hu and Lazar, 1999; Xu et al., 2002 ]. Moreover, the prototypic N-CoR molecule encodes three CoRNR box motifs (N1, N2, and N3) whereas the prototypic SMRT encodes only two (S1 and S2) (Figure 1), further differentiating the specificities of these two paralogs for different nuclear receptors [ Cohen et al., 2001; Makowski et al., 2003; Webb et al., 2000 ]. SMRT and N-CoR also differ in their response to kinase pathways that regulate their function. SMRT is phosphorylated in response to growth factor receptor/MAP kinase cascades; this phosphorylation results in release of SMRT from TRs, RARs, and PLZF, derepression of target genes serviced by these transcription factors, and the redistribution SMRT from the nucleus to the cytoplasm [ Hong and Privalsky, 2000; Hong et al., 1998; Jonas and Privalsky, 2004 ]. N-CoR, in contrast, is refractory to this form of direct intervention by growth factor/ kinase cascades, although N-CoR appears to be regulated indirectly by cytokines through a MEKK1/TAB2 intermediate [ Baek et al., 2002 ]. Conversely, N-CoR, but not SMRT, can be negatively regulated by Akt in the control of the NF-κB transcriptional response [ Hermanson et al., 2002 ]. Expression levels of SMRT and N-CoR differ in different tissues and at different times in development, indicating that organisms adapt their corepressor repertoire for distinct biological purposes [ Jepsen et al., 2000 ].
Corepressor diversification through alternative mRNA splicing: the search for the “true” SMRT N-terminus
In addition to the evolutionary gene duplication and divergence events that led to the generation of the N-CoR and SMRT paralogs, a second, equally important source of corepressor diversity arises from alternative mRNA splicing. The first suggestion that SMRT might be alternatively-spliced was foreshadowed by differences in the predicted N-terminal SMRT sequence as published by various rival laboratories [ Chen and Evans, 1995; Chen et al., 1996; Ordentlich et al., 1999; Park et al., 1999; Sande and Privalsky, 1996 ]. Three SMRT species differing in their N-terminal initiation sites, were identified, denoted from longest to shortest as SMRTα (also known as SMRTe), s-SMRT (originally denoted SMRT or TRAC-2), and TRAC-1 (also known as C-SMRT) (Figure 1 and Figure 2). The longest version, SMRTα is roughly colinear with the longest known N-CoR clone, and encompasses a similar (but non-identical) set of repression and transcription factor tethering domains (Figure 2) [ Horlein et al., 1995; Ordentlich et al., 1999; Park et al., 1999 ]. The two smaller variants, s-SMRT and TRAC-1, encompass overlapping C-terminal domains, but lack some or most of the N-terminal repression/cofactor docking sites found in SMRTα (Figure 2) [ Chen and Evans, 1995; Chen et al., 1996; Ordentlich et al., 1999; Park et al., 1999; Sande and Privalsky, 1996 ]. s-SMRT is able to repress when experimentally tethered to a promoter, whereas TRAC-1 cannot repress and operates in transfections as a dominant-negative inhibitor of corepressor function [ Chen and Evans, 1995; Chen et al., 1996; Sande and Privalsky, 1996 ]. SMRTα is well documented as a physiologically relevant, widely expressed splice variant that can mediate repression by a wide spectrum of transcription factor partners. The biological relevance of s-SMRT and TRAC-1 is less clear; these N-terminal truncated variants may potentially mediate a subset of the repression functions of SMRTα, or may even serve to as "anti-corepressors" [ Chen et al., 1996; Sande and Privalsky, 1996 ]. Unfortunately, both the s-SMRT and TRAC-1 clones, as initially published, display chimeric characteristics indicative of a joining of sequences from normally separate chromosomes [ Chen and Evans, 1995; Sande and Privalsky, 1996 ]. It is possible that s-SMRT and TRAC-1 are the products of authentic, if uncharacterized chromosomal rearrangements in the aneuploid cell lines from which they were isolated, or they may simply be artifacts created during generation of the corresponding cDNA libraries. Nonetheless, mRNAs and/or immunoreactive polypeptides approximating these truncated SMRTs in length, and a cDNA possibly encoding a version of N-CoR similar to TRAC-1, have been detected in several contexts [e.g. [ Chen et al., 1996; Cote et al., 2004; Hollenberg et al., 1996; Meng et al., 2005 ]), and it is possible that these N-terminal corepressor variants may ultimately prove of some physiological significance.
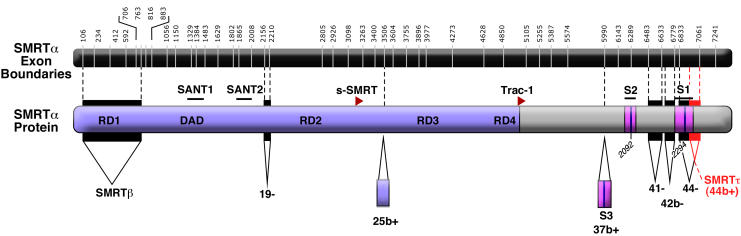
Figure 2. Alternative mRNA splicing of SMRT.
The murine SMRTα exon/intron organization is depicted on top, with exons shown in black. The locations of introns are indicated by white lines and numbered by base position within the mature mRNA. The encoded murine SMRTα protein is shown below from N- to C- terminus. General features are as in Figure 1. Exonic sequences spliced out of the murine SMRTα sequence are indicated by black or red backgrounds, by downward pointed triangles, and by a negative sign next to their ordinal number; exonic sequences inserted relative to the murine SMRTα sequence are indicated by boxes below the main schematic, by upward pointing arrows, and by positive signs next to their ordinal number [Goodson et al., 2005; Malartre et al., 2004; Short et al., 2005]. The splicing event deleting exon 44b is also denoted SMRTτ [Goodson et al., 2005].
Corepressor diversification through alternative mRNA splicing: custom fitting of SMRT and N-CoR to their transcription factor partners
In contrast to the ambiguities as to the authentic SMRT N-terminus, analysis of the effect of alternative mRNA splicing on internal SMRT sequences has yielded significantly more definitive and provocative results. The first hints of this type of modification were embedded, but little noticed, in the first two published sequences of SMRT, which differed by the presence or absence of a 47 codon "insert" just downstream of the S1 receptor interaction domain (Figure 2, highlighted in red) [ Chen and Evans, 1995; Chen et al., 1996; Sande and Privalsky, 1996 ]. Genomic sequence subsequently confirmed that this insert had all the characteristics of an alternative exon (exon 44b; Figure 2), and mRNAs both possessing and lacking this exon can be readily detected in EST data bases from a broad variety of species and cell types [ Goodson et al., 2005; Malartre et al., 2004; Short et al., 2005 ]. For lack of a better nomenclature, the SMRT isoform lacking this exon was denoted SMRTτ (tau), whereas the SMRTα isoform was defined as containing this insert [ Goodson et al., 2005 ]. The SMRTτ variant retains many of the same biochemical and molecular biological features seen for SMRTα but differs notably in its affinity for different nuclear hormone receptors relative to the SMRTα form [ Goodson et al., 2005 ]. This is most clearly manifested as a much higher affinity of SMRTα for TRs relative to SMRTτ, whereas both SMRTα and SMRTτ have virtually equal affinities for RARs [ Goodson et al., 2005 ]. This result is consistent with the location of this alternative exon immediately flanking the S1 CoRNR box motif (Figure 2), and therefore in an appropriate position to modulate the corepressor/nuclear receptor interaction [ Cohen et al., 2001; Hu and Lazar, 1999; Xu et al., 2002 ]. Ratios of SMRTα to SMRTτ differ in different tissues, suggesting that organisms can adapt the receptor specificity of the corepressor to the biological requirements of different cell types [ Goodson et al., 2005; Short et al., 2005 ].
Further mining of EST databases, combined with wet laboratory dissections, has revealed that the C-terminal region of SMRT is subject to an aggressive array of alternative mRNA splicing events, many of which are predicted to modify or adapt the interactions of SMRT with its transcription factor partners (Figure 2) ([ Malartre et al., 2004; Short et al., 2005 ], M. Goodson, unpublished observations). Perhaps the most remarkable of these additional splicing events introduces into SMRT a novel exon (exon 37b) containing a third CoRNR box (indicated in purple and blue in Figure 2); this generates a (S3+S2+S1) SMRT variant closely resembling the previously established (N3+N2+N1) N-CoR in its triple CoRNR box valency [ Malartre et al., 2004; Short et al., 2005 ]. Although first characterized for Xenopus, where it is expressed in almost all adulttissues at equal levels with (S2+S1) SMRTα, this trivalent SMRT species is also expressed in a variety of vertebrates, including mice, where it is observed primarily in brain ([ Malartre et al., 2004; Short et al., 2005 ], and M. Goodson, unpublished observations]. It remains to be established if the nuclear receptor interaction properties of this newly recognized (S1+S2+S3) SMRT resemble those of N-CoR, or if it instead displays its own, idiosyncratic set of receptor interactions and affinities. Still other C-terminal variants of SMRT have been detected that lack the S1 CoRNR box, or that modify SMRT sequences outside of the CoRNR boxes themselves (typified by loss of exons 41, 42b, or 44 relative to the SMRTα epitome; Figure 2) [ Short et al., 2005 ]. A number of these alternative splice variants differ in their abundance at different times of development or in different tissues [ Malartre et al., 2004; Short et al., 2005 ]. Although additional investigation will be required to fully understand the biochemical and biological properties of these newly recognized SMRT variants, it appears safe to predict that these alternative splicing events represent a mechanism by which cells can custom tailor repression to specific subsets of transcription factors and their target genes. In considering the possibilities, it should be remembered that many non-receptor transcription factors bind outside of the CoRNR motifs employed by the nuclear receptors (Figure 1), and thus splicing events that do not impinge directly on the S1, S2, or S3 domains may nonetheless alter the ability of SMRT to interact with specific subsets of its transcription factor repertoire.
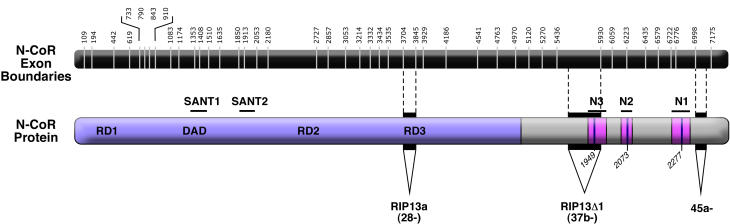
Turning to the other corepressor paralog, we find that the C-terminal domain of N-CoR is also modified by alternative mRNA splicing (Figure 3). Although the archetypic N-CoR contains three CoRNR boxes, early studies presciently established the existence of an alternatively spliced form of N-CoR, denoted RIP13Δ1, that deletes the N3 box (Figure 3) [ Horlein et al., 1995; Seol et al., 1996 ]. Thus both SMRT and N-CoR are expressed as variants possessing either two or three nuclear receptor interaction domains. The absence or inclusion of the third CoRNR box in N-CoR/RIP13Δ1 alters the affinity of the corepressor for different nuclear receptor partners. For example, "classical" (N3+N2+N1) N-CoR displays unusually high affinity for TRs, and this has been mapped to the cooperative actions of N3 and N2; in contrast RIP13Δ1, limited to the N2 and N1 interaction surfaces, has significantly lower affinity for TRs [ Cohen et al., 2001; Makowski et al., 2003; Webb et al., 2000 ]. Conversely, the (N2+N1) RIP13Δ1 clone appears to display a higher relative affinity for the Rev-Erb Aα, RVR, and COUP-TFII orphan receptors than does the (N3+N2+N1) form of N-CoR [ Bailey et al., 1997; Downes et al., 1996 ]. A second alternative splice near the very C-terminus of N-CoR, removing most of exon 45, has been identified in mice (Figure 3); no function has yet been assigned to this corepressor region and the levels of this N-CoR variant appear constant in a variety of tissues [ Short et al., 2005 ]. Apart from these two examples, alternative splicing events in the C-terminal portion of N-CoR appear to be significantly less frequent than in the equivalent regions of SMRT, perhaps indicative of a less diverse evolutionary program for N-CoR relative to SMRT [ Short et al., 2005 ].
Figure 3. Alternative mRNA splicing of N-CoR.
The human N-CoR exon/intron organization is depicted on top, with exons shown in black and the locations of introns indicated by white lines and numbered as to their base positions within the mature mRNA. The encoded human N-CoR protein is shown below from N- to C- terminus. General features are as in Figure 1. Exonic sequences spliced out of the human N-CoR sequence are indicated by black backgrounds, by downward pointed triangles, and by a negative sign next to their ordinal number [Horlein et al., 1995; Malartre et al., 2004; Seol et al., 1996; Short et al., 2005]. The splicing event that removes exon 28 has also been denoted RIP13a; the splicing event that removes exon 37b has also been denoted RIP13Δ1 [Seol et al., 1996].
In summary, both N-CoR and SMRT have their affinity for different transcription factors adjusted "on the fly" through mRNA splicing events that either add, remove, or adjust the interaction surfaces on the corepressors. Several additional layers of complexity should be considered as to the potential effects of these splicing events. Nuclear receptors typically bind to DNA as protein homo- or heterodimers, with each receptor molecule thought to contact one of the CoRNR boxes in the SMRT or N-CoR corepressor [ Privalsky, 2004 ]). The ability of a given corepressor splice variant to recognize a given receptor dimer may therefore depend not only on the identity of each receptor in the dimer, but also on the orientation and spacing of the receptors as they assemble on a given target promoter (e.g. [ Zamir et al., 1997 ]). It is also conceivable, if not yet experimentally proven, that interactions with different transcription factors might be able to induce different allosteric states in SMRT and N-CoR so as to produce distinct functional outcomes; these allosteric effects, if they occur, might differ for the different SMRT splice variants. Finally, it should be noted that the C-terminal domain of these corepressors not only tethers SMRT and N-CoR to their transcription factor partners, but also binds a number of other proteins of incompletely understood function (e.g. Sharp, Figure 1), and is an important site of post-translational modifications that regulate corepressor function [ Hermanson et al., 2002; Hoberg et al., 2004; Hong and Privalsky, 2000; Jonas and Privalsky, 2004; Shi et al., 2001; Zhou et al., 2001 ]. At least several of the alternative mRNA splicing events discussed here may prevent, enhance, or otherwise alter the recruitment of these additional proteins and/or the effects of these regulatory modifications.
Corepressor diversification through alternative mRNA splicing: modifications of the SMRT and N-CoR repression domains
In contrast to the extensive splice diversity discovered in the C-terminal domain of SMRT, there are somewhat fewer documented examples of alternative exon utilization within the N-terminal repression domains of either SMRT or N-CoR (putting aside the unresolved tale of s-SMRT and TRAC-1). One interesting corepressor variant that is well documented in this regard is SMRTβ, which by excising exons 2 to 6 has lost repression domain 1 and deleted potential interaction surfaces for mSin3, SAP30, and TBL/R-1 (Figure 1 and Figure 2) [ Ordentlich et al., 1999 ]. SMRTβ displays weaker repression properties in transfection assays than does SMRTα, and if overexpressed can paradoxically enhance RAR-mediated gene expression and promote hormone-responsiveness in retinoid-resistant leukemias [ Cote et al., 2004; Ordentlich et al., 1999 ]; SMRTβ may be mediating these positive transcriptional effects by counteracting the stronger repression functions of SMRTα present in the same cells, or more heretically, alternative splicing may confer on SMRTβ genuine coactivator capabilities.
Looking further down the corepressor sequence, exon 19 of SMRT is also subject to alternative splicing, corresponding to a region flanking repression domain 2 (Figure 2) ([ Short et al., 2005 ] and M. Goodson, unpublished data]. This alternative use of exon 19 may represent a species difference rather than a developmentally-regulated splicing event and its functional consequences are not known [ Short et al., 2005 ]. More extensively described in the literature is a splicing variant of N-CoR, denoted RIP13a, that lacks exon 28 and therefore a portion of repression domain 3 (Figure 3) [ Muscat et al., 1998; Seol et al., 1996 ]. The loss of exon 28 of N-CoR removes sequences that have been implicated (to varying degrees of confidence) as interaction sites for mSin3, TFIIB, and/or the class II histone deacetylases HDAC4 and 5 (compare Figure 1 and Figure 3) [ Ordentlich et al., 2001; Privalsky, 2004 ]. The repression properties of RIP13a therefore might be expected to differ from those of N-CoR, particularly on different promoters or in different cell contexts; this prediction remains to be experimentally verified. Notably, a nearby, but not-quite identical location in SMRT is also subject to an alternative splicing event that, in the mouse, introduces an insert from exon 25b into the published SMRTα sequence [ Short et al., 2005 ]; as with RIP13a the SMRT exon 25b variant appears likely to possess modified repression properties, although this has not been experimentally documented.
Additional splicing events may eventually be discovered within the N-terminal repression domains of SMRT and N-CoR that have remained undetected thus far due to the 3' bias of the current EST data bases. Nonetheless, it appears clear that alternative splicing is particularly diverse in the C-terminal portion of SMRT, the region containing the bulk of the known transcription factor interaction sites, rather than in the more N-terminal regions responsible for assembly of the remainder of the corepressor holo-complex. In fact, many of the alternative mRNA splicing events that do map within the N-terminal two-thirds of SMRT and N-CoR encompass not only known repression domains, but also impact tethering surfaces for MyoD, Pit-1, PLZF, BCL-6, and/or ETO, additional members of the transcription factor cohort that utilize SMRT and N-CoR to mediate repression (Figure 1). The data to date, therefore, favor a particularly prominent role for mRNA splicing in defining and diversifying the “afferent” targeting of SMRT (and N-CoR) to particular subsets of transcription factor partners, and a lesser role in modifying the “efferent” transcriptional repression properties of the corepressor complex once recruited to a promoter.
Where to next?
Clearly the crucial future task will be to fully define the specific functions of each of the spliced variants alluded to in this overview. Good progress has recently been achieved in one key aspect of this goal: cataloging which spliced forms are expressed in different species, in different tissues and at different times in development [ Goodson et al., 2005; Malartre et al., 2004; Short et al., 2005 ]. Detailed characterizations of the biochemical and biological properties of the individual SMRT and N-CoR are also in progress, but will need to further expanded both in vitro and in vivo. The reader browsing this overview will undoubtedly (and justifiably) propose still another crucial objective: the development of a practical and definitive nomenclature to describe the various corepressor variants. It is evident that the current nomenclature has evolved to be both confusing and awkward. A unified and rationale approach to naming the different spliced variants will be essential. One suggestion: retain N-CoR and SMRT to distinguish the two paralogs, but further identify the various spliced forms originating from each locus by a simple numerical nomenclature (e.g. SMRT-1 and SMRT-2 instead of SMRTα and SMRTβ; N-CoR-1 and N-CoR 2 instead of N-CoR and RIP13Δ1.
In conclusion, SMRT, and to a lesser extend, N-CoR have joined the ranks of a flourishing horde of vertebrate gene products whose functions are elaborated, adjusted, and custom-fitted by alternative mRNA splicing. In looking backward, it is essential to re-examine much of the data already generated on corepressor function in light of this new found diversity, making mental note of which spliced variant was examined for each study, and evaluating which variant yielded which result. In looking forward, a most interesting and productive future appears to lie ahead as the properties and biological roles of the various spliced corepressors are revealed.
Acknowledgments
The research attributed to our laboratory was supported by the Public Health Service/National Institutes of Health through awards R37CA53394 (MLP), RO1DK53538 (MLP), 5F32DK062654 (MG), and T32GM07377 (BAJ).
Abbreviations
- BCL-6
B-cell leukemia protein-6
- B-Myb
B locus of cellular myeloblastosis oncogene
- CBF-1
CCAAT-box binding factor
- CoRNR box
CoR-nuclear receptor interaction box
- DAD
deacetylase activating domain
- ETO-1/2
eight/twenty one translocation protein-1/2
- GPS-2
G protein suppressor-2
- HDAC
histone deacetylase
- MAPK
mitogen-activated protein kinase
- MEKK1
MAP/ERK protein kinase kinase-1
- MyoD
myoblast determination protein
- N-CoR
nuclear receptor corepressor
- NF-κB
nuclear transcription factor κ B
- Oct-1
octamer-element binding factor-1
- Pit-1
pituitary transcription factor-1
- PLZF
promyelocytic zinc-finger protein
- RAR
retinoic acid receptor
- RD
repression domain
- RIP13
receptor interacting protein 13
- SANT
SWI3, ADA2, N-CoR, and TFIIB related domain
- SAP30
Sin3 associated protein 30
- Sharp
SMRT/HDAC-1 associated protein
- Siah-2
seven in absentia homolog-2
- SMAD
SMA,/MAD related
- SMRT
silencing mediator for retinoid-thyroid hormone receptors
- SRF
serum response factor
- TAFII-32/70
TATA-binding protein associated factor-32/70
- TBL-1
transducin B-like protein-1
- TBLR-1
TBL-related protein-1
- TFIIB
transcription factor IIB
- TR
thyroid hormone receptor
- TRAC
TR/RAR associated cofactor
References
- Baek S. H., Ohgi K. A., Rose D. W., Koo E. H., Glass C. K., Rosenfeld M. G. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Bailey P. J., Dowhan D. H., Franke K., Burke L. J., Downes M., Muscat G. E. Transcriptional repression by COUP-TF II is dependent on the C-terminal domain and involves the N-CoR variant, RIP13delta1. J Steroid Biochem Mol Biol. 1997;63:165–74. doi: 10.1016/s0960-0760(97)00079-4. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Evans R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Umesono K., Evans R. M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci U S A. 1996;93:7567–71. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina A., Love J. D., Li Y., Lazar M. A., Neuhaus D., Schwabe J. W. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc Natl Acad Sci U S A. 2005;102:6009–14. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. N., Brzostek S., Kim B., Chorev M., Wondisford F. E., Hollenberg A. N. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol Endocrinol. 2001;15:1049–61. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- Cote S., McNamara S., Brambilla D., Bianchini A., Rizzo G., del Rincon S. V., Grignani F., Nervi C., Miller W. H., Jr. Expression of SMRTbeta promotes ligand-induced activation of mutated and wild-type retinoid receptors. Blood. 2004;104:4226–35. doi: 10.1182/blood-2003-10-3583. [DOI] [PubMed] [Google Scholar]
- Downes M., Burke L. J., Bailey P. J., Muscat G. E. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA α and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 1996;24:4379–86. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R. N., Tjian R. Regulating the regulators: lysine modifications make their mark. Cell. 2003;112:11–7. doi: 10.1016/s0092-8674(02)01278-3. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Goodson M. L., Jonas B. A., Privalsky M. L. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J Biol Chem. 2005;280:7493–503. doi: 10.1074/jbc.M411514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Barak O., Lazar M. A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O., Jepsen K., Rosenfeld M. G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–9. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hoberg J.E., Yeung F., Mayo M.W. SMRT derepression by the IkappaB kinase alpha: a prerequisite to NF-kappaB transcription and survival. Mol Cell. 2004;16:245–55. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Hollenberg A. N., Monden T., Madura J. P., Lee K., Wondisford F. E. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem. 1996;271:28516–20. doi: 10.1074/jbc.271.45.28516. [DOI] [PubMed] [Google Scholar]
- Hong S. H., Privalsky M. L. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20:6612–25. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H., Wong C. W., Privalsky M. L. Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol Endocrinol. 1998;12:1161–71. doi: 10.1210/mend.12.8.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Hermanson O., Onami T. M., Gleiberman A. S., Lunyak V., McEvilly R. J., Kurokawa R., Kumar V., Liu F., Seto E., Hedrick S. M., Mandel G., Glass C. K., Rose D. W., Rosenfeld M. G. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–63. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Rosenfeld M. G. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–98. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jonas B. A., Privalsky M. L. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J Biol Chem. 2004;279:54676–86. doi: 10.1074/jbc.M410128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton E. K., Brown M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-α and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol Endocrinol. 2005;19:1543–54. doi: 10.1210/me.2004-0395. [DOI] [PubMed] [Google Scholar]
- Liu Z., Auboeuf D., Wong J., Chen J. D., Tsai S. Y., Tsai M. J., O'Malley B. W. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc Natl Acad Sci U S A. 2002;99:7940–4. doi: 10.1073/pnas.122225699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard D. M., O'Malley B. W. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–32. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Makowski A., Brzostek S., Cohen R.N., Hollenberg A.N. Determination of nuclear receptor corepressor interactions with the thyroid hormone receptor. Mol Endocrinol. 2003;17:273–86. doi: 10.1210/me.2002-0310. [DOI] [PubMed] [Google Scholar]
- Malartre M., Short S., Sharpe C. Alternative splicing generates multiple SMRT transcripts encoding conserved repressor domains linked to variable transcription factor interaction domains. Nucleic Acids Res. 2004;32:4676–86. doi: 10.1093/nar/gkh786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Webb P., Yang Y.F., Shuen M., Yousef A.F., Baxter J.D., Mymryk J.S., Walfish P.G. E1A and a nuclear receptor corepressor splice variant (N-CoRI) are thyroid hormone receptor coactivators that bind in the corepressor mode. Proc Natl Acad Sci. 1998;102:6267–72. doi: 10.1073/pnas.0501491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat G. E., Burke L. J., Downes M. The corepressor N-CoR and its variants RIP13a and RIP13Delta1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70. Nucleic Acids Res. 1998;26:2899–907. doi: 10.1093/nar/26.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–16. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordentlich P., Downes M., Evans R. M. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–16. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Downes M., Xie W., Genin A., Spinner N. B., Evans R. M. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci U S A. 1999;96:2639–44. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. J., Schroen D. J., Yang M., Li H., Li L., Chen J. D. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc Natl Acad Sci U S A. 1999;96:3519–24. doi: 10.1073/pnas.96.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Bioinformatics. Gene counters struggle to get the right answer. Science. 2003;301:1040–1. doi: 10.1126/science.301.5636.1040. [DOI] [PubMed] [Google Scholar]
- Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Aggarwal A., Glass C., Rose D.W., Rosenfeld M. K. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;12:511–26. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Privalsky M. L. Regulation of SMRT and N-CoR corepressor function. Curr Top Microbiol Immunol. 2001;254:117–36. doi: 10.1007/978-3-662-10595-5_6. [DOI] [PubMed] [Google Scholar]
- Privalsky M. L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–15. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Sande S., Privalsky M. L. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–25. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- Schulz M., Eggert M., Baniahmad A., Dostert A., Heinzel T., Renkawitz R. RU486-induced glucocorticoid receptor agonism is controlled by the receptor N terminus and by corepressor binding. J Biol Chem. 2002;277:26238–43. doi: 10.1074/jbc.M203268200. [DOI] [PubMed] [Google Scholar]
- Seol W., Mahon M. J., Lee Y. K., Moore D. D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–55. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- Shang Y., Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shi Y., Downes M., Xie W., Kao H.Y., Ordentlich P., Tsai C.C., Hon M., Evans R.M. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–51. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short S., Malartre M., Sharpe C. SMRT has tissue-specific isoform profiles that include a form containing one CoRNR box. Biochem Biophys Res Commun. 2005;334:845–52. doi: 10.1016/j.bbrc.2005.06.175. [DOI] [PubMed] [Google Scholar]
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T. A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Takimoto G. S., Graham J. D., Jackson T. A., Tung L., Powell R. L., Horwitz L. D., Horwitz K. B. Tamoxifen resistant breast cancer: coregulators determine the direction of transcription by antagonist-occupied steroid receptors. J Steroid Biochem Mol Biol. 1999;69:45–50. doi: 10.1016/s0960-0760(98)00148-4. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Fondell J. D. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm. 2004;68:93–122. doi: 10.1016/S0083-6729(04)68003-4. [DOI] [PubMed] [Google Scholar]
- Urnov F. D., Wolffe A. P., Guschin D. Molecular mechanisms of corepressor function. Curr Top Microbiol Immunol. 2001;254:1–33. doi: 10.1007/978-3-662-10595-5_1. [DOI] [PubMed] [Google Scholar]
- Wagner B. L., Norris J. D., Knotts T. A., Weigel N. L., McDonnell D. P. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol. 1998;18:1369–78. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Anderson C.M., Valentine C., Nguyen P., Marimuthu A., West B.L., Baxter J.D., Kushner P.J. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14:1976–85. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., Parks D. J., Moore J. T., Kliewer S. A., Willson T. M., Stimmel J. B. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–7. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Zamir I., Zhang J., Lazar M. A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–46. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- Zhang X., Jeyakumar M., Petukhov S., Bagchi M. K. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol Endocrinol. 1998;12(4):513–24. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Gross W., Hong S.H., Privalsky M.L. The SMRT corepressor is a target of phosphorylation by protein kinase CK2 (casein kinase II) Mol Cell Biochem. 2001;220:1–13. doi: 10.1023/a:1011087910699. [DOI] [PMC free article] [PubMed] [Google Scholar]