Abstract
Here, we describe the identification and characterization of the cytokinesis-deficient mutant cell line 17HG5, which was generated in a restriction enzyme–mediated integration mutagenesis screen designed to isolate genes required for cytokinesis in Dictyostelium discoideum. Phenotypic characterization of the 17HG5 cell line revealed no apparent defects in the global functionality of the actomyosin cytoskeleton except for the observed cytokinesis defect when grown in suspension culture. Plasmid rescue was used to identify the disrupted gene locus (pats1; protein associated with the transduction of signal 1) that caused the cytokinesis defect. Disruption of the pats1 locus was recreated through homologous recombination in several independent cell lines, each recapitulating the cytokinesis-defective phenotype and thereby confirming that this gene locus is important for proper cytokinesis. Sequence data obtained by analysis of the genomic region flanking the inserted restriction enzyme–mediated integration plasmid revealed an 8892-bp genomic open reading frame encoding a 2964-amino-acid protein. The putative pats1 protein contains 3 regulatory domains (RI-phosphatase, RII-GTP–binding, R-III protein kinase), 13 leucine-rich repeats, and 8 WD-40 repeats. These regulatory domains coupled with the protein–protein interacting domains suggest that pats1 is involved in signal transduction during cytokinesis in Dictyostelium.
INTRODUCTION
Cytokinesis, the final step in cellular division, has been an area of intense study for several years (for review, see Glotzer, 1997; Robinson and Spudich, 2000). Recently, our understanding of cytokinesis has improved, but the overall regulation of this highly orchestrated event still remains a mystery. For example, in late anaphase, several proteins involved in the formation of the contractile ring begin to align at the cellular midbody, and by the end of telophase, the well-developed contractile ring begins to constrict through an actin-myosin–mediated interaction (for review, see Robinson and Spudich, 2000). Finally, the dividing cell is cleaved into two separate daughter cells. Although many of the proteins associated with cytokinesis have been identified, how these proteins assemble at the proper time and in the correct orientation to achieve cytokinesis is not well understood.
Taking advantage of the power of molecular genetic approaches afforded by the model organism Dictyostelium discoideum, De Lozanne and Spudich (1987) and Knecht and Loomis (1987) demonstrated the requirement for myosin II in cytokinesis. Cells in which the heavy chain of the myosin II protein (MHC) was knocked out, either through homologous recombination (De Lozanne and Spudich) or through antisense RNA (Knecht and Loomis), became large and multinucleated when grown in suspension culture yet appeared normal as attached cultures. Both wild-type and myosin II–null cells are able to propagate as stationary cultures when either attachment-assisted mitotic cleavage (Neujahr et al., 1997) or traction-mediated cytofission (Fukui et al., 1990) is used. This phenotype has since served as a paradigm for subsequent searches for cytokinesis-specific genes (Larochelle et al., 1996; Vithalani et al., 1996). These screens were based on the use of restriction enzyme–mediated integration (REMI) (Kuspa and Loomis, 1992) to randomly mutagenize the Dictyostelium genome and then screen the resulting mutant cells for defects in cytokinesis by comparing their ability to propagate in suspension versus stationary culture.
Through this REMI screen, racE, a unique member of the rho family of small GTPases, was identified as being required for cytokinesis in Dictyostelium (Larochelle et al., 1996). Furthermore, racE was localized to the cell cortex through GFP tagging (Larochelle et al., 1997), and it was shown that racE-null cells have a reduced level of cortical tension, leading to their failure in cytokinesis when grown in suspension culture (Gerald et al., 1998). The REMI screen was also used in the identification of LvsA, a protein required for cytokinesis in Dictyostelium (Kwak et al., 1999). More recently, LvsA was classified as a member of the class II BEACH protein family (Wang et al., 2002). The function of this novel protein family has not been elucidated, but it is predicted that class II members are involved in cytokinesis.
In addition to the REMI screen, other approaches have also identified genes involved in cytokinesis in Dictyostelium. For example, deletion of coronin, an actin-binding protein that localizes to the polar edges of dividing cells (de Hostos et al., 1991), results in cells that become large and multinucleated when grown as stationary cultures (de Hostos et al., 1993). Cortexillin I and II, actin-bundling proteins that localize to the leading edges in nondividing cells, require double knockouts to observe a cytokinesis defect (Faix et al., 1996). Furthermore, both cortexillins are recruited to the nascent cleavage furrow, with cortexillin I arriving before myosin II. In myosin II–null cells, cortexillin I recruitment is further enriched, suggesting that cortexillin I and myosin II play synergistic roles in cytokinesis (Weber et al., 2000).
Here, we describe the identification of a novel gene, pats1 (protein associated with the transduction of signal 1), that is required for cytokinesis in Dictyostelium. Using REMI mutagenesis, we disrupted the pats1 locus near its 3′ end, resulting in the formation of the cytokinesis-defective cell line 17HG5. 17HG5 cells grown in suspension culture become large and multinucleated, similar to myosin II–null cells. Further analysis of the pats1 locus revealed several putative regulatory domains, and the overexpression of the RIII domain fused to GFP resulted in a severe cytokinesis defect in cells when grown as stationary cultures. Our analysis suggests that pats1 is a key regulator of cytokinesis in Dictyostelium.
MATERIALS AND METHODS
REMI and Plasmid Rescue
REMI was used to generate Dictyostelium mutants, which were subsequently screened for cytokinesis defects as described previously by Larochelle et al. (1996). In short, parental DH-1 Dictyostelium cells were electroporated in the presence of both BglII linearized pRHI30 and the restriction endonuclease Dpn II. The transformed cells were plated into 96-well plates and grown in FM media. Uracil-selected colonies were screened for cytokinesis defects by duplicate plating into 24-well plates. One cell culture was kept stationary, and the duplicate was maintained in suspension by shaking at 225 rpm. Transformants capable of growing in stationary culture but not in suspension were considered cytokinesis mutants and were investigated further. The transformant 17HG5 was characterized further because of its strong cytokinesis-deficient phenotype. Southern blot analysis of 17HG5 genomic DNA (gDNA) was performed by digesting gDNA with restriction enzymes that cut outside of pRHI30. The digested gDNA was electrophoresed, transferred to a nylon membrane, and probed with 32P-labeled pRHI30 to determine which enzymes would be useful for plasmid rescue (anything larger than 4.5 kilobases [kb], the size of pRHI30, but less than ∼10 kb).
For plasmid rescue, 17HG5 cells were grown in five 15-cm plates until confluent. Genomic DNA was isolated and independently digested with each of the following restriction enzymes: BclI, HpaI, or HindIII (enzymes determined from Southern blot analysis; L. Kuchnicki, D. Larochelle; unpublished data). The digested samples were then ligated with T4 DNA ligase and transformed into Escherichia coli JM 109 cells via electroporation. Ampicillin-resistant bacterial colonies were screened for gDNA insert by restriction endonuclease digestion, and any recovered plasmids with inserts of correct size were sequenced.
Recapitulation
The genetic disruption in 17HG5 cells was recreated in the DH-1 parental cell line by homologous recombination (Larochelle et al., 1996). Briefly, 1.0 × 105 DH-1 cells in 125 μl of electroporation buffer (50 mM sucrose, 10 mM sodium phosphate, pH 6.1) were placed in a 0.1-cm gap electroporation cuvette along with 5 μg of HindIII-linearized pLC4B2 DNA (from HindIII plasmid rescue). The cells were electroporated two times at 0.85 kV, 25 μF with a Bio-Rad electroporator with a 5-min interval on ice between electroporations. The cells were plated into three 96-well plates and selected in FM media lacking uracil. Cells were fed weekly until colonies appeared. Colonies were then screened for cytokinesis defects as described above. Multiple cytokinesis-defective cell lines were isolated, and the clones 6AD8 and 6BE9 were investigated further by Southern blotting. Genomic DNA from 6AD8, 6BE9, 17HG5, and DH-1 cells was digested with HindIII and analyzed by Southern blotting with 32P-labeled SLA-465 as the probe.
Sequence Analysis
All DNA sequencing reactions were carried out using the Applied Biosystems BigDye terminator cycle sequence ready reaction mix with appropriate primers and template and separated on a Perkin Elmer-Cetus ABI Prism 377 DNA Sequencer. Analysis of the sequence was performed using Sequencher software. DNA sequence was used as a template to search the Dictyostelium genomic database from the DGP Dictyostelium web site at the University of California, San Diego, La Jolla, CA; http://www-biology.ucsd.edu/others/dsmith/dictydb.html#A, and the Dictyostelium Genome Sequencing project web site at The Baylor College of Medicine, http://dictygenome.bcm.tmc.edu/bd/dicty_blast.html. Also, the NCBI database at http://www.ncbi.nlm.nih.gov/was used to find homologous sequences and proteins as well as conserved domains. The Motif Scan in the Protein Sequence web site, http://hits.isb-sib.ch/cgi-bin/PFSCAN, was used to scan the protein sequence for any conserved motifs. WD-40 repeats were identified using the BioMolecular Engineering Research Center (BMERC) PSA server at http://bmerc-www.bu.edu/psa/.
GFP Transformants
GFP fusion proteins were made using the cloning vector pTX-GFP supplied by Tom Egelhoff (Levi et al., 2000). The following four primers were designed to incorporate the R-III domain and carboxy-terminal WD-40 repeat region, respectively, into the BamHI site of pTX-GFP: JA-13, 5′-CGCGGATCCTTCGTACCATGTTTCCATTG; JA-14, 5′-CGCGGATCCTGATTTACTTGGAGAGGATTG; JA-6, 5′-GGGGATCCACTGCAGCTCCTTCTACAA; and JA-15, 5′-CGCGGATCCTGAATTACTACTACCACC. PCR products were band-purified and cloned into the pTX-GFP expression vector. The recombinant plasmids were then transfected into DH-1, 17HG5, and MHC-null cells as described above.
The GFP-MHC vector was obtained from Arturo De Lozanne and transfected as described above into DH-1 and 17HG5 cells. Colonies were collected and clones were pseudosynchronized as described previously (Gerald et al., 1998), with the following modifications. Log-phase cells were diluted into fresh media, passed onto coverslips, placed into six-well plates, and fixed at different time intervals.
Fixation
All cells were fixed for microscopic observation as described previously (Gerald et al., 2001). Briefly, cells were grown in six-well plates on 22 × 22-mm glass coverslips in HL-5 medium. The cells were fixed in 30% picric acid, 2% paraformaldehyde, 10 mM PIPES, pH 6.5, for 30 min, followed by a 10-min bath in 70% ethanol. Finally, the cells were stained with 0.08 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in PBS, pH 7.3, containing 25 mM magnesium chloride for 20 min and washed 2 times in PBS, pH 7.3, for 5 min each wash. The coverslips were then mounted onto slides in 25% glycerol, 100 mg/ml 1,4-diazabicylclo-[2.2.2]octane (DABCO), and sealed with nail polish.
All images were captured on a Nikon E600 microscope fitted with phase-contrast, DIC, and epifluorescence optics and a Spot RT Slider cooled CCD camera. Images were initially captured using SPOT software and were later processed using Adobe Photoshop software.
Triton-Insoluble Cytoskeleton
Triton-insoluble cytoskeletons were collected as described by Kolman et al. (1996). Briefly, 1.5 × 106 cells were pelleted by microcentrifugation at 2500 rpm for 2 min and resuspended in 150 μl 100 mM MES, pH 6.8, 2.5 mM EDTA, 5 mM MgCl2, and 2 mM ATP. An equal volume of the same buffer containing 1% Triton X-100, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 17.42 μg/ml phenylmethylsulfonyl fluoride was added to each sample before vortexing for 15 s. The samples were then centrifuged for 2 min at 14,000 rpm at 4°C, and the soluble supernatant was removed from the insoluble pellet. The pellet was resuspended in 25 μl SDS-PAGE loading buffer and boiled for 3 min, and the supernatant was first acetone-precipitated and then resuspended in 25 μl SDS-PAGE loading buffer and boiled. The samples were run on duplicate SDS-PAGE gels; one gel was processed for Western blotting and the other for Coomassie blue staining.
Rapid Amplification of cDNA Ends
Both three-prime and five-prime rapid amplification of cDNA ends reactions were carried out as described by Frohman et al. (1988) using the respective gene-specific primers: JA-4, 5′-GTCCAAATCAAGCTTCTCAAAGTGC-3′ and JA-24, 5′-TATATCATTGAAAGT-GGTTATTTCTG-3′.
Cell Culture
All cells were grown in HL-5 media as stationary cultures unless otherwise noted. DH-1 cells were supplemented with uracil at 40 μg/ml. GFP control, GFP R-III, GFP WD-40 repeat domain, and GFP MHC transformants were grown in HL-5 plus G418 at 10 μg/ml.
Concanavalin A Capping
Cell-surface capping was assayed using FITC-labeled concanavalin A as previously described (Larochelle et al., 1996; Kwak et al., 1999)
Sodium Azide
Cells were grown in HL-5 media in 3-cm plates until confluent. The medium was then removed, and the cells were placed in 20-mM sodium azide in HL-5. Cells were viewed under phase-contrast microscopy at 1-min intervals for 5 min and every 5 min after that until 30 min had elapsed.
Antibodies
Rabbit GFP antibodies were purchased from Molecular Probes (Eugene, OR) and used for GFP and GFP-fusion protein detection in Western blotting. Goat anti-rabbit HRP-conjugated antibodies from Bio-Rad (Hercules, CA) were used as secondary antibodies in Western blot detection.
RESULTS
Phenotypic Characterization
The cytokinesis-defective cell line 17HG5 was isolated from a REMI screen designed to identify cytokinesis-specific genes. Wild-type Dictyostelium cells are able to undergo cytokinesis when grown as stationary or suspension cultures and remain mononucleated or binucleated. However, cytokinesis mutants are unable to divide in suspension culture and become large and multinucleated. They are able to propagate as stationary cultures through alternative mechanisms. To confirm that the 17HG5 cell line was a cytokinesis mutant, cells were grown on coverslips (stationary) or in shaking flasks (suspension) then fixed and stained with DAPI. Parallel cultures of DH-1 cells were fixed and stained as controls. DAPI staining revealed that 17HG5 cells become large and multinucleated when grown in suspension culture, but DH-1 cells are mononucleated and binucleated when grown under either stationary or suspension conditions (Figure 1).
Figure 1.
Dictyostelium pats1 mutant cells become large and multinucleated when grown in suspension culture. The nuclear stain DAPI was used to stain DH-1 (wild-type), 17HG5 (pats1 mutant), and 6BE9 (recapitulant) cells grown as either stationary or suspension cultures. Both DIC and fluorescent images of each cell line grown under both conditions were captured at the same magnification. Bottom right, bar, 10 μm for all images.
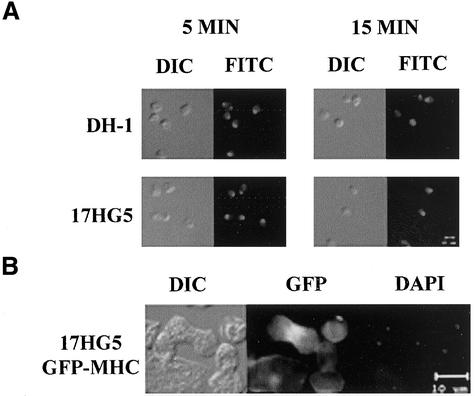
Since the actomyosin cytoskeleton plays a large role in cytokinesis (for review, see Robinson and Spudich, 2000), its global functionality in the 17HG5 cell line was assayed. First, we tested the ability of 17HG5 cells to cap FITC-concanavalin A. When wild-type Dictyostelium cells are pulsed with FITC–concanavalin A, they cap the bound cell surface receptors through a myosin-dependent process (Fukui et al., 1990). We found 17HG5 cells are able to cap cell-surface receptors normally (Figure 2A). The ability of Dictyostelium cells to react when placed in a sodium azide solution by cringing is also dependent on a functional actomyosin cytoskeleton (Pasternak et al., 1989). Normally, cells react to sodium azide by rounding up and lifting off the substrate. Like wild-type cells, 17HG5 cells are also capable of cringing when placed in a sodium azide solution (data not shown).
Figure 2.
Concanavalin A capping in DH-1 (wild-type) and 17HG5 cells and MHC-GFP localization in a dividing 17HG5 cell grown in stationary culture. (A) DH-1 and 17HG5 were briefly pulsed with FITC-conjugated concanavalin A and fixed after either 5 or 10 min. Both DIC and fluorescent images of each cell line were captured at the same magnification. (B) 17HG5 cells transformed with GFP-MHC were fixed and stained with DAPI. The dividing cell can be distinguished from the others by its characteristic dumbbell shape and condensed nuclei (as seen in the DAPI panel). All images were captured at the same magnification.
When Dictyostelium cells are placed in starvation conditions, they enter into a developmental life cycle that culminates in the formation of fruiting bodies composed of a stalk and a spore head. Development also requires an intact cytoskeleton, as demonstrated with MHC II–null cells, which arrest at the mound stage and are unable to form fruiting bodies (De Lozanne and Spudich, 1987). To test the ability of 17HG5 cells to complete the developmental program, cells were placed in starvation buffer and allowed to develop on 0.45-μm HA filters in 3-cm plates while being maintained in a dark environment. Like wild-type cells, 17HG5 cells are capable of forming fruiting bodies with viable spores (J. Abysalh, L. Kuchnicki, D. Larochelle; unpublished data). Taken together, these results suggest that the disruption of the pats1 locus in the 17HG5 cell line does not globally impair the actomyosin cytoskeleton.
Cytokinesis depends on the correct placement of the actomyosin ring at the cellular midbody to obtain proper nuclear segregation. During cytokinesis, wild-type Dictyostelium cells recruit myosin II to the cleavage furrow, and this can be seen either by immunohistochemical staining of fixed cells (Fukui, 1990) or by expressing GFP-MHC (Zang and Spudich, 1998). To determine whether the localization of myosin II to the cleavage furrow of dividing cells was altered in the 17HG5 cell line, cells were transformed with GFP-MHC. Figure 2B shows proper localization of myosin II to the cleavage furrow during cytokinesis in 17HG5 cells grown as stationary cultures, suggesting that myosin II localization in attached cultures is not affected in the 17HG5 cell line.
Pats1 Identification
Identification of the pats1 locus began by mapping of the integration site by Southern blotting of 17HG5 gDNA, independently digested with several restriction enzymes and probed with radiolabeled pRHI30 (the REMI plasmid). The observed banding patterns from Southern blots revealed that pRHI30 had integrated only once within the 17HG5 genome, made evident by the detection of a single band for each restriction enzyme used (data not shown). The Southern blot data allowed us to map several restriction sites around the integration site of pRHI30 (Figure 3A), and three of these sites were subsequently used for plasmid rescue (BclI, HpaI, and HindIII). One plasmid from each digest was isolated for cloning and sequencing of the gDNA flanking the pRHI30 integration site. The three plasmids were pLC4A2, from the BclI digest (which contained 400 base pairs (bp) of the pats1 sequence upstream of pRHI30 and 240 bp downstream of pRHI30); pLC4C2, from the HpaI digest (which contained 2.2 kb of the pats1 downstream sequence); and pLC4B2, from the HindIII digest (which contained 800 bp of the pats1 upstream sequence and 700 bp of the pats1 downstream sequence). All three plasmid gDNA inserts were subsequently sequenced. The plasmid pLC4B2 was also used to recreate the genetic disruption
Figure 3.
A map of the putative pats1 protein, including a restriction map of the pRHI30 insertion site and a Southern blot showing the disruption of the Dictyostelium pats1 locus through REMI and homologous recombination. (A) Through analysis of the 17HG5 Southern blots, we were able to physically map several restriction sites around the insertion site of pRHI30 within the pats1 locus. (B) A map of the putative pats1 protein. Comparison of the sequence obtained from plasmid rescue with data from the Dictyostelium genome project revealed pRHI30 inserted within the partial cDNA clone SLA-465 (accession No. AU 060157). Additional sequence comparisons revealed two more cDNA clones, ddS30o23 (accession No. GJ39335), which aligns with the protein kinase domain of the pats1 open reading frame, and LC-AL19 (accession No. C22973), which aligns with the 5′ region of the pats1 locus. Also, three putative domains (RI-phosphatase, RII-GTP binding domain, RIII-protein kinase domain), an LRR region containing 13 LRRs, and two WD-40 repeat regions, each containing four WD-40 repeats, were identified. KIAA 1790 is a currently uncharacterized human locus that shares homology with the GTP-binding/protein kinase domains of the pats1 protein. (C) A Southern blot of HindIII-digested genomic DNA isolated from DH-1(wild-type) and several mutant cell lines was probed with the cDNA clone SLA-465 diagrammed in B. The pats1 REMI mutant (17HG5) and the mutants created by homologous recombination (6AD8 and 6BE9) contain the same gene disruption as illustrated in B. The pats1 locus in DH-1 cells is intact.
Recreation of the pats1 Mutation
To demonstrate that the cytokinesis-defective phenotype of 17HG5 cells was actually a result of a disruption of the pats1 locus, we set out to recreate the disruption using homologous recombination. To accomplish this, parental DH-1 cells were transformed with HindIII-linearized pLC4B2 and grown in FM media lacking uracil. Cell lines that incorporated the linearized plasmid through homologous recombination were selected for, and colonies were grown as both stationary and suspension cultures. Any clones that could not grow in suspension culture were considered cytokinesis-defective. From this screen, we isolated multiple cell lines that exhibited the same cytokinesis defect as observed in 17HG5 cells, and of those, the cell lines 6AD8 and 6BE9 were kept for further observation. Southern blot analysis was performed on the two recapitulated cell lines to demonstrate that pLC4B2 had integrated into the same region of the pats1 locus as did pRHI30 in the 17HG5 cell line. To do this, gDNA from the DH-1, 17HG5, 6AD8, and 6BE9 cell lines was digested with HindIII and probed with the cDNA clone SLA-465 (Figure 3B), a clone that spans the integration site of pRHI30 in the pats1 locus. A single band of ∼2.0 kb was detected in the DH-1 lane, whereas this band shifted to ∼6.5 kb in all three of the other lanes (Figure 3C). This shift of 4.5 kb corresponds to the size of pRHI30, indicating that pRHI30 integrated within the same region in the two recapitulated cell lines compared with the 17HG5 cell line. Furthermore, these cell lines exhibited the same cytokinesis-defective phenotype (Figure 1 and data not shown) as observed in the 17HG5 cell line, suggesting that it is indeed the disruption of the pats1 locus that is responsible for this phenotype.
Sequence Analysis
The three plasmids obtained from plasmid rescue (pLC4A2, pLC4C2, and pLC4B2) were used as templates in sequencing reactions using the T7 and A0118 primers contained in the plasmid pRHI30. Using the obtained sequence as a template to search the Dictyostelium genomic and cDNA databases from the DGP Dictyostelium web site at the University of California, San Diego, we found an identical match to the cDNA clone SLA-465 (accession No. AU060157) from the Tsukuba cDNA sequencing project. Also, analysis of the cDNA clone sequence revealed a Dpn II restriction site, consistent with our sequence data, that was the point of integration of pRHI30 during the REMI mutagenesis. Using SLA-465 as a template in further searches and linking overlapping clones, we were able to extend the sequence 9070 bp upstream of the integration site and 4446 bp downstream of the integration site. Within the genomic sequence, we identified an 8892-bp open reading frame encoding a 2964-amino-acid protein (accession No. AY170918). Using 3′ rapid amplification of cDNA ends, we were able to determine the 3′ end of the transcript to be 825 bp from the insertion site. The 5′ end of the coding sequence has not yet been determined. However, the 8892-bp pats1 open reading frame encompasses, in frame, the cDNA clone SLA-465 (7624–8384 bp). In addition, we have identified two cDNA clones (ddS30o23, accession No. GJ39335 and FC-AL19, accession No. C22973) (Figure 3B) that align with the RIII domain of the pats1 open reading frame from 6194 to 6760 bp and with the 5′ region from 1 to 255 bp, respectively. That these cDNA clones align with the large, single open reading frame of the pats1 locus is consistent with the fact that the pats1 open reading frame we have identified is indeed coding sequence.
We also compared the amino acid sequence we obtained with all of the genomic databases at NCBI and found several matches. First, the sequence from 1628 to 2343 aa shares 26% amino acid identity and 45% conserved amino acid sequence to the human open reading frame KIAA-1790, a currently uncharacterized human locus (Figure 3B). Also, there are at least three conserved sequence domains within the pats1 locus, each with homologies to different regulatory proteins. The RI domain (Figures 3B and 4), extending from 607 to 1145 aa, shares 36% amino acid identity and 58% conserved amino acid sequence homology with myotubularin, a member of the protein tyrosine phosphatase superfamily of proteins that is necessary for proper myotube development (Taylor et al., 2000). The RII domain (Figures 3B and 4), from amino acid 1627 to 1747, contains homology to the Rho family of small GTPases. The RIII domain (Figures 3B and 4) extending from amino acid 2087 to 2351, shares homology with a multitude of kinases, including 35% nucleotide identity and 51% conserved sequence with human MAPKKK 10, a serine/threonine kinase. Additional comparison of the RIII-domain amino acid sequence revealed strong homology to the catalytic domain of the protein tyrosine kinase superfamily. These lines of evidence strongly suggest that the RIII domain is a kinase, although its physiological substrate has not yet been determined.
Figure 4.
Alignment of the myotubularin, rho, and protein kinase domains. Using the NCBI protein Blast program and conserved-domain analysis, we show the conservation between the three pats1 domains and their appropriate homologues. The RI domain is aligned with the myotubularin-related protein 1 (accession No. NP 058681). The RII domain is aligned with the Rho conserved domain (CDD: smart00174.5.RHO, PSSM-id: 151) from the conserved-domain analysis website. The RIII domain is aligned with the tyrosine kinase active site motif (CDD: smart00219.5.TyrKc, PSSM-id: 7320), which was also obtained from the conserved-domain analysis. In the figure, capital letters between the two sequences represent identical matches, and + signs represent conserved amino acids.
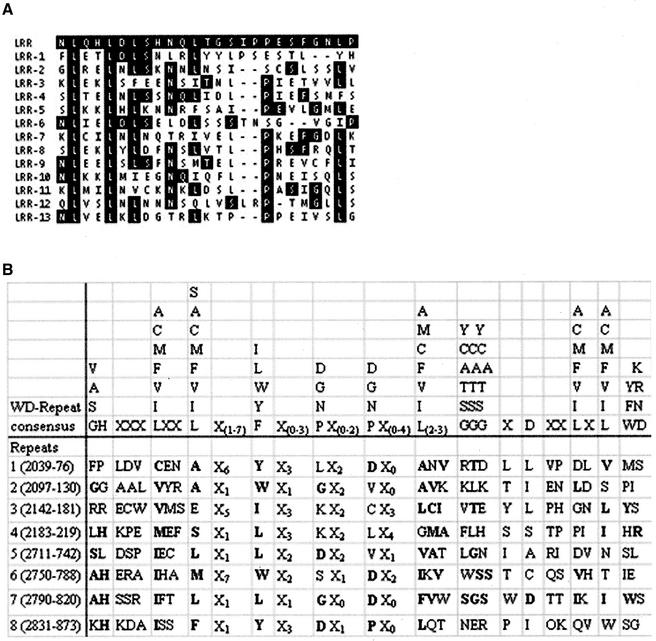
Further protein sequence analysis revealed two additional motifs. The amino acid sequence from 1232 to 1554 contains 13 leucine-rich repeats (LRRs), a motif thought to be involved in protein–protein interactions (Kobe and Deisenhofer, 1999) (Figures 3B and 5A). Also, eight WD-40 repeats were found near the C-terminus, and these repeats have also been implicated in protein–protein interactions (Neer et al., 1994) (Figures 3B and 5B). These repeats include the following amino acids: 2039–2076, 2097–2130, 2142–2181, 2183–2219, 2711–2742, 2750–2788, 2790–2820, and 2831–2873. Furthermore, the last four WD-40 repeats share homology with the Dictyostelium MHCK B protein, a member of the WD-40 repeat protein family (Clancy et al., 1997).
Figure 5.
Alignment of the WD-40 repeats and conservation between the LRRs. (A) The LRR consensus sequence is highlighted in black at top of table. The 13 pats1 repeats are listed below the consensus sequence. Conserved amino acids appear in black boxes. (B) The WD consensus sequence was obtained from Konzok et al. (1999), and bold letters show conservation between the consensus and the predicted repeats.
Domain Analysis
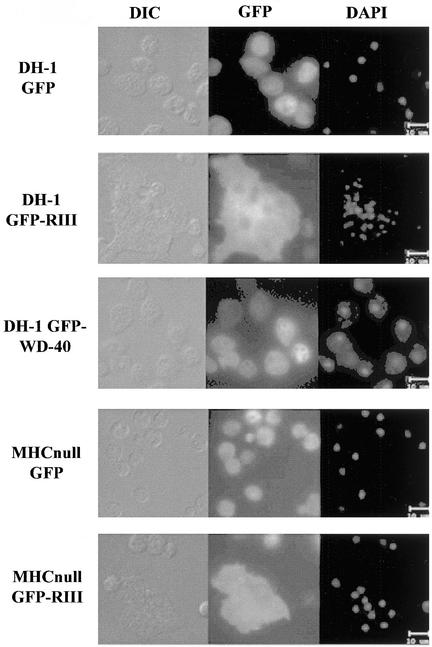
To begin to understand the importance of the identified domains (see above) in the putative pats1 protein, we overexpressed the GFP control, the GFP-RIII domain (2087–2351 aa), and the GFP–C-terminal WD-40 repeat domain (2542–2873 aa) fusion proteins independently in both DH-1 and 17HG5 cells. Interestingly, all transformants (in DH-1 and 17HG5 cells) expressing the GFP-RIII fusion protein contained a substantial population of very large and multinucleated cells when grown as attached cultures (Figure 6 and J. Abysalh, D. Larochelle, unpublished data). On the basis of GFP fluorescence, we observed considerable variability in the expression levels within a population of cells. Nonetheless, the large-cell phenotype observed in stationary cultures was seen consistently in cells expressing the greatest amounts of fusion protein (based on GFP fluorescence). DH-1 cell lines expressing GFP alone or the GFP–C-terminal WD-40 repeat domain grew normally and remained mononucleated or binucleated regardless of GFP expression levels (Figure 6). Also, when the cell lines expressing GFP-RIII were grown as suspension cultures, the extremely large multinucleated cells were no longer observed. The absence of large cells is probably the result of their inability to withstand the shear forces applied from maintaining the cells in suspension culture by shaking, causing them to lyse. Furthermore, both GFP-fusion proteins and GFP alone lacked distinct cellular localization (on the basis of GFP fluorescence) and were predominantly cytosolic (Figure 6).
Figure 6.
Severe cytokinesis defect in both DH-1 and MHC-null cells overexpressing the GFP-RIII fusion protein. Both DH-1 (wild-type) and MHC-null cells were transformed with either pTX-GFP or pTX-GFP-RIII. In addition, DH-1 cells were transformed with pTX-GFP–C-terminus WD-40 repeat. Cells grown on coverslips were fixed and the nuclei were visualized with the nuclear stain DAPI. DIC, GFP fluorescence, and DAPI fluorescence images were captured at the same magnification for all of the transformants.
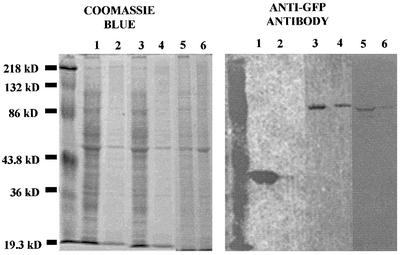
Because of the strong cytokinesis defect observed in attached cells overexpressing the RIII domain and the cytokinesis defect observed in cell lines that have a disruption within the WD-40 repeat region, we set out to determine whether these domains associate with the cytoskeleton. This was done by performing Triton-insoluble cytoskeleton assays as described by Kolman et al. (1996). Briefly, Triton-insoluble supernatants and pellets from DH-1 cell lines expressing GFP, GFP-RIII, and GFP-WD40 repeat domain were prepared and run on duplicate gels. Essentially all of the GFP (as visualized with an anti-GFP antibody) was found to be in the soluble fraction, whereas appreciable amounts of both the GFP-RIII protein and the GFP-WD40 repeat protein coprecipitated with the Triton-insoluble cytoskeletal fraction (Figure 7). This result suggests that there is significant interaction between both the RIII domain and the WD-40 repeat domain with some component of the cytoskeleton.
Figure 7.
GFP-RIII and GFP-WD40 repeat domains partially fractionate with the Triton-insoluble cytoskeletal pellet. Triton-soluble and Triton-insoluble samples of DH-1 GFP, DH-1 GFP-RIII, and GFP-WD40 repeat transformants were prepared and run on duplicate SDS-PAGE gels. One gel was stained with Coomassie blue, and the other was subjected to Western blot analysis probed with an anti-GFP antibody. Lanes: 1, DH-1 GFP Triton-soluble sample; 2, DH-1 GFP Triton-insoluble sample; 3, DH-1 GFP-RIII Triton-soluble sample; 4, DH-1 GFP-RIII Triton-insoluble sample; 5, DH-1 GFP-WD40 repeat Triton-soluble sample; 6, DH-1 GFP-WD40 Triton-insoluble sample.
Because the Triton-insoluble cytoskeleton assay showed localization of the RIII and WD-40 repeat fragments to the actomyosin cytoskeleton, we decided to transform MHC-null cells with the same recombinant vectors as above. Surprisingly, the very large multinucleated cells observed in DH-1 and 17HG5 cells expressing GFP-RIII were also seen consistently in attached cultures of MHC-null cells transformed with the GFP-RIII fusion protein (Figure 6). Once again, the large multinucleated cells overexpressing the GFP-RIII fusion protein were those that were brightest (on the basis of GFP fluorescence). Conversely, MHC-null cells transformed with the GFP control vector grew normally as attached cultures, regardless of GFP expression levels (Figure 6). These results suggest that the cytokinesis defect observed in stationary cells overexpressing the GFP-RIII domain is independent of myosin II.
DISCUSSION
The use of D. discoideum as a model organism for the study of cytokinesis has resulted in the discovery of many genes shown to be required for proper cellular division. The identification of these genes has come about primarily through two methods. One method involves the standard genetic approach, in which proteins capable of interacting with known components of cytokinesis are isolated and their associated genes are identified and subsequently knocked out by gene replacement (Robinson and Spudich, 2000). In addition to standard genetic approaches of targeting known genes, REMI has been useful for identifying novel genes required for cytokinesis. Continuing with the REMI screen, we have identified a novel locus, pats1, whose disruption results in a cytokinesis defect. The putative protein encoded by this locus contains several regulatory and protein–protein interacting domains, making it an excellent candidate for linking signal transduction and the actomyosin cytoskeleton during cytokinesis.
The RI domain is homologous with myotubularin, a member of the protein tyrosine phosphatase superfamily, and this homology includes the presence of the Cys-X5-Arg active site motif (Taylor et al., 2000). This active site motif is the hallmark for all members of the superfamily of protein tyrosine phosphatases (Taylor et al., 2000), and the strong myotubularin homology, coupled with the presence of the active site motif, suggests that the RI domain is a phosphatase. Myotubularin, although a member of the protein tyrosine phosphatase superfamily, has the ability to dephosphorylate the inositol phospholipid PI(3)P in vitro and in vivo (Taylor et al., 2000), suggesting that the physiological substrate of the RI domain may be either a protein tyrosine residue or an inositol phospholipid.
The RII domain shares homology with both rho and ras small GTPases. Included within the conserved regions are parts of the highly conserved GTP-binding domains, suggesting that the RII domain has the ability to bind GTP (Bourne et al., 1991). It is believed that GTPases, like rac, rho, and ras, work as molecular switches activated by the binding of GTP and inactivated through the hydrolysis of the bound GTP to GDP (Chisholm, 1997). The binding of GTP to the RII domain may provide pats1 with a similar regulatory ability.
The RIII domain has significant overall sequence homology to serine threonine kinases, but it also contains strong homology with the catalytic domain of the tyrosine kinase family. Furthermore, the overexpression of the RIII domain results in a severe cytokinesis defect in cell lines when grown as attached cultures (Figure 6). This phenotype is similar to that of coronin-null (de Hostos et al., 1993) and cortexillin I and II double-knockout cell lines (Faix, et al, 1996). Coronin and the cortexillins are actin cross-linking proteins, suggesting that pats1 may also be involved in modulation of the actin cytoskeleton. The strong likelihood that the pats1 protein functions as a protein kinase, coupled with the cytokinesis-deficient phenotype observed when the RIII domain is overexpressed, indicates that the pats1 protein may be a key regulator of cytokinesis. Whether this regulation is through direct or indirect interaction with the actomyosin cytoskeleton has not yet been determined.
There are several interesting protein–protein interacting domains within the pats1 protein sequence as well. There are 13 LRRs, a motif predicted to form nonglobular structures that are capable of interacting with other proteins (Kobe et al., 1999). These repeats give pats1 the ability to either target downstream effectors or interact with upstream regulators. There are also eight WD-40 repeats, four of which are found in the protein kinase domain (RIII). The other four WD-40 repeats are located in the C-terminus. Together, the eight WD-40 repeats may participate in the targeting of the pats1 protein to effector proteins (Neer et al., 1994). Also, the region containing the four C-terminus WD-40 repeats shares homology with the Dictyostelium MHCK B protein, an MHC kinase containing seven WD-40 repeats (Clancy et al., 1997). Together, the presence of a phosphatase domain, a GTP-binding domain, a kinase domain, and LRRs and WD-40 repeats, coupled with the strong cytokinesis defect observed when this gene locus is disrupted, suggests that the pats1 protein is a multifunctional protein essential for proper cytokinesis. Although cytokinesis is the only defect we have observed thus far in cells containing a disruption in the pats1 locus, it is possible that the pats1 protein may function in other cellular processes. If this is the case, however, the cells must have compensatory mechanisms for these other cellular processes, because we have observed no defects other than in cytokinesis.
Our data further suggest that the pats1 protein may be involved in myosin II localization during cytokinesis. The Triton-insoluble cytoskeleton assay revealed an interaction between the WD-40 repeat domain and the actomyosin cytoskeleton. Furthermore, the WD-40 repeat domain has homology to MHCK B. Steimle et al. (2001) showed that the WD-40 repeats in D. discoideum MHCK B are essential for the proper targeting of the kinase to myosin II. Preliminary observations of binucleated 17HG5 cells grown in suspension culture and expressing GFP-MHC showed no localization of GFP-MHC. Conversely, DH-1 cells raised similarly, of similar appearance, and also expressing GFP-MHC had distinct GFP-MHC localization to the presumptive cleavage furrow (J. Abysalh, D. Larochelle; unpublished data). Together, these results are suggestive of a role for the pats1 protein in the proper localization of MHC to the cleavage furrow in cells raised in suspension culture.
The discordance of phenotypes observed when the pats1 locus is disrupted versus when the RIII domain is overexpressed may be a manifestation of the multifunctionality of the pats1 protein and overlapping functions shared by other proteins. The dominant negative effect observed with the overexpression of the RIII kinase domain may arise from the blocking of phosphorylation of the downstream target of RIII (and possibly compensatory kinases). This results in a severe cytokinesis defect that can be observed when these cell lines are grown as attached cultures. Conversely, cells containing a disruption of the pats1 locus, because of compensating kinases, are able to propagate as attached cultures and manifest their cytokinesis defect only when placed in suspension culture. In essence, therefore, the pats1 protein is required for cytokinesis in suspension culture but not for attachment-dependent mechanisms (i.e., traction-mediated cytofission and attachment-assisted mitotic cleavage), in which compensating pathways are functioning. These pathways are disrupted when the RIII domain is overexpressed. Alternatively, the disruption of the pats1 locus at the 3′ end, as seen in the 17HG5 cell line, could result in the translation of a truncated form of the pats1 protein. This truncated form could retain some function capable of maintaining cytokinesis in cell lines when grown as attached cultures but could be unable to maintain cytokinesis in cell lines when grown as suspension cultures.
To conclude, we have identified a novel gene locus, pats1, that is required for cytokinesis in D. discoideum. The pats1 locus encodes a putative protein with 3 potential regulatory domains, 8 WD-40 repeats, and 13 LRRs. Sequence analysis shows strong evidence for a protein tyrosine phosphatase, a protein kinase domain, and a GTP-binding domain, suggesting that the pats1 protein may be a multifunctional protein the function of which is mediated, in part, through interactions with the cytoskeleton. Finally, overexpression of the RIII domain in stationary cultures results in a severe cytokinesis defect. Taken together, this leads us to believe that pats1 is an essential regulator of cytokinesis in D. discoideum.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Tom Egelhoff for providing the pTX-GFP expression vector, Arturo De Lozanne for the GFP-MHC vector and Takahiro Morio for the cDNA clone SLA465. We also thank members of the Larochelle laboratory and the Biology Department at Clark University for helpful comments, assistance, and support throughout the period of this work. This work was supported by Faculty Development Awards and start-up funds from Clark University to D.A.L. Finally, the authors wish to acknowledge the assistance of the National Science Foundation in the purchase of a microscopy imaging facility through the multi-user equipment and instrumentation resources (ref: DBI-0070241).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0335. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0335.
REFERENCES
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Chisholm RL. Cytokinesis: a regulatory role for ras-related proteins? Curr Biol. 1997;7:R648–R650. doi: 10.1016/s0960-9822(06)00325-3. [DOI] [PubMed] [Google Scholar]
- Clancy CE, Mendoza MG, Naismith TV, Kolman MF, Egelhoff TT. Identification of a protein kinase from Dictyostelium with homology to the novel catalytic domain of myosin heavy chain kinase A. J Biol Chem. 1997;272:11812–11815. doi: 10.1074/jbc.272.18.11812. [DOI] [PubMed] [Google Scholar]
- de Hostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta-subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Rehfueβ C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Faix J, Steinmetz M, Boves H, Kammerer RA, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y. Actomyosin organization in mitotic Dictyostelium amoebae. Ann NY Acad Sci. 1990;582:156–165. doi: 10.1111/j.1749-6632.1990.tb21676.x. [DOI] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald N, Dai J, Ting-Beall HP, De Lozanne A. A role for Dictyostelium racE in cortical tension and cleavage furrow progression. J Cell Biol. 1998;141:483–492. doi: 10.1083/jcb.141.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald N, Damer CK, O'Halloran TJ, De Lozanne A. Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cell Motil Cytoskeleton. 2001;48:213–223. doi: 10.1002/1097-0169(200103)48:3<213::AID-CM1010>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cytokinesis. Curr Biol. 1997;7:R274–R276. doi: 10.1016/s0960-9822(06)00135-7. [DOI] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1999;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Kolman MF, Futey LM, Egelhoff TT. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J Cell Biol. 1996;132:101–109. doi: 10.1083/jcb.132.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility J. Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E, Gerald N, Larochelle DA, Vithalani KK, Niswonger ML, Maready M, De Lozanne A. LvsA, a protein related to the mouse beige protein, is required for cytokinesis in Dictyostelium. Mol Biol Cell. 1999;10:4429–4439. doi: 10.1091/mbc.10.12.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani K, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1330. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle DA, Vithalani KK, De Lozanne A. The role of Dictyostelium racE in cytokinesis: mutational analysis and localization studies by use of green fluorescent protein. Mol Biol Cell. 1997;8:935–944. doi: 10.1091/mbc.8.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerisch G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system, and formation of the cleavage furrow. J Cell Sci. 1997;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- Pasternak C, Spudich JA, Elson EL. Capping of surface receptors and concomitant cortical tension are generated by conventional myosin. Nature. 1989;341:549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Spudich JA. Towards a molecular understanding of cytokinesis. Trends Cell Biol. 2000;10:228–237. doi: 10.1016/s0962-8924(00)01747-5. [DOI] [PubMed] [Google Scholar]
- Steimle PA, Naismith TV, Licate L, Egelhoff TT. WD repeat domains target Dictyostelium myosin heavy chain kinases by binding directly to myosin filaments. J Biol Chem. 2001;276:6853–6860. doi: 10.1074/jbc.M008992200. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol-3-phosphate. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithalani KK, Shoffner JD, De Lozanne A. Isolation and characterization of a novel cytokinesis-deficient mutant in Dictyostelium discoideum. J Cell Biochem. 1996;62:290–301. doi: 10.1002/(sici)1097-4644(199608)62:2<290::aid-jcb16>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Wang, N., Wu, W.-I., and De Lozanne, A. (2002). The BEACH family of proteins: phylogenetic and functional analysis of six Dictyostelium BEACH proteins. J. Cell. Biochem. In press. [DOI] [PubMed]
- Weber I, Neujahr R, Du A, Kohler J, Faix J, Gerisch G. Two-step positioning of a cleavage furrow by cortexillin and myosin II. Curr Biol. 2000;10:501–506. doi: 10.1016/s0960-9822(00)00452-8. [DOI] [PubMed] [Google Scholar]
- Zang JH, Spudich JA. Myosin II localization during cytokinesis occurs by a mechanism that does not require its motor domain. Proc Natl Acad Sci USA. 1998;95:13652–13657. doi: 10.1073/pnas.95.23.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]