Abstract
A series of data has accumulated over the past five years that raises questions about our current understanding of the transcriptional process and its regulation. Following the discovery of coactivators for nuclear receptors (NRs), a large number of these molecules have been reported in the literature. This perspective will summarize some opinions on the significance of this large number of factors.
Sequentiality and Processivity of Nuclear Receptor Coregulators
A series of data has accumulated over the past five years that raises questions about our current understanding of the transcriptional process and its regulation. Following the discovery of coactivators for nuclear receptors (NRs), we have been amazed at the plethora of these molecules that have been reported in the literature [] McKenna et al., 1999 ; [ McKenna and O'Malley, 2002 ]. A cursory inspection, not attempting to critique and eliminate those that may not be true coactivators, sets the number at over 100. The impressive size of this number begs an explanation – and the explanation appears to reside in the many diverse cellular roles for coactivators.
The large number of coactivators is explained in part by the fact that our concept of coactivator function has expanded considerably. As we might have guessed, there is receptor selectivity for coactivators [ Li et al., 2003 ], although how many of the 48 human nuclear receptors use distinct or unique coactivators is unknown at present. Evidence for ligand-specific recruitment of coactivators is in the literature [ Korzus et al., 1998 ], and promoter-gene specific requirements for coactivators probably occur in certain instances [ Puigserver et al., 1998 ]. Interestingly, there is evidence for cell specific expression for only a limited number of coactivators .
Data are accumulating rapidly that suggest that coactivators play major roles in many diverse cellular processes such as the coordination of expression of gene sets that regulate metabolic processes [ Picard et al., 2002 ; Puigserver et al., 1998 ]. These molecules have the capacity to bind to functional sets of DNA-bound transcription factors and coordinately enhance the expression of groups of genes required for certain metabolic functions. Coactivators are known to play a role in regulating cell growth, and consequently are often overexpressed in cancers [ Anzick et al., 1997 ]. Coactivators also act as intracellular targets for signaling from membrane receptors [ Wu et al., 2002 ], providing homeostatic sensing to the internal NR pathways from membrane receptor regulated pathways.
In terms of transcriptional regulation, we initially considered coactivators as simply ‘power boosters′ for DNA-binding transcription factors such as NRs, capable of accelerating the process of transcription initiation. We now understand that coactivators are involved in more diverse steps of transcription than only the step of ‘initiation′ of transcription. This new information leads us to reconsider some of our ideas on hormonal regulation of the transcription process.
There are a few newly available pieces of data that must be taken into account in any theory of NR mediated gene activation. Firstly, it recently has been shown that coactivators (in addition to NRs themselves) have a very short residence time at promoter sites during real time imaging of the nucleus [ McNally et al., 2000 ; Stenoien et al., 2001 ]. Secondly, in steady state conditions in cells, coactivators exist in high molecular weight complexes (~4-6 proteins), and thus are unlikely to enter the regulatory complex one by one [ McKenna et al., 1998 ]. Our data indicates that these high molecular weight complexes are very heterogeneous and contain many combinations of different coactivators. Thirdly, it is likely that in most instances, a member of the SRC-1 family forms the initial primary bond with steroid receptors to initiate transcription. Fourthly, NR coactivators play a role in alternative RNA splicing [ Auboeuf et al., 2002 ; Monsalve et al., 2000 ] and in termination of regulated transcription [ Lonard et al., 2000 ; Nawaz et al., 1999 ], and perhaps other subreactions of the transcriptional process. Fifthly, most of the published coactivators interact directly with NRs and do so at only two major sites, the AF-2 region in the ligand binding domain and the AF-1 region in the N-terminus.
Most certainly, coactivator binding to receptor AFs is governed by a combination of the intracellular concentration of each of the coactivators and their individual affinities for a given receptor AF. The product of these two parameters governs occupancy of a given receptor binding site. Thus, many coactivators may not come into play at a given receptor, when occupied by a given ligand at a given promoter. Biochemical reactions in a cell are in equilibrium and are thereby open to shifts in this equilibrium. For example, a cell may increase its concentration of a specific coactivator under certain conditions. An example is the demonstration that fasting increases the concentration, and thus function, of PGC-1 in the liver. This is not the norm for coactivators, however, since most do not change dramatically in concentration during the lifetime of a normal differentiated cell. Perhaps more commonly, post-translational modifications of coactivators increase or reduce the affinities of a coactivator for a given set of transcription factors. Post-translational modifications of coactivators, such as phosphorylation or acetylation/methylation have been reported to increase or decrease the affinity of the modified coactivator for target receptors or other coactivators [ Dutertre and Smith, 2003 ; Rowan et al., 2000 ].
Nevertheless, despite the above explanations which could account for a significant proportion of coactivators, the extensive number of reported coactivators that stimulate NR mediated transcription is still very large, and the fact remains that the vast majority of these coactivators bind at the same AF-1 and AF-2 sites of receptors. These two facts force us to ‘think out of the box′ in terms of their function.
Thus, we and others have searched for additional novel functions not easily predicted for coactivators at the time of their discovery. Early on, coactivators were discovered that appear to dictate ubiquitinylation of the coactivator complex and were predicted to play a role in short-term or long-term turnover of the active complex by the 26S proteasome [ Lonard et al., 2000 ; Nawaz et al., 1999 ]. A subset of coactivators has been implicated in the regulation of alternative mRNA processing; receptor-coactivator regulation of RNA splicing occurs coordinately with transcriptional regulation [ Auboeuf et al., 2002 ]. Perhaps future studies even will extend their functions to the other intermediate reactions of RNA chain elongation, RNA 5′ - and 3′ - processing, and transport of mRNA to the cytoplasmic ribosomes. These considerations might account for the presence of many more coactivators in cells, but only if they do not act simultaneously, because they all must enter AF-2 and AF-1 regions to perform their functions.
Consequently, an argument can be made for sequential kinetics for coactivator-complex entry to AF-1/AF-2 sites. This explanation is not as radical as it sounds, since it already has been suggested as an explanation to resolve the conflict as to whether SRC/p160 family members are the important coactivators for NRs such as TR and VDR, or whether the SMCC-type (TRAPs/DRIPs) complex of coactivators (TRAPs/DRIPs) represent the functional complex [ Oda et al., 2003 ; Sharma and Fondell, 2002 ]. If they act in sequence, then both are necessary but not sufficient.
Once we turn to kinetics for an explanation of the AF-dependent interaction, why not take the hypothesis one step further- and predict sequential interactions for multiple additional coactivator complexes in each target gene activation. It is now only one additional leap of faith to suggest that there also may be many additional nonproductive interactions for coactivator complexes that currently are beyond our ability to monitor with time-dependent ChIP analyses, or to prove even with technologies such as real- time live imaging. It might be remembered that gene expression may not be effected by the ′exact′ series of reactions that are indicated in our diagrams of the process. RNA polymerase II makes many false starts and forms multiple short incomplete RNA chains in the course of gene transcription. In other words, each RNA start by the enzyme is not necessarily productive.
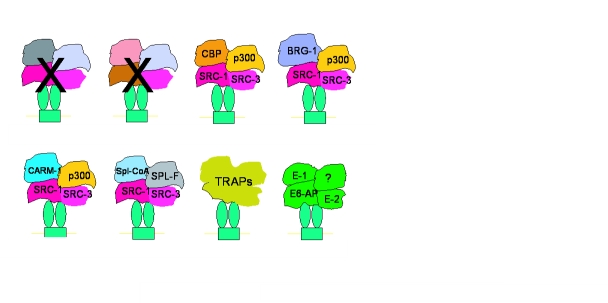
In Figure 1 , we speculate that a sequence of stochastic nonproductive interactions of diverse coactivator complexes leads to an eventual productive interaction with a steroid receptor dimer in place at the promoter of a gene. The exact complexes and sequence are less important than the concept. When a specific and productive interaction occurs, transcription advances one functional step forward; the receptor next must seduce a new productive interaction with a different requisite coactivator complex that advances the gene through the next subreaction needed for effective transcription. Each individual step in this process is likely replete with additional nonproductive interactions, but since the half-life of coactivator interactions at promoters has been estimated to be less than 20 seconds, little time is lost in discharging an inappropriate complex and recruiting the next correct coactivator complex required to produce a translatable mRNA.
Figure 1 . Model for sequential interactions of diverse coactivator complexes with a steroid receptor dimer at the promoter of a gene.
See text for more details.
Such a sequence of events is not illogical. In fact, it may be illogical to propose a precise vectoral movement of coactivator complexes directly to their sites of action. There is no doubt that subcellular targeting is possible using peptides and other types of signals, but it is difficult to accept this as an explanation for the myriad individual gene activities in the eukaryotic nucleus, especially in the face of the minimal and rate-limiting cellular concentrations of coactivators. How the correct receptor-coactivator complex for a substep of transcription might signal its validity when it enters the promoter is unknown. Suspicion centers on the many recently discovered posttranslational modifications capable for proteins on the chromosome, including phosphorylation, methylation, acetylation, ubiquitinylation/sumoylation and glycosylation [ McKenna and O'Malley, 2002 ].
When finally defined, the series of steps in decoding a gene is likely not to be as linear and precise as we once thought. The rapid kinetics inherent to simple diffusion and concentration may lie at the heart of the process. In any event, as usual in biology, it will appear beautiful in its evolutional simplicity when unraveled.
Abbreviations
- AF-1
activation function 1
References
- Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. IB1, a steroid receptor coactivator amplified in breast and ovarian cancer . Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Auboeuf D., Honig A., Berget S. M., O'Malley B. W. Coordinate regulation of transcription and splicing by steroid receptor coregulators . Science. 2002;298:416–9. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- Dutertre M., Smith C. L. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-α: regulation by phosphorylation sites in the A/B region depends on other receptor domains . Mol Endocrinol. 2003;17:1296–314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., Rosenfeld M. G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions . Science. 1998;279:703–7. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- Li X., Wong J., Tsai S. Y., Tsai M. J., O'Malley B. W. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification . Mol Cell Biol. 2003;23:3763–73. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard D. M., Nawaz Z., Smith C. L., O'Malley B. W. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation . Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., Lanz R. B., O'Malley B. W. Nuclear receptor coregulators: cellular and molecular biology . Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., Nawaz Z., Tsai S. Y., Tsai M. J., O'Malley B. W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo . Proc Natl Acad Sci U S A . 1998;95:11697–702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators . Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- McNally J. G., Muller W. G., Walker D., Wolford R., Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells . Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B. M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1 . Mol Cell. 2000;6:307–16. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Nawaz Z., Lonard D. M., Smith C. L., Lev-Lehman E., Tsai S. Y., Tsai M. J., O'Malley B. W. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily . Mol Cell Biol. 1999;19:1182–9. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Sihlbom C., Chalkley R. J., Huang L., Rachez C., Chang C. P., Burlingame A. L., Freedman L. P., Bikle D. D. Two distinct coactivators, DRIP/mediator and SRC/p160, are differentially involved in vitamin D receptor transactivation during keratinocyte differentiation . Mol Endocrinol. 2003;17:2329–39. doi: 10.1210/me.2003-0063. [DOI] [PubMed] [Google Scholar]
- Picard F., Gehin M., Annicotte J., Rocchi S., Champy M. F., O'Malley B. W., Chambon P., Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues . Cell. 2002;111:931–41. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis . Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rowan B. G., Garrison N., Weigel N. L., O'Malley B. W. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein . Mol Cell Biol. 2000;20:8720–30. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Fondell J. D. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo . Proc Natl Acad Sci U S A . 2002;99:7934–9. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D. L., Nye A. C., Mancini M. G., Patel K., Dutertre M., O'Malley B. W., Smith C. L., Belmont A. S., Mancini M. A. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α-coactivator complexes in living cells . Mol Cell Biol. 2001;21:4404–12. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S. Y., Tsai M. J., O'Malley B. W. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase . Mol Cell Biol. 2002;22:3549–61. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]