Abstract
A major goal of the Nuclear Receptor Signaling Atlas (NURSA) is to elucidate the biochemical and physiological roles of nuclear receptors in vivo. Characterizing the tissue expression pattern of individual receptors and their target genes in whole animals under various pharmacological conditions and genotypes is an essential component of this aim. Here we describe a high-throughput quantitative, real-time, reverse-transcription PCR (QPCR) method for the measurement of both the relative level of expression of a particular transcript in a given tissue or cell type, and the relative change in expression of a particular transcript after pharmacologic or genotypic manipulation. This method is provided as a standardized protocol for those in the nuclear receptor field. It is meant to be a simplified, easy to use protocol for the rapid, high-throughput measurement of transcript levels in a large number of samples. A subsequent report will provide validated primer and probe sequence information for the entire mouse and human nuclear receptor superfamily.
Introduction
Characterizing the tissue expression pattern of individual receptors and their target genes in whole animals under various pharmacological conditions and genotypes is one of the aims of the Nuclear Receptor Signaling Atlas (NURSA). Here we describe a high-throughput quantitative, real-time, reverse-transcription PCR (QPCR) method for the measurement of both the relative level of expression of a particular transcript in a given tissue or cell type, and the relative change in expression of a particular transcript after pharmacologic or genotypic manipulation.
Reagents and Instruments
Thermal cycler
Although several instruments are now available, we chose to use the Applied Biosystems (ABI, Foster City, CA) 7900HT instrument because of its high-throughput capacity, its potential for use with robotics, and its relatively widespread availability in the field. The 7900HT is a rapid cycling instrument, with a single run lasting approximately 2 hours. The instrument allows the use of either 96-well or 384-well plates. The protocols employed herein use the 384-well format. The instrument also allows for the addition of a robotic plate loader arm, which enables unattended operation throughout the day.
RNA and cDNA preparation
RNA STAT-60 was obtained from Tel-Test, Inc., Friendswood, TX; cat. #CS-502). RNase-free DNase I was obtained from Roche Molecular Biochemicals, Indianapolis, IN; cat. #776785. 1X First Strand Buffer, 10mM DTT and SuperScript RTII reverse transcriptase, were obtained as a kit from Invitrogen, Carlsbad, CA, cat. #18064-014. 2mM dNTPs were obtained from ABI, cat. #N808-007, and 0.08µg/µl random hexamers from Roche Molecular Biochemicals, cat. #1034731.
QPCR Assay
384-well Optical Reaction Plate are obtained from ABI (cat. #4309849). Sybr® Green PCR MasterMix is obtained from Sigma (St. Louis, MO; cat. #S4438). 8-well PCR tube-strips are obtained from CLP, San Diego, CA; cat. #3426.8A) and the optical adhesive cover from ABI (cat. #4313663 or 4311971).
Methods
SYBR® Green I versus TaqMan® -based Assays
There are two types of fluorescence monitoring chemistries available for use on the ABI 7900HT, SYBR® Green, and TaqMan® [Applied Biosystems, 2003]. The first employs the DNA-intercalating dye, SYBR® Green I as the reporter fluorophore. It works like ethidium bromide by binding double-stranded DNA, which is the product of the PCR. As the reaction cycle progresses, the instrument monitors and records the increase in fluorescence over time. The SYBR® Green assay only requires a validated primer pair in addition to the regular PCR components.
The TaqMan® chemistry utilizes FRET (fluorescence resonance energy transfer) technology. It requires the use of an additional type of primer, called a probe, which is labeled with a fluorescent reporter dye on its 5′ end and a 3′ quencher dye. While the probe is still intact, the 5′ and the 3′ dyes are in close proximity, and the fluorescent signal is quenched via FRET. As the PCR product is synthesized, the probe, which sits on a specific sequence in a region between the forward and reverse primers, is cleaved by the nuclease activity of the Taq polymerase. As the 5′ reporter dye is released, it continuously fluoresces. One probe is cleaved for every PCR product made during the reaction, and the machine records the concomitant fluorescence increase over time.
There are pros and cons to each of the chemistries employed for QPCR. One advantage of SYBR® Green over TaqMan® is that the initial assay preparation only requires a few days for primer design and validation. TaqMan® requires the additional synthesis of the dual-labeled probe after the validation of the potential primer set using SYBR® Green. The probe synthesis usually takes 2-3 weeks, and is expensive relative to the cost of the primers. The increasing availability of validated TaqMan® primer-probe sets from several vendors may eventually bring the costs of this method down.
In general, TaqMan® has been considered to be more sensitive when detecting low copy numbers (<10 copies) because of its ability to resolve the signal of a single copy of template [Wittwer et al., 1997]. However, in most cases the sensitivity of the instrument is limited to >10 copies of template due to stochastic effects [Morrison et al., 1998]. SYBR® Green may have a slight edge in sensitivity at >10 copies because the reporter dye binds to any double-stranded DNA present in the reaction, and does not require a probe-cleavage event for the fluorescence detection, as does TaqMan®. The result is detection of the PCR product at earlier cycles [Wittwer et al., 1997]. This is especially important in the case of low-abundance transcripts (>10 copies), where the number of PCR cycles required for fluorescence detection above background might be beyond the range of cycles in the TaqMan®, but not the SYBR® assay. The double-stranded DNA binding property is also a disadvantage of SYBR® Green over TaqMan® because non-specific products and mRNAs with high sequence identity may be detected. One additional advantage of TaqMan® is that the probe offers an added layer of specificity in addition to the forward and reverse primers. The probe sequence must exactly match the target sequence to which it binds, as a single nucleotide difference in the probe sequence will prevent the cleavage event necessary to generate a reporter signal.
Endogenous Normalizer
One of the experimental controls included in a gene-expression assay is the loading, or internal control. It is used to normalize the signal value of each sample so that the differences between samples are the result of a real biological difference and not because of inconsistent loading. Housekeeping genes are the typical choice due to their mostly consistent expression levels in all cell types. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-actin, cyclophilin, and 18S rRNA are commonly used. Both GAPDH and β-actin have been shown to vary with numerous conditions [Suzuki et al., 2000], and are not the best choice. Cyclophilin expression is equal among most tissues, with the exception of heart and muscle (unpublished observation). These tissues show lower levels of the transcript relative to other tissues, so the normalized values for the transcript under study may be exaggerated. 18S may be the preferred standard because of its relatively invariant level among tissues and treatment conditions (also observed in northern blots). However, because of its very high expression, the concentration of starting template in a QPCR assay must be low, and the detection of the gene of interest may be lost at the lower template quantity. The choice of the endogenous reference, therefore, should be determined empirically and based on the system under study.
Primers
Perhaps the most important factor of QPCR analysis is the choice and validation of primers. The oligonucleotide sequences must be highly specific, which may be difficult when dealing with isoforms or splice-variants. A primer set that has been used in conventional, end-point analysis PCR may not be sufficient for quantitative measurements. The primer set must pass a set of validation parameters, as described later. One drawback of QPCR over northern analysis is that QPCR cannot detect the quality (i.e., the size) of an RNA message, the number of differently sized messages (e.g., splice-variants), or their expression levels relative to each other. One way around this problem is to design sub-type-specific primer sets, but unfortunately this strategy still does not result in any information concerning the overall quality of the transcripts. For this reason, it is worth investing the time to confirm the quality of the template RNA by northern analysis against a known transcript.
Quantitative Assay Types
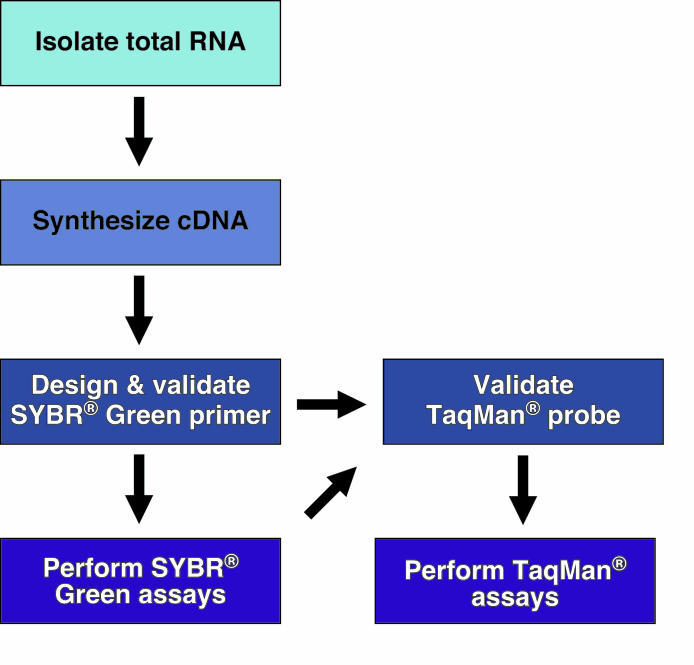
Each of the following sections describes the detailed methodology for performing both SYBR® Green and TaqMan® assays. Figure 1 depicts the order of events necessary to employ these techniques.
Figure 1. Typical work-flow for designing and implementing Real-Time PCR assays.
See text for more details
There are three quantitative methods: the absolute standard curve, the relative standard curve, and the comparative cycle time (Ct) methods [Applied Biosystems, 2003]. Herein, we describe methods for the relative standard curve and the comparative Ct (also called ΔCt) methods. For discussion concerning the absolute standard curve method [Applied Biosystems, 1997].
The relative standard curve method is used to assess the amount of a particular transcript in a group of samples. It is comparable to a multiple-tissue northern blot. It requires that a template dilution series of a cDNA standard be included for each gene on each plate. For relative quantitation, the template concentration values are arbitrary. Quantities interpolated from the resulting standard curve are used to calculate relative mRNA levels in each unknown sample.
The comparative Ct method (or ΔCt) is used to assess relative changes in mRNA levels between two samples, and does not require the use of the template dilution series. It does, however, require that the amplification efficiencies of the genes to be compared (described later) be the same [Applied Biosystems, 1997]. Cycle times, rather than interpolated template quantities, are used for the calculations. An example of employing the comparative Ct method would be the comparison of the expression of a nuclear receptor target gene in wild-type vs. receptor-null animals, or ligand-treated vs. untreated animals.
One-step vs. Two-step Reverse Transcriptase PCR
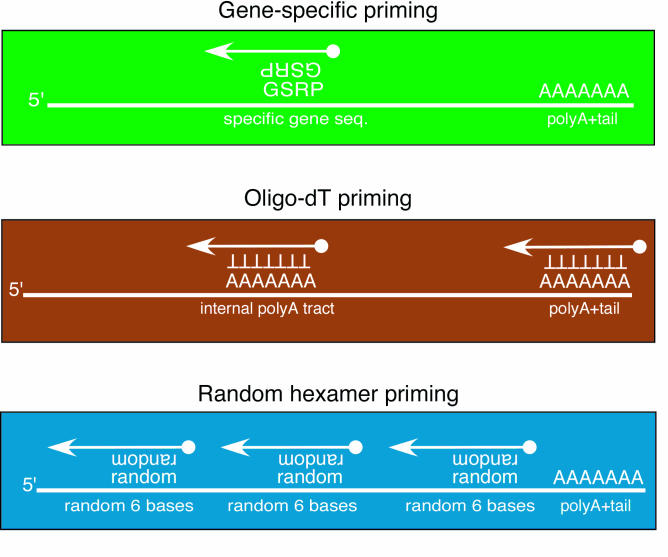
Reverse transcription (RT) is the enzymatic process by which complementary DNA (cDNA) is synthesized by an RNA-dependent DNA polymerase, i.e. reverse transcriptase. It requires a small oligonucleotide DNA primer. There are several types of primers, including gene-specific reverse primers (GSRP), oligo-(d)Ts, and random hexamers, also called p(dn)6. The GSRP is specific to the cDNA to be synthesized, and may be used to selectively RT a specific RNA transcript. Oligo-(d)Ts consist of short stretches of thymines and selectively target RNAs with a polyA+ tail, or any polyA tract. Random hexamers are mixtures of 6 base-pair primers with varying sequences. They randomly bind their complementary RNA sequences. A schematic representation of the various types of priming for reverse transcription is shown in Figure 2.
Figure 2. Representation of RNA priming for reverse transcription.
See text for more details
cDNA is synthesized in one or two steps. One-step RT-PCR is done in a single reaction in which the reaction mixture contains the reverse transcriptase and the PCR polymerase. Here, both the RT primer and one of the PCR primers is a GSRP. Two-step RT-PCR involves two separate reactions for the RT and the PCR, and the RT primer is either oligo-(d)Ts, or random hexamers. The RT reaction is carried out at 42°C to reduce or inhibit the occurrence of RNA secondary structure.
For quantitative RT-PCR (QPCR), random hexamers are used to ensure the efficient amplification of all RNAs in a sample. Because the primer sequences are random and, therefore, can bind anywhere along the RNA, there is a very high probability that all transcripts will be reverse transcribed at near 100% RT efficiency (ABI, personal communication). When using oligo-(d)Ts, only RNAs containing a polyA sequence will be amplified, and incomplete, or partially degraded RNA will not be detected in the subsequent QPCR assay. Use of the GSRP appears attractive at first, since a specific target will be amplified. This can be potentially problematic, however, because the RT efficiency of each transcript has to be taken into consideration. Also, RNA secondary structure may prevent the efficient RT of a specific target, and would also have to be taken into consideration.
RNA and cDNA Preparation
Please refer to the sections “Q-PCR supplies, reagents and stock solutions” and “Preparation of cDNA Standards” in the Supplementary File 1 for the detailed protocols and worksheets for this section.
Total RNA is prepared from cells or frozen tissues with RNA STAT-60 according to the manufacturer′s instructions. For a typical assay, 4µg of total RNA is treated with a 1:5 dilution of RNase-free DNase I in the presence of 4.2mM MgCl2 in a final volume of 20µl. This is performed in 0.2ml thin-walled PCR tubes in a standard thermocycler at 37°C, 30 min., 75°C, 10 min., and 4°C hold. The reverse-transcription mix consisting of 1X First Strand Buffer, 10mM DTT, 200U of SuperScript RTII reverse transcriptase, 2mM dNTPs and 0.08µg/µl random hexamers is then added directly to the tubes with the DNAse-treated RNA for a final volume of 100µl. The cDNA synthesis is carried-out in the thermocycler at 25°C for 10 min., 42°C for 50 min., 72°C for 10 min., and 4°C hold. Following the reverse-transcription, DEPC-treated H2O is added to the unknown samples to bring the volume to 200µl, and the cDNA concentration to 20ng/µl. (Note that samples used as cDNA standards are not diluted prior to making the 5-fold dilution series used in primer validation and standard curve assays.) This will result in enough template cDNA to test approximately 40 QPCR targets.
QPCR Primer Design for Assays using the ABI 7900HT
QPCR assays rely on a set of universal cycling conditions. The thermocycle, as well as the buffer conditions (MgCl2, salts, dNTPs, Taq DNA polymerase) will always be the same. One of the few parameters that does vary is the QPCR primer set. Because of this, the design and the pre-validation of the primers are essential to generate reliable data.
The nucleotide sequence and mRNA exon structure for each gene of interest are obtained from the NCBI Locus Link database [NCBI, 2003]. Ideally, the reference sequence (RefSeq) is obtained and used for primer design. If a RefSeq is not available, the GeneBank entry that gives an “NM_” or “complete cds” is used. If the mRNA to genomic alignment is not available from Locus Link, a BLAST search of the mRNA against the proper genome can be used to determine intron/exon boundaries.
For both SYBR® Green and TaqMan® -based assays, Primer Express™ Software (Applied Biosystems) is used to design the TaqMan®MGB Probe and Primer sets. The software returns a list of primer and probe sequences as matched primer/probe sets, and primers are chosen based on their binding sites. In order to amplify only mRNA, and not genomic DNA, the PCR product, or amplicon, should span the intron junction between two exons [Ambion, 2001]. When using TaqMan®, ideally, the probe should sit across the junction. Minimally, each primer should sit in completely separate exons. In some cases, the software may not return any sets that meet these criteria initially. In this case, two design parameters in the software may be adjusted, amplicon length and amplicon melting temperature (Tm). The amplicon length should be a minimum of 50 base-pairs, and maximum of 150 base-pairs, and if necessary, should be adjusted in 10 base-pair increments. Amplicon Tm′s usually start at 85C and can be adjusted upward to a maximum of 95C, especially in the case of GC-rich target sequences. Once the primers have been chosen, a general BLAST of each primer sequence is run to ensure their unique specificity.
Results
Initial Validation of Primer Sets
Please refer to the sections “SYBR® Green Template Titration Assay” and “TaqMan® Template Titration Assay” in Supplementary File 1 for the detailed protocols and worksheets for this section.
Oligonucleotides are purchased from a commercial vendor at the small-scale synthesis with the minimum purification. We purchase our primers from Integrated DNA Technologies at 25nM scale with standard desalting. We have found that additional purification, such as HPLC, is not necessary and only adds to the cost of each primer. The small-scale synthesis also adds to the savings, especially in the case that the primers do not work initially.
The oligonucleotides are resuspended to 100µM in sterile-filtered ddH2O, and an aliquot is diluted to 2.5µM. The forward and reverse 2.5µM primers are then mixed at a 1:1 ratio for use in subsequent assays. In the past, matrix tests, in which several combinations of different forward and reverse primer concentrations are assayed, were used to determine optimal final primer concentrations [Morrison et al., 1998]. We have found that this is no longer necessary, as 150nM final concentration each of forward and reverse primers has proven to be a universal condition in our assays.
To validate the primers, a template titration assay is done. For human transcripts, we use the Universal Reference Total RNA from BD Clontech (Palo Alto, CA). For mouse transcripts, a mixture of equal quantities of RNA from each of the following tissues is used: adrenal, whole brain, embryo, heart, intestine, kidney, liver, lung, muscle, ovary, pancreas, placenta, macrophage, skin, testis, and white adipose tissue. The assay consists of a 5-fold dilution series of cDNA reverse-transcribed from the universal RNA (50ng, 10ng, 2ng, 0.4ng, 0.08ng, 0.016ng), and 2 control samples: a no template control (NTC), and a no reverse transcriptase (-RT) control. Amplification of the NTC sample indicates the presence of primer-dimer formed during the reaction. The difference in cycle time between the NTC and the experimental samples must be greater than seven to ensure that the assay is monitoring specific fluorescence of the experimental sample (personal communication, ABI). The -RT sample is included to confirm the absence of genomic amplification.
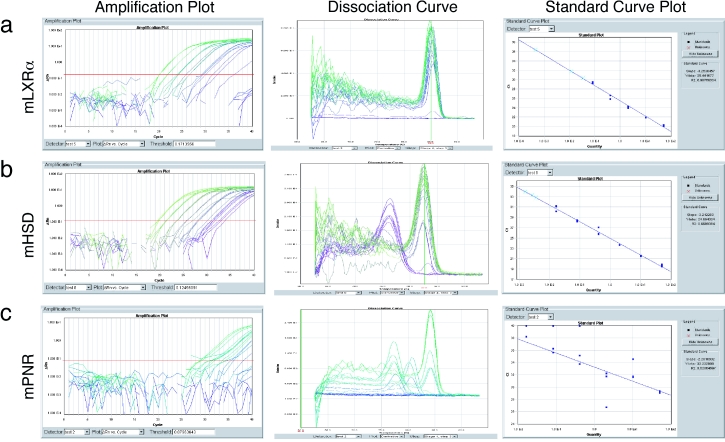
After the template titration assay run is complete, the ABI 7900HT instrument software, SDS2.1, will plot a standard curve and a dissociation curve for each target gene. A valid primer set should have a slope of –3.3 and a correlation coefficient (R2-value) >0.95 for the standard curve. In addition, the dissociation curve should appear as a single “stacked” peak at the amplicon Tm determined by the Primer ExpressTM software. Examples of a typical standard curve plot for a valid primer set (e.g., using mouse LXR-α), an invalid primer set due to non-specific priming (e.g., using mouse hydroxysteroid dehydrogenase [HSD]), and an invalid primer set due to poor amplification (e.g., using mouse PNR) are shown in Figure 3. The slope of the standard curve is indicative of PCR efficiency (formula 1, Figure 4), given by formula 2, Figure 4 [Applied Biosystems, 2001]. The slope can be affected by template quality, pipetting errors, etc., and for this reason, a standard curve for the endogenous reference gene is always run on the same plate. The difference in slopes of the reference gene and the gene of interest should be less than 0.1 [Applied Biosystems, 1997].
Figure 3. Primer set validation.
(a) Example of a valid primer set for mouse LXR- α using SYBR® Green. The presence of a single peak in the dissociation curve and the -3.3 slope and 0.99 R2 value of the standard curve plot are indicative of a good set of primers. (b) Example of an invalid primer set for mouse HSD due to the amplification of non-specific products, as indicated by the presence of multiple peaks in the dissociation curve. This may be an effect of non-specific priming or primer dimerization. Note that the slope of the standard curve plot (-3.2) is within the acceptable range of a valid primer set, but the dissociation curve renders this set of primers unacceptable. (c) Example of an invalid primer set for mouse PNR due to poor amplification. Note the unacceptable slope (-2.28) in the standard curve plot, and the presence of multiple peaks in the dissociation curve.
Figure 4. Formulas for Q-PCR calculations.
See text for more details.
Once a primer set has been validated using the SYBR® Green chemistry, switching to the TaqMan® chemistry is as simple as adding the probe at 250nM final concentration, and switching to the buffer optimized for use with TaqMan® probes (ABI cat. #4324018).
QPCR Assay Preparation
The QPCR assay is run in a 384-well Optical Reaction Plate with 10µl final volume per well. Each sample is run in triplicate for each gene to be assayed. The reaction consists of 25ng template cDNA, 1X SYBR® Green PCR MasterMix and 150nM each of forward and reverse primer. Reaction components are pre-mixed in 8-well PCR tube-strips and then pipetted with a multi-channel pipette into the reaction plate. Premixing the components decreases the amount of user-introduced variation by ensuring a homogeneous mix of template and primers, and also by minimizing the number of pipettes, which can introduce bubbles and/or aerosol contaminants, into the optical plate itself [Applied Biosystems, 2001]. The plate is then covered with an optical adhesive cover and centrifuged to bring the liquid to the bottom of the wells of the plate.
Data Analysis
Standard Curve Method
Please refer to the sections “SYBR® Green Standard Curve QPCR Assay Worksheet” and “TaqMan® Standard Curve QPCR Assay Worksheet” in Supplementary File 1 for the detailed protocols and worksheets for this section.
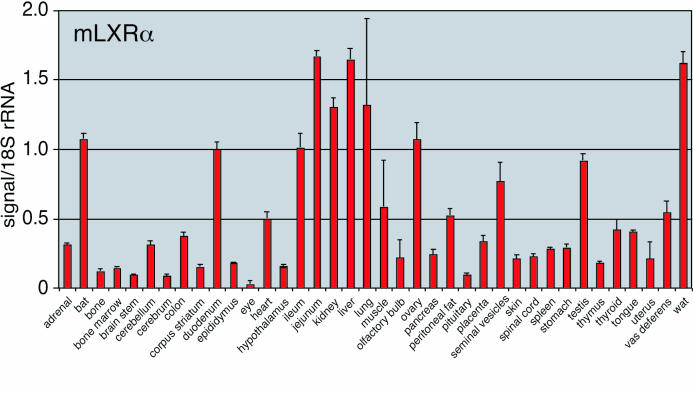
The instrument software calculates the quantity of transcript in each unknown sample based on the linear regression formula of the standard curve, and data are exported as a tab-delimited text file. Further data analyses are done using Microsoft Excel, or another comparable program. For each sample, the quantity of the gene of interest (GOI) and the reference gene (reference) are determined in triplicate, and from these values, the average transcript quantity (avg), the standard deviation of the average (stdev), and the coefficient of variation (formula 3, Figure 4) of the average is determined, given by formula 4, Figure 4. A single outlier point that has a value >17%CV may be removed from the calculations. To determine the mRNA level in each unknown sample, the gene of interest is normalized to the reference gene, e.g. 18S rRNA, to account for cDNA loading differences and calculated using formula 5, Figure 4. The resulting normalized values are plotted as a bar graph ± the standard deviation (formula 6, Figure 4), given by formula 7, Figure 4. An example of LXR- α expression in 38 mouse tissues is shown in Figure 5.
Figure 5. Expression profile for mouse LXR- α generated using the standard curve method.
The relative levels of the mRNA transcript are shown (S.D. (from triplicate readings). 18S rRNA was used as the normalizer gene so that the level of LXR- α may be compared between tissue types.
Comparative Ct, or ΔΔCt Method
Please refer to the sections “SYBR® Green ΔΔCt QPCR Assay Worksheet” and “TaqMan® ΔΔCt QPCR Assay Worksheet” in Supplementary File 1 for the detailed protocols and worksheets for this section.
For each of the 3 replicates of a sample, the average cycle time (Ct), the standard deviation (stdev), and the coefficient of variation (formula 3, Figure 4) is calculated, given by formula 4, Figure 4. Outlier points >4%CV are removed from the calculations. For each sample, the average Ct of the GOI (avg. CtGOI) is normalized to the average Ct of the reference gene (avg. Ctref) for the same sample to calculate the normalized Ct for the GOI (formula 8, Figure 4) using formula 9, Figure 4. The standard deviation of the ΔCt is calculated using formula 10, Figure 4. The calibrator is chosen to be the sample, the tissue, the gene, or the control group to which the others will be compared. For example, in an experiment in which wild-type mice are compared to knockout mice, the calibrator would be the wild-type mouse sample. The ΔΔCT, or calibrated value, for each sample is given by formula 11, Figure 4. The stdev ΔΔCt is the same as stdev ΔCt. The fold-induction for each sample relative to the calibrator = 2(-ΔΔCt). The resulting induction values are usually plotted as a bar graph. The measure of experimental error is the standard deviation of the mean (stdevfold change), given by formula 12, Figure 4. If there are multiple samples in multiple treatment groups, the average the fold induction for each group is plotted. The biological variance is the standard error of the mean, given by formula 13, Figure 4. An example is shown in Figure 6, where the induction of SREBP-1c expression is compared between wild-type mice fed control vehicle or one of a number of typical nuclear receptor ligands.
Figure 6. Typical experiment in which wild-type male mice were treated with several different nuclear receptor agonists.
The relative changes in mRNA expression for known receptor target genes, in this case SREBP-1c, an LXR target gene, were measured using the comparative Ct method. (a) SREBP-1c expression plotted for each of the animals (S.D. Each color represents a different experimental group (n=4 animals). (b) SREBP-1c expression plotted as averages of the fold changes for the 4 animals in each treatment group shown in (a) (SEM. Note that the SREBP-1c transcript increases relative to control (VEH) in the animals treated with an RXR agonist (LG268), an LXR agonist (T1317), or both LG269 and T1317 together (T+LG). Note that ligands for PPAR- α (fenofibrate), PPAR- γ (troglitazone), FXR (CDCA), PXR (PCN), and CAR (TCPOBOB) have no effect on the level of the SREBP-1c message relative to control.
Discussion
Here, we have described a simple to use, yet effective real time QPCR methodology. The protocols and the worksheets provided are intended to introduce and standardize the assay for researchers in the nuclear receptor field, but may be applied to any field of study. In a subsequent publication, a complete set of QPCR primers and TaqMan® probes for all mouse and human nuclear receptors will be reported.
Supplementary Material
Acknowledgments
We thank all current and former lab members for invaluable critique and discussions concerning the development of these techniques, in addition to the labor involved with the animal studies. We also thank Applied Biosystems for the technical support. Any questions or suggestions concerning the downloadable materials are encouraged and should be addressed to Angie Bookout at abookout@biochem.swmed.edu. DJM is an investigator of the Howard Hughes Medical Institute. This work was funded in part by the NIH NURSA grant number NIDDK: U19DK62434.
Abbreviations
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GOI
Gene of interest
- GSRP
Gene-specific reverse primer
- PCR
polymerase chain reaction
- QPCR
Quantitative real-time PCR
- Tm
Amplicon melting temperature
References
- Technotes Newsletter. Vol. 8. Ambion; 2001. The Top 10 Most common quantitative PCR pitfalls. [Google Scholar]
- Applied Biosystems: 1997. Relative Quantitation Of Gene Expression: ABI PRISM 7700 Sequence Detection System: User Bulletin #2: Rev B. [Google Scholar]
- Applied Biosystems: 2001. ABI Prism 7900HT User Manual. [Google Scholar]
- Applied Biosystems; 2003. Essentials of Real Time PCR. [Google Scholar]
- Morrison T. B., Weis J. J., Wittwer C. T. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–8. [PubMed] [Google Scholar]
- Locus Link. NCBI; 2003. [Google Scholar]
- Suzuki T., Higgins P. J., Crawford D. R. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- Wittwer C. T., Herrmann M. G., Moss A. A., Rasmussen R. P. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–1. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.