Abstract
Survivin, a member of the inhibitor of apoptosis protein family, has attracted growing attention due to its expression in various tumors and its potential application in tumor therapy. However, its subcellular localization and function have remained controversial: Recent studies revealed that survivin is localized at the mitotic spindle, binds caspases, and could thus protect cells from apoptosis. The cell cycle-dependent expression of survivin and its antiapoptotic function led to the hypothesis that survivin connects the cell cycle with apoptosis, thus providing a death switch for the termination of defective mitosis. In other studies, survivin was detected at kinetochores, cleavage furrow, and midbody, localizations being characteristic for chromosomal passenger proteins. These proteins are involved in cytokinesis as inferred from the observation that RNA interference and expression of mutant proteins led to cytokinesis defects without an increase in apoptosis. To remedy these discrepancies, we analyzed the localizations of a survivinDsRed fusion protein in HeLa cells by using confocal laser scanning microscopy and time-lapse video imaging. SurvivinDsRed was excluded from the interphase nucleus and was detected in centrosomes and at kinetochores. It dissociated from chromosomes at the anaphase/telophase transition and accumulated at the ends of polar microtubuli where it was immediately condensed to the midbody. Overexpression of both survivinDsRed and of a phosphorylation-defective mutant conferred resistance against apoptosis-inducing reagents, but only the overexpressed mutant protein caused an aberrant cytokinesis. These data characterize in detail the dynamics of survivin in vertebrate cells and confirm that survivin represents a chromosomal passenger protein.
INTRODUCTION
Inhibitor of apoptosis proteins (IAPs) were first identified as baculoviral proteins that were capable of suppressing apoptosis in infected insect cells (Crook et al., 1993). This class of proteins contains one to three zinc-binding motifs, termed baculoviral IAP repeats (BIRs). Recently, IAP homologues were found in different species, including yeast, nematodes, drosophila, and vertebrates (Deveraux and Reed, 1999). Survivin is a short-lived mammalian protein (Zhao et al., 2000) that contains a single BIR motif but lacks a carboxy-terminal RING motif (Ambrosini et al., 1997). It resembles the BIR-containing proteins from yeasts and nematodes (Uren et al., 1998) and shows homology to IAPs, which are responsible for controlling apoptosis (for review, see Deveraux and Reed, 1999). Survivin is expressed at the G2/M transition of the cell cycle (Li et al., 1998; Kobayashi et al., 1999) and crystal structure analysis revealed that survivin dimerizes by amino acid regions close to the BIR domain (Verdecia et al., 2000). In addition, a C-terminal alpha helix has been suggested to form an oligomerization domain (Muchmore et al., 2000; Shi, 2000). Inhibition of human survivin has been associated with cell cycle defects (Li et al., 1998), and disruption of its genetic locus led to an embryonic lethal phenotype in homozygous mice, suggesting a key role in mitosis (Uren et al., 2000). Furthermore, survivin expression was found in embryonic tissue and in proliferating hematopoietic stem cells as well as in reproductive tissues (Konno et al., 2000; Endoh et al., 2001), confirming its cell cycle-specific function.
Human survivin became of particular interest because it was identified as a tumor-associated antigen highly expressed in various tumors (Ambrosini et al., 1997). It is, therefore, a suitable target for immunotherapeutic and gene therapy approaches to defeat cancer (Grossman et al., 1999, 2001; Schmitz et al., 2000; Mesri et al., 2001). Recent studies on human survivin showed that ectopic expression of survivin can protect cells against spindle toxin-induced apoptosis (Li et al., 1998, 1999). The protective effect of survivin was supposed to be due to its caspase-binding capacity and to depend on its spindle association. The types of caspases involved are still disputed (Banks et al., 2000; Conway et al., 2000; O'Connor et al., 2000; Shin et al., 2001).
On the other hand, survivin has been classified as a chromosomal passenger protein that interacts with Aurora-B and INCENP (Skoufias et al., 2000; Uren et al., 2000; Wheatley et al., 2001) to build a chromosomal passenger complex that is supposed to play a crucial role in the execution of cytokinesis. Revealing the exact localization and the biological function of survivin during mitosis became even more difficult due to the identification of two novel splice variants of survivin in human and in murine cells (Mahotka et al., 1999; Conway et al., 2000). These isoforms may interfere with the detection of the full-length human 142-amino acid survivin by using monoclonal antibodies. To remedy the discrepancies concerning the localization and function of human survivin, we generated HeLa cell lines stably expressing survivinDsRed and enhanced green fluorescent protein (EGFP)-α-tubulin fusion proteins as well as HeLa cell lines expressing a survivin mutant with a defective p34cdc2 phosphorylation site fused to DsRed. By using time-lapse video imaging we gained further insight into the trafficking of this protein during mitosis. Furthermore, we analyzed the effects of the overexpressed survivinDsRed and the mutant fusion protein with respect to its antiapoptotic function and its role in the execution of cytokinesis.
MATERIALS AND METHODS
Construction of Retroviral Vectors
For the generation of dsRed-tagged human survivin the coding region of survivin (Schmitz et al., 2000) was modified by polymerase chain reaction (PCR) by using the survivin forward primer Surv5′-EcoRI (5′-GAGAGAGAATTCACAACCATGGGTGCCCCGACGTT-GCCC-3′) and survivin reverse primer Surv-deltaStop-BamHI (5′-GAGAGAGGATCCTCCATGGC AGCCAGCTGCTC-3′), resulting in additional EcoRI and BamHI restriction sites, respectively, to facilitate directional cloning. The survivin reverse primer contained a GGA at codon position 143, thus eliminating the stop codon. The PCR amplification was carried out as described previously (Schmitz et al., 2000). The PCR product was ligated into the corresponding restriction sites of pIRES2-EGFP (BD Biosciences Clontech, Heidelberg, Germany), generating pSurvivindeltaStop-IRES2-EGFP.
For the generation of a survivindsRed fusion protein a 705-base pair BamHI/NotI restriction fragment was excised from pDsRed (BD Biosciences Clontech) and ligated into the corresponding restriction sites of pSurvivindeltaStop-IRES2-EGFP, replacing the internal ribosome entry site (IRES)-EGFP cassette. The resulting pSurvivinDsRed was restricted with BglII/HpaI and the survivinDsRed fragment was ligated into the corresponding restriction sites of the retroviral vector pzc-CFG5.1-MCS, a vector that was derived by replacement of the IRES-EGFP-ZeozinR cassette from pcz-CFG5-IEGZ (Berberich-Siebelt et al., 2000) (This vector originally described as pEGZ/MCS has been renamed to pcz-CFG5-IEGZ.) with EcoRI/Kpn2I digestion and ligation of a 43-bp polylinker containing a multiple cloning site. Pcz-CFG5.1-SurvDsRed was digested with BamHI/HpaI and an EGFP-fragment was ligated into the vector, creating pcz-CFG5-SurvEGFP. Full-length human α-tubulin from pEGFP-Tub (BD Biosciences Clontech) was excised with BgI II/BamHI restriction and ligated into the BamHI-site of pcz-CFG2-fEGFPf, described previously (Berberich-Siebelt et al., 2000), generating the vector pzc-CFG2-fEGFPf-Tub. Full-length Survivin was cut with EcoRI/BamHI and ligated into the EcoRI/BamHI restriction sites of pcz-CFG5-IEGZ, producing pcz-CFG5-Survivin-IEGZ. A retroviral vector coding for dsRed was generated by inserting the BamHI/HpaI dsRed-fragment from pSurvivinDsRed into pcz-CFG5.1-MCS, resulting in pcz-CFG5.1-DsRed. All vector insert boundaries and inserts were verified by sequencing.
Site-directed Mutagenesis of Survivin at Position 34
Survivin Threonin34→Alanin (T34A) mutation was introduced by site-directed mutagenesis of the pcz-CFG5.1SurvivinDsRed vector by using the oligonucleotide selection primer SelektNruI 5′-GCGCTGCTTCGGGATGTACGGGCCAG-3′ and mutation primer MutPos34 5′-GCTGCGCCTGCGCCCCGGAGCGGATG-3′ and the Transformer site-directed mutagenesis kit (BD Biosciences Clontech) according to the manufacturer's instructions, generating the vector pcz-CFG5.1-SurvM34DsRed. Mutation at position 34 in the coding region of Survivin was confirmed by sequencing.
Transduction of Retroviral Vectors into HeLa Cells and Generation of Stable Cell Lines
Retroviral particles were generated as described previously (Soneoka et al., 1995). Briefly, 293T cells (DuBridge et al., 1987) were cotransfected with an expression construct for gag-pol (pHIT60), the MoMuLV-based retroviral vectors and the vesicular stomatitis virus G-protein (pcz-VSV-Gwt; described by Kalajzic et al., 2001). Viral supernatants were harvested 48 and 72 h after transfection, pooled, and filtered (0.45-μm pore size filter). Polybrene was added to a final concentration of 8 μg/ml, and the supernatants were used immediately or stored at −80°C until use. HeLa target cells (105) were plated in 30-mm dishes a day before transduction and were transduced with titrated retroviral titers of 5 and 50 multiplicity of infection (MOI). Forty-eight hours after transduction the cells were subcultured. After limiting dilution, single clones were analyzed by fluorescence microscopy, picked, and cultured. In some experiments, an additional transduction with retroviral particles coding for EGFP-α-tubulin fusion proteins with 10 MOI was performed.
Preparation of Recombinant Survivin and Generation of an Anti-Survivin Monoclonal Antibody (mAb)
Recombinant survivin was prepared as described previously (Schmitz et al., 2000). Affinity-purified survivin was used to immunize BALB/c mice (1 × 100 μg and 3 × 50 μg). Hybridomas were generated by the fusion of spleen cells with X-63AG8 myeloma cells according to standard procedures. Hybridomas were screened for reactivity with recombinant survivin by enzyme-linked immunosorbent assay and Western blotting. Positive wells were established and cloned twice by limiting dilution. The anti-survivin mAb 9B1D9 recognized recombinant human survivin as well as endogenous survivin expressed in HeLa cells.
Preparation of Recombinant DsRed and SurvivinDsRed Protein
Recombinat DsRed and survivinDsRed were generated by digesting pSurvDsRed with HpaI/SacI, generating a 1150-base pair SurvDsRed fragment that was ligated into corresponding SmaI/SacI restriction sites of pQE31 (QIAGEN, Hilden, Germany), generating pQE31-SurvDsRed. A DsRed control plasmid was generated by BamHI digestion of pQE31-SurvDsRed and excision of the coding region for survivin, thereby generating pQE31-DsRed. The plasmids were used to transform Escherichia coli strain M15 (QIAGEN). Protein expression was induced by 0.1 mM isopropyl β-d-thiogalactoside and the bacteria were incubated for 16 h on a shaker at room temperature (RT). Cells were spun down and resuspended in 50 mM, H2NaPO4, 300 mM NaCl, pH 8.0, in the presence of chicken egg white lysozyme (Sigma Chemie, Dreeich, Germany). After sonification lysed cells were spun down at 15,000 × g (30 min, 4°C), and the supernatant containing the recombinant proteins was collected and purified using metal chelate chromatography columns (Ni2+-nitrilotriacetic acid-agarose) (QIAGEN). Purified protein samples were dialyzed against phosphate-buffered saline (PBS), pH 7.4.
Immunofluorescence and Confocal Laser Scanning Microscopy
Cells were fixed in ice-cold paraformaldehyde for 20 min and incubated 30 min in PBS. They were permeabilized with 1% sodium citrate/0.1% Triton X-100 and washed three times with PBS. Then the cells were washed three times with PBS containing 0.1% bovine serum albumin. Cells were directly used for microscopy or were incubated for 1 h at room temperature with polyclonal anti-survivin (1:120 diluted; R & D Systems, Wiesbaden, Germany), monoclonal anti-survivin (clone 9B1D9, 1:80 diluted in PBS/0.1% bovine serum albumin [BSA]), or mouse anti-AIM-1 antibody (1:120 diluted; Tatsuka, unpublished data). In other experiments cells were incubated with TO-PRO-3 dye, diluted 1:105 (Molecular Probes, Leiden, The Netherlands) for detection of DNA. After washing again with PBS/0.1% BSA, the cells were incubated for 1 h with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit-IgG or anti-mouse IgG (both stock solutions, 1:80 diluted, as recommended by the supplier; Dianova, Hamburg, Germany). Finally, cells were washed three times in PBS/0.1% BSA and once in double distilled water, before being examined by confocal laser scanning microscopy (NCS-NT; Leica, Wetzlar, Germany) by using the filters SP590, DD488/568, RSP580, and BF530/30 for detection of EGFP and SP590, DD588/568, RSP580, and LP590 for detection of DsRed, respectively. TO-PRO3 nuclear staining was detected with the filters BP647, RSP660, and LP665. Image files were digitally processed for presentation by using Adobe Photoshop (Adobe Systems, San Jose, CA).
Immunoblot Analysis
For protein analysis of survivin expression in HeLa wild-type and in a HeLa cell line transduced with 50 MOI of the survivin construct pcz-CFG5.1-SurvivinDsRed, total protein lysates were prepared. Protein concentration was determined according to Bradford (1976). Equal amounts of protein (50 μg) were subjected to electrophoresis (Laemmli, 1970) and blotted onto polyvinylidene difluoride membranes (PALL, Dreieich, Germany). Survivin and survivinDsRed immunostaining were performed with the monoclonal anti-mouse antibody 9B1D9 or with a polyclonal rabbit anti-survivin antibody (R & D Systems). The secondary rabbit anti-mouse antibody and secondary goat anti-rabbit antibody both coupled to horseradish peroxidase (both 1:2500; DAKO, Hamburg, Germany) were visualized with enhanced chemoluminescence (Roche Diagnostics, Mannheim, Germany) as recommended by the supplier. In another experiment HeLa cell lines with high or low expression of survivinDsRed and survivinM34DsRed, respectively, were used for the immunoblot. In control experiments using only secondary antibodies, no reactivity was detected in HeLa cell lysates. For densitometric quantification of endogenous and transgenic survivin expression the enhanced chemoluminescence-exposed films were scanned and analyzed by the Phoretix 1D Advanced version 4.01 software (Nonlinear Dynamics; Biostep, Jahnsdorf, Germany).
Terminal Deoxynucleotidyl Transferase dUTP Nick-End Labeling (TUNEL) Assay and Annexin V Staining of Apoptotic Cells
HeLa cells (2 × 105) were cultivated overnight on coverglasses and the analysis of apoptosis in adherent cell lines was carried out by terminal dUTP-FITC desoxynucleotidyl transferase nick-end labeling (Roche Diagnostics) as recommended by the supplier. For determination of apotosis in HeLa wild-type or HeLa cells with high expression of survivinDsRed, survivinM34DsRed, and DsRed FACS assisted analysis of annexin V staining was performed as recommended by the supplier (Roche Diagnostics). Cells (105) were plated in 30-mm dishes and incubated either with 0.1 μg/ml nocodazole, 0.1 μg/ml colchicine, 0.01 μg/ml cisplatin, 0.1 μg/ml doxorubicine, or 50 μM C2 ceramide (N-acetyl-d-sphingosine; Sigma Chemie). After 24 h cells were trypsinized and analyzed. The apoptotic index was calculated according to the formula apoptotic index = mean percentage of experimental apoptosis − mean percentage of spontaneous apoptosis. Spontaneous apoptosis was defined as apoptosis caused by transgenic protein expression. Statistical analysis was performed with Student's t test.
Preparation of Chromosomes and Staining with Recombinant DsRed and SurvivinDsRed Protein
For the preparation of chromosomes 4 × 106 HeLa cells were incubated with 10 μM colchicine for 1.5 h (37°C, 5% CO2), washed twice with PBS, and trypsinized. Cells were collected with PBS and spun down at 250 × g at 4°C for 8 min. The supernatant was discarded and the cell pellet was resuspended in 4 ml of 0.56% KCl for osmolysis. After 10 min at RT the cells were again spun down at 250 × g. Cells were fixed in ice cold methanol/acetic acid (3:1) and washed twice with PBS and resuspended in 250 μl of methanol/acetic acid. Cells were dropped on an ethanol-washed cover slide and dried. The dried preparation was washed three times with PBS/0.1% BSA. Staining of metaphase chromosomes on cover slides was performed with 1 μg of recombinant DsRed or survivinDsRed diluted in 500 μl of PBS and incubation for 1.5 h in a humidified chamber. Then, cover slides were washed three times with PBS/0.1% BSA. Counterstaining of chromosomes was performed by 15-min incubation at RT with SYTOX Green (5 × 10−6 diluted; Molecular Probes), in a humidified chamber.
Time-Lapse Video Imaging
HeLa cells were grown in Lab-Tek chambered coverglasses (NUNC, Wiesbaden, Germany) The chambered coverglasses were mounted on a heated microscope stage of an inverted Axiovert 100 microscope (Carl Zeiss, Jena, Germany). The microscope stage was covered with an incubation system (Zeiss M 200; Carl Zeiss), including a humidifier, a CO2 controller, and a temperature controller. Images were acquired at 30- or 60-s intervals with a Plan-Apochromat 100× (numerical aperture 1.4) objective (Carl Zeiss) by using an Orca II cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ) and the Openlab software (Improvision, Coventry, United Kingdom). Image acquisitions were managed by an automation program, including changes of filters for green fluorescent protein (488-nm excitation/530-nm emission) and for DsRed (558-nm excitation/583-nm emission), respectively. Exposure time, electronic dimming, and contrast were adjusted for EGFP and DsRed signals before monitoring the cell cycle. Single layers were extracted from the Openlab layered image files and converted to TIF documents. In other experiments the different cell lines were transferred into a special microscopy chamber (Hofer et al., 1999) and analyzed by video-enhanced contrast microscopy (Leica DMIRB with Nomarski interference-contrast, objective Leica PL APOx100, oil). The behavior of the cells was documented by capturing 180 sequential frames (1 frame/30 s). Documentation and analysis were performed with the public domain NIH Scion Image 1.61 program. Image files were digitally processed for presentation using Adobe Photoshop (Adobe Systems). Statistical analysis was performed with Student's t test.
RESULTS
Generation of HeLa Cell Lines Stably Expressing DsRed-tagged Survivin: Immunochemical Characterization of Fusion Protein and Monitoring Its Localization during Cell Cycle
For a continuous monitoring of survivin in single cells during the entire cell cycle and for the analysis of its colocalization with structural cell proteins, we constructed retroviral vectors coding for survivin fusion proteins that are tagged with living color fluorescent proteins. Experiments revealed that survivin tagged at the C terminus with DsRed (survivinDsRed) or EGFP (survivinEGFP) gave specific mitotic signals in transduced HeLa cells, whereas vectors coding for DsRed or EGFP failed to stain mitotic structures (our unpublished data). Because of their brilliant fluorescence emission we decided to use survivinDsRed constructs for further analysis. In addition, survivinDsRed allowed the simultaneous monitoring of the mitotic spindle by coexpression of an EGFP-α-tubulin construct.
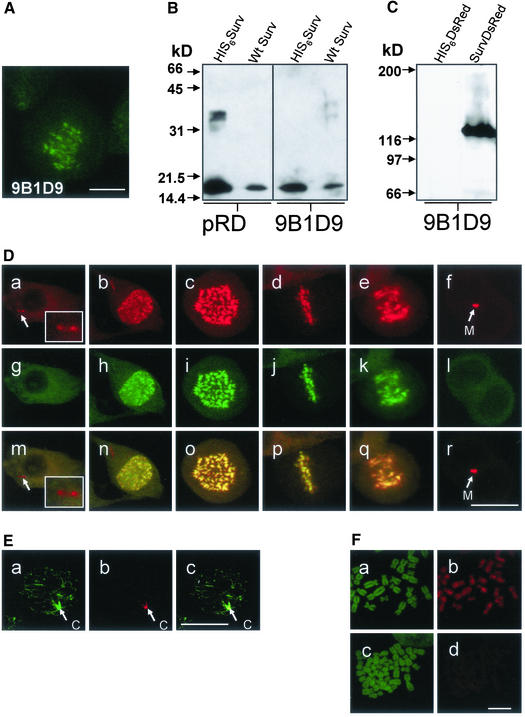
To detect native survivin and recombinant survivinDsRed constructs within cells and on Western blots, we prepared a panel of survivin-specific monoclonal antibodies by immunizing mice with full-length recombinant survivin. One antibody, mAb 9B1D9, proved to be particularly useful to recognize both native survivin in cells and survivin in immunoblots. As shown by laser scan microscopy in Figure 1A mAb 9B1D9 stained survivin located at the metaphase plate in dividing cells. When the antibody was used to characterize recombinant HIS6survivin and endogenous survivin in cell lysates by immunoblot analysis a 16.5-kDa band was obtained (Figure 1B). The mAb that specifically detected SurvivinDsRed was compared with the polyclonal anti-survivin antibody (pRD; Figure 1B). Both antibodies bound to recombinant as well as endogenous survivin. Survivin-DsRed protein expressed in HeLa cells as detected with the 9B1D9 antibody revealed a molecular mass of ∼120 kDa, indicating a tetrameric structure (Figure 1C). Tetramerization of DsRed has been shown to be a prerequisite for its fluorescence activity (Baird et al., 2000; Yarbrough et al., 2001). The antibody did not react with recombinant DsRed protein included as control.
Figure 1.
Expression and localization of tetrameric survivinDsRed and detection of endogenous and transgenic survivin proteins with the mAb 9B1D9. (A) Immunofluorescence analysis of a HeLa metaphase cell by using the monoclonal anti-survivin antibody 9B1D9 and FITC-labeled rabbit anti-mouse Fab as secondary antibody. Bar, 8 μm. (B) Immunoblot analysis of purified His-tagged survivin and HeLa cell lysates (12.5% SDS-PAGE) by using a polyclonal anti-survivin antibody (pRD; R & D Systems) or the monoclonal mAb 9B1D9 antibody. (C) Immunoblot analysis of purified His-tagged survivinDsRed fusion protein and His6DsRed protein as control (6% SDS PAGE) by using the anti-survivin mAb 9B1D9. Molecular weight markers are indicated on the left. (D) SurvivinDsRed localizations during cell cycle. Confocal laser scanning microscopy analysis of HeLa cells highly expressing survivinDsRed during mitosis (a–f), additionally detected with the 9B1D9 mAb in indirect FITC-immunofluorescence (g–l). Merge of survivinDsRed signals and anti-survivin mAb 9B1D9 FITC-fluorescence signals(m–r). a, g, and m, interphase. Note the two distinct red spots of survivinDsRed in the cytoplasm, which are not detected by the 9B1D9 mAb (arrows, inlay). b, h, and n, prophase. c, i, and o, prometaphase. d, j, and p, metaphase. e, k, and q, anaphase. f, l, and r, late telophase. Note that the 9B1D9 mAb failed to detect survivinDsRed at the midbody. Midbodies (M) are marked by arrows. Bar, 20 μm. (E) Detection of survivinDsRed at centrosomes. a–c, survivinDsRed-expressing interphase cell stained with an anti-γ-tubulin mAb in indirect immunofluorescence (a). Note that survivinDsRed (b) is localized in centrosomes (c, merge). Bar, 20 μm. (F) Binding of survivinDsRed to chromatin structures is mediated by the survivin portion of the fusion protein. a–c, Prepared metaphase chromosomes were stained with SYTOX green (a and c) and incubated either with recombinant survivinDsRed (b) or DsRed protein (d). Bar, 6 μm.
To trace the localization of survivin during the cell cycle, HeLa cell lines were produced that stably expressed survivinDsRed. High and low expression levels were obtained by transduction with an MOI of 50 and 5, respectively. Control cell lines were generated by transduction with vectors coding for DsRed, EGFP, and with pcz-CFG5-survivin-IEGZ, a construct that caused a bicistronic expression of survivin and of an EGFP-ZeoR-fusion protein via an internal ribosomal entry site. The latter served to test whether retroviral overexpression of wild-type survivin induces apoptosis, an effect that has been observed for murine survivin, polo-like kinase, or Aurora- and IpP1-like midbody-associated protein (AIM-1) (Tatsuka et al., 1998; Kobayashi et al., 1999; Conn et al., 2000). In none of the cell lines expressing DsRed, EGFP, survivin, or survivin fusion proteins an increased rate of apoptosis was observed by TUNEL analysis in comparison with HeLa wild-type cells (our unpublished data).
Figure 1D shows the intracellular localization of highly expressed survivinDsRed in HeLa cells at various phases of the cell cycle. During interphase it was excluded from the nucleus and was diffusely distributed in the cytoplasm (Figure 1D, a) with the exception of distinct spots that were identified as centrosomes (Figure 1D, a, arrows; inlay) by simultaneous staining with a monoclonal anti-γ-tubulin antibody in indirect FITC-immunofluorescence (Figure 1E, a–c). The survivinDsRed signal disappeared from centrosomes when the cell entered prophase. At beginning of chromatin condensation in early prophase survivinDsRed appeared in the nucleus (Figure 1D, b) where it was found associated with condensing chromosomes. In prometaphase (Figure 1D, c) intensive fluorescence of survivinDsRed associated with chromosomes was seen. This chromosomal association was also found in the metaphase plate (Fig, 1D, d, equatorial view). During late anaphase and at the beginning telophase, survivinDsRed was lost from chromosomes and appeared in the midbody (Figure 1D, e and f). When the mAb 9B1D9 was used to stain the survivinDsRed-expressing cells in indirect immunofluorescence it specifically recognized survivin associated with chromatin (Figure 1D, g–l), but did not detect survivin localized in centrosomes or midbodies. This failure of the mAb 9B1D9 to recognize survivinDsRed at some of its localizations becomes particularly evident when the DsRed fluorescent signals were merged with the FITC signals (Figure 1D, m–r).
The striking concentration and uniform distribution of overexpressed survivinDsRed on chromosomes during prometaphase and metaphase suggested a particular binding capacity of survivin to chromosomal structures. To exclude that this localization was mainly due to the fusion partner dsRed, recombinant survivinDsRed and dsRed proteins were purified and tested for their ability to bind to prepared chromosomes. As shown by Figure 1F only survivinDsRed bound to chromosomes, whereas the DsRed protein failed to bind, suggesting that the survivin portion determines the chromatin-binding capacity of survivinDsRed.
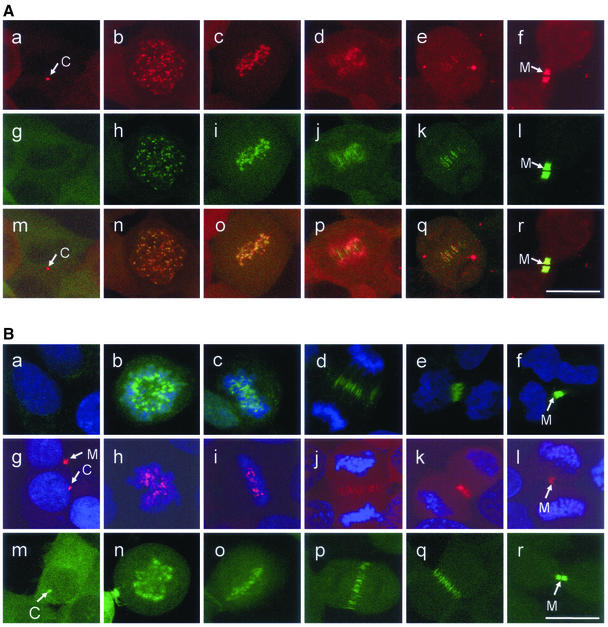
Because the 9B1D9 antibody failed to detect telophase localization of the endogenous survivin we used a polyclonal anti-survivin antibody to detect the fusion protein as well as the endogenous protein. Staining of HeLa cells with moderate expression of survivinDsRed (Figure 2A, a–f) and of HeLa wild-type cells (Figure 2B, a–f) with the polyclonal survivin antiserum (R & D Systems) revealed an intracellular localization of survivin indistinguishable from that of the fusion proteins (Figure 2A, a–f), except for the localization at centrosomes (Figure 2A, a and g). To further exclude localization artifacts due to the complex structure of the tetrameric survivinDsRed fusion protein, survivinEGFP was stably expressed in HeLa cells. It showed the same intracellular distribution during the cell cycle as survivinDsRed, including localization in centrosomes and midbodies (Figure 2B, m–r).
Figure 2.
Localizations of survivinDsRed during the cell cycle are indistinguishable from localizations revealed with endogenous survivin or survivinEGFP. (A) Confocal laser scanning microscopy analysis of HeLa cells moderately (transduction with 5 MOI) expressing survivinDsRed during mitosis (a–f), additionally detected with a polyclonal anti-survivin antibody in indirect FITC-immunofluorescence (g–l). Merge of survivinDsRed signals and anti-survivin antibody FITC-fluorescence signals (m–r). a, g, and m, interphase. Note the distinct red spot of survivinDsRed in the cytoplasm (a), which is not detected by the polyclonal antibody (g, m, arrow). b, h, and n, prophase. c, i, and o, metaphase. d, j, and p, early anaphase. e, k, and q, late anaphase/telophase. f, l, and r, late telophase. Midbodies (M) and centrosomes (C) are marked by arrows. Bar, 20 μm. (B) Localization of endogenous survivin, survivinDsRed, and survivinEGFP during the cell cycle. a–f, detection of endogenous survivin with a polyclonal anti-survivin antibody and staining of DNA with TO-PRO-3. g–l, HeLa cells transduced with 5 MOI survivinDsRed and stained with TO-PRO-3. m–r, localization of survivinEGFP protein in transduced HeLa cells. a, g, and m, interphase. b, h, and n, prometaphase. c, i, and o, metaphase. d, j, and p, anaphase/telophase. e, k, and q, telophase. f, l, and r, late telophase. Midbodies (M) and centrosomes (C) are marked by arrows. Bar, 20 μm.
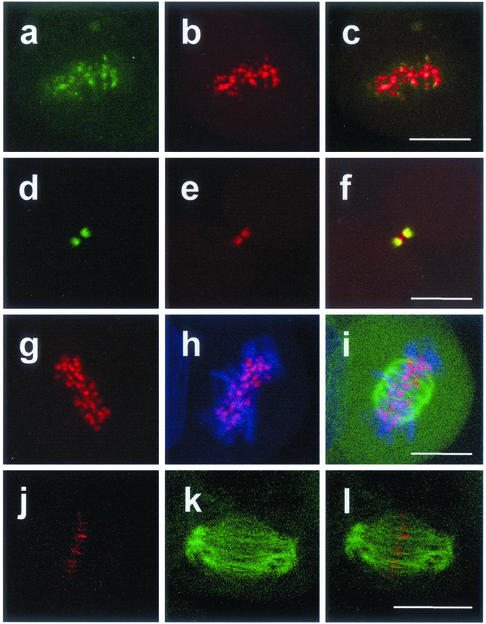
Staining with an antibody specific for the chromosomal passenger protein Aurora-B kinase/AIM-1 revealed that survivinDsRed colocalizes at kinetochores during metaphase (Figure 3, a–c). At late telophase colocalization with Aurora-B kinase/AIM-1 was also seen in midbodies (Figure 3, d–f). To analyze the localization of survivin during the cell cycle in more detail HeLa cells with low expression of survivinDsRed (transduction with 5 MOI) were additionally transduced with 10 MOI of particles coding for EGFP-α-tubulin (Figure 3, g–l) and were labeled with the DNA stain TO-PRO-3 (Figure 3, g–i). The survivinDsRed protein was detected at the centromeric region of the chromosomes. Furthermore, survivinDsRed signals were found accumulated at the ends of polar microtubuli in anaphase/telophase, demarcating the cleavage furrow (Figure 3, j–l). At variance with recent reports describing the detection of survivin at the mitotic spindle (Li et al., 1998, 1999), we were unable to find SurvivinDsRed at this localization.
Figure 3.
SurvivinDsRed colocalizes with Aurora B-kinase and with the ends of microtubules at kinetochores and the central spindle. Analysis by confocal laser scanning microscopy. a–f, HeLa cells transduced with 5 MOI pcz-CFG5.1-survivinDsRed and stained with an anti-AIM-1/Aurora B-kinase antibody in indirect FITC-immunofluorescence. a, b, and c, metaphase. a, Aurora-B kinase, b, survivinDsRed; c, merge of a and b. Note that Aurora-B kinase and survivinDsRed are colocalized at kintetochores. d–f, late telophase. d, Aurora-B kinase, e, survivinDsRed; f, merge of d and e. Note that AIM-1/Aurora-B-kinase and survivinDsRed are colocalized at the midbody. g–l, HeLa cells were double transduced with 5 MOI of pcz-CFG5.1-survivinDsRed and 10 MOI of pcz-CFG2-EGFP-tubulin and stained with TO-PRO-3 dye. g, survivinDsRed. h, survivinDsRed/TO-PRO-3 merge. i, survivinDsRed/TO-PRO-3/EGFP-α-tubulin triple merge. j–l, late anaphase/early telophase. HeLa cells were double transduced with 5 MOI of pcz-CFG5.1-survivinDsRed and 10 MOI pcz-CFG2-EGFP-α-tubulin. j, survivinDsRed; k, EGFP-α-tubulin; l, merge of j and k. Note that survivinDsRed is colocalized with the ends of the polar microtubules. Bar (a–i), 10 μm; (g–l), 15 μm.
Trafficking of SurvivinDsRed from Kinetochores to Cleavage Furrow Is Microtubuli Independent
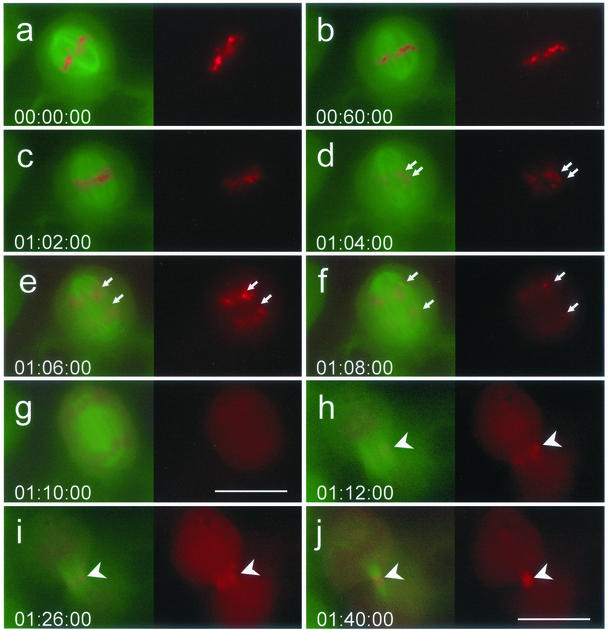
To analyze the localization of survivin during development of the cleavage furrow and subsequent midbody formation we performed time-lapse video imaging of HeLa cells that simultaneously expressed survivinDsRed and EGFP-α-tubulin. It was of particular interest to find out how survivinDsRed reaches the cleavage furrow and whether survivin assembly at this structure contributes to the initiation of cell cleavage. There are two possible ways how survivin can traffic from kinetochores to the cleavage furrow. First, survivin is released from kinetochores and diffuses to the cleavage furrow to reassemble at the ends of the polar microtubules. Second, it leaves kinetochores during anaphase and moves onto the central microtubules, thus marking a premature cleavage furrow before onset of telophase. As illustrated in Figure 4, at early anaphase survivinDsRed signals in cells transduced with 50 MOI continuously disappeared from the kinetochores (Figure 4, c and d), but there was still a faint survivinDsRed staininig on the chromosomes that faded away as anaphase proceeded (Figure 4, e and f). A stepping over microtubular structures to the anaphase polar spindles was not observed; instead, survivinDsRed was found uniformly distributed in the cytoplasm, suggesting free diffusion of survivinDsRed proteins (Figure 4g). In late anaphase, survivinDsRed was found to reassemble at the telophase disk of the cleavage furrow (Figure 4h) where it immediately condensed into the midbody formed by the contractile actin/myosin ring (Figure 4, i and j).
Figure 4.
Release of survivinDsRed from chromosomes, its equal distribution in the cytoplasm and reaggregation at the telophase disk. Time-lapse video imaging analysis of a HeLa cell line stably expressing survivinDsRed and EGFP-α-tubulin. a and b, metaphase; c–g, anaphase; and h–j, telophase. Exposure time in h–j was three times longer than in a–g. Arrows mark the progression of chromatide separation during mitosis. Arrowheads mark the cleavage furrow. Bars (a–g), 20 μm; h–j, 30 μm. Time is indicated as hours:minutes:seconds.
SurvivinDsRed Mutated at p34cdc2 Phosphorylation Site Causes Aberrant Execution of Cytokinesis
To get further insight into the function of survivin, we generated HeLa cell lines stably expressing a survivin mutant with a defective p34cdc2 phosphorylation site at amino acid position 34, named survivinM34DsRed. Transduction of the cells was performed with 5 or 50 MOI of particles coding for the mutant survivin fusion protein.
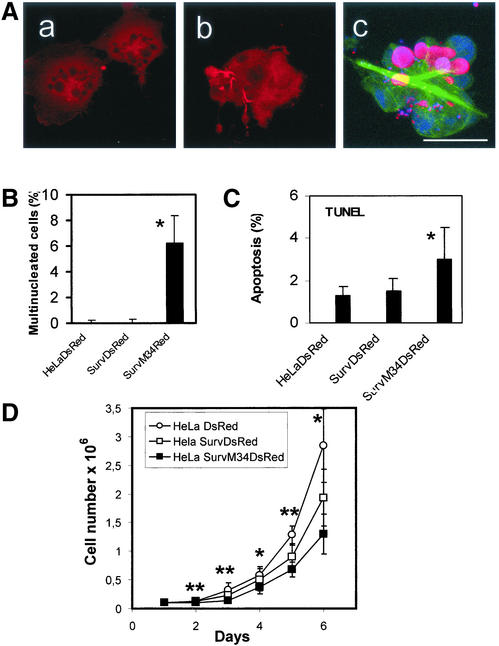
A striking finding was that a high expression level of the transgenic survivinM34DsRed resulted in a high incidence of defective cytokinesis, leading to an increased frequency of multinuclear cells (Figure 5, A and B), whereas low expression of this mutant had no effects (our unpublished data). The overexpression of the mutated survivinDsRed resulted in elongated and abnormal midbodies, but also in a defective cleavage of cells. This often resulted in formation of membrane-embedded vesicles containing DNA and survivinM34DsRed, which appeared as cell debris in the cell culture supernatant (Figure 5A, c). Also, detached multinuclear cells were detected in the cell culture supernatant, which accounted for 16.7 ± 4.1% of the whole cell population. The incidence of multinuclearity among adherent cells amounted to 6% in cells overexpressing the mutant survivinM34DsRed, whereas the DsRed-transduced control cells showed 0.13% and the cells expressing survivinDsRed 0.15% multinuclearity, respectively (Figure 5B). The enhanced frequency of multinuclearity in survivin-M34DsRed-transduced cells probably resulted in a slightly increased rate of apoptosis, determined by TUNEL analysis (Figure 5C). Because cultures of cells expressing the mutant survivin fusion protein showed a great number of detached cells and cellular debris the determination of apoptotic indices was restricted to adherent cells. The higher incidence of apoptosis and especially the effects of aberrant cytokinesis are reflected by the reduced rate of cell proliferation seen in cultures of survivinM34DsRed-expressing cells compared with cells tranduced with DsRed only (Figure 5D). Proliferation of cells expressing survivinDsRed was not significantly different from the DsRed-transduced control cells.
Figure 5.
Overexpression of survivinM34DsRed causes defective cytokinesis, apoptosis, and impaired cellular growth. (A) Multinuclearity of HeLa cells caused by high-level expression of the survivinM34DsRed mutant. a and b, multiple nuclei and aberrant cleavage of cell bodies. C, triple merge of a cell provirally coexpressing mutant survivinM34DsRed and α-tubulin and being stained with TO-PRO3. DNA-staining (blue), α-tubulin (green), and merge of survivinM34DsRed and DNA (pink). Note the irregular appearance of the microtubular cytoskeleton and cleavage of membranes containing DNA or DNA-bound survivinM34DsRed. Bar, 20 μm. (B) Incidence of multinuclearity in adherent HeLa cells transduced with 50 MOI of particles coding for survivinDsRed, survivinM34DsRed, or as control, DsRed. Of each cell line 2500 cells were analyzed. Bars, ±SD. p < 0.05 for HeLa survivinM34DsRed cells compared with HeLa DsRed cells and HeLa survivinDsRed cells, respectively. (C) TUNEL analysis of HeLa cells with high-level expression of DsRed, survivinDsRed, and survivinM34DsRed. Of each cell line 2000 cells were analyzed. Bars, ±SD. *p < 0.05 for HeLa survivinM34DsRed compared with HeLa DsRed and HeLa survivinDsRed, respectively. (D) Impaired cellular growth of HeLa cells overexpressing survivinM34DsRed. Cells (105) were plated per well in culture dishes. At the indicated time points cells were trypsinized and counted. Data represent means of five independent experiments. Bars, ±SD. *p < 0.05 or **p < 0.01 when cells overexpressing survivinM34DsRed were compared with the DsRed-transduced control cell line.
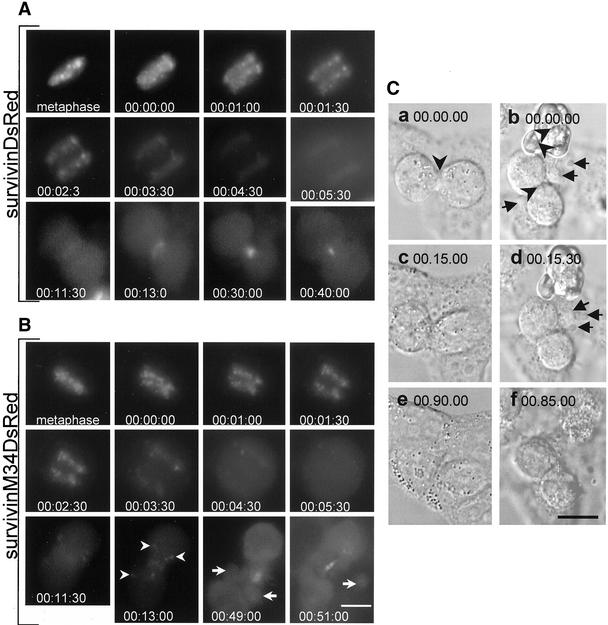
To identify the principal effects of the highly expressed mutant protein we performed time-lapse video imaging of HeLa cells stably overexpressing survivinDsRed and survivinM34DsRed, respectively. SurvivinDsRed and survivinM34DsRed showed the same spatial distribution throughout mitosis. We found no differences in the mean duration of the metaphase between cells with high expression of survivinDsRed (37 ± 16 min, n = 7) and survivinM34DsRed (35 ± 18 min, n = 11), but both cell lines showed a significantly increased mean duration of metaphase compared with DsRed-transduced control cells (15 ± 6 min, n = 4, p < 0.05). The mean duration of anaphase was ∼10 min in all cell lines analyzed. In contrast to the wild-type survivin fused to DsRed, the mutant survivinM34DsRed often was not completely concentrated at the telophase disk (Figure 6B, arrowheads). During late telophase all cells expressing p34cdc2 phosphorylation-defective survivinM34DsRed showed membrane blebbing, especially close to the cleavage furrow and midbody (Figure 6B, arrows). This defective cytokinesis could be clearly demonstrated by time-lapse differential interference contrast imaging. Some multinucleated cells failed to be cleaved and then became apoptotic (Figure 6C, b arrowhead), but all cells analyzed showed formation of blebs in the area of the cleavage furrow, causing loss of membrane-embedded cytoplasm during cytokinesis (Figure 6C, b and d, arrows). Despite the marked membrane alterations these cells were often able to finally execute cleavage of daughter cells.
Figure 6.
High-level expression of p34cdc2 phosphorylation-defective survivin causes aberrant cleavage of HeLa cells. (A) Time-lapse video imaging of a HeLa cell line with high-level expression of survivinDsRed fusion protein. Images were acquired at intervals of 30 s. Start of chromosomal segregation was set as time zero. (B) HeLa cell line with high level expression of mutant survivinM34DsRed. Note that not all survivinM34DsRed signals accumulated at the cleavage furrow. Arrowheads indicate localization of signals in the cytoplasm of the daughter cells. Arrows depict blebbing of the daughter cells in the vicinity of the premature midbody at telophase. (C) Differential interference contrast microscopy of HeLa cells with high expression of survivinDsRed (a, c, and e) or mutant survivinM34DsRed (b, d, and f). Arrows indicate membrane blebbing at the cleavage furrow at telophase. Arrowheads depict cleavage furrows. Bars (A and B), 15 μm; and C, 20 μm.
Both Overexpressed SurvivinDsRed and SurvivinM34DsRed Mutated at p34cdc2 Phosphorylation Site Confer Resistance to Apoptotic Reagents
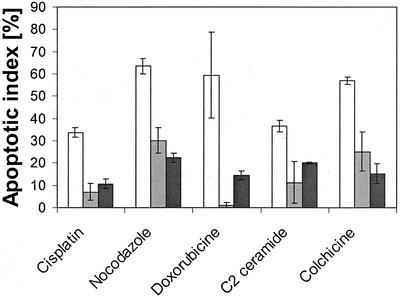
As reported by Li et al. (1998, 1999), the antiapoptotic effect of survivin depends on its association with the mitotic spindle. Because in our studies strongly expressed survivin-DsRed was not associated with microtubules, it was of interest to test whether it was able to prevent apoptosis. To this end, HeLa cells overexpressing survivinDsRed, or, as a control, DsRed were treated with several apoptosis-inducing reagents. Colchicine and nocodazole, both interacting with microtubuli, induced apoptosis in ∼60% of DsRed-transduced HeLa control cells, whereas HeLa cells overexpressing survivinDsRed seemed to be protected (Figure 7). Neither of the two drugs was found to influence the localization of survivinDsRed on chromosomes and kinetochores (our unpublished data). As further shown in Figure 7, the DNA-damaging drug cisplatine and the reagent doxorubicine, which binds into the minor groove of the DNA helix and leads to mutations and subsequently to apoptosis, caused apoptosis in 35 and 60%, respectively, of DsRed-transduced HeLa cells. Again, overexpression of the survivin fusion protein resulted in enhanced resistance to apoptosis induced by both reagents. The proapoptotic effect of doxorubicine was even completely abolished. Finally, C2 ceramide has been described to cause a dysregulated mitochondrial membrane potential, leading to cytochrome c efflux and to activation of caspase 9, resulting in apoptosis (Gudz et al., 1997). Approximately 36% of HeLa cells stably transduced with DsRed underwent apoptosis after treatment with C2 ceramide, whereas apoptosis was markedly reduced in cells with high expression of survivinDsRed (11%).
Figure 7.
Overexpressed survivinDsRed and p34cdc2 phosphorylation-defective mutant survivinM34DsRed are inhibitors of apoptosis. Annexin V analysis of HeLa cells treated with various proapoptotic reagents. HeLa cells with high-level expression of DsRed (white columns), survivinDsRed (gray columns) and mutant survivinM34DsRed (black columns) were analyzed. Data represent means of three independent experiments. Bars, ±SD. *p < 0.05 and **p < 0.01.
Previously, it has been reported that a p34cdc2 phosporylation-defective mutant of survivin (survivinM34) displayed a dominant negative phenotype that resulted in apoptosis in transfected melanoma and HeLa cells (Grossman et al., 1999; Li et al., 1999; Mesri et al., 2001). In this study, we stably overexpressed survivinM34DsRed in a HeLa cell line. Surprisingly, it not only failed to directly induce apoptosis, it even protected the cells against the effect of the various proapoptotic reagents to a degree similar to that observed with the wild-type survivin fused to DsRed (Figure 7).
p34cdc2 Phosphorylation-defective SurvivinM34DsRed Fails to Localize at Kinetochores
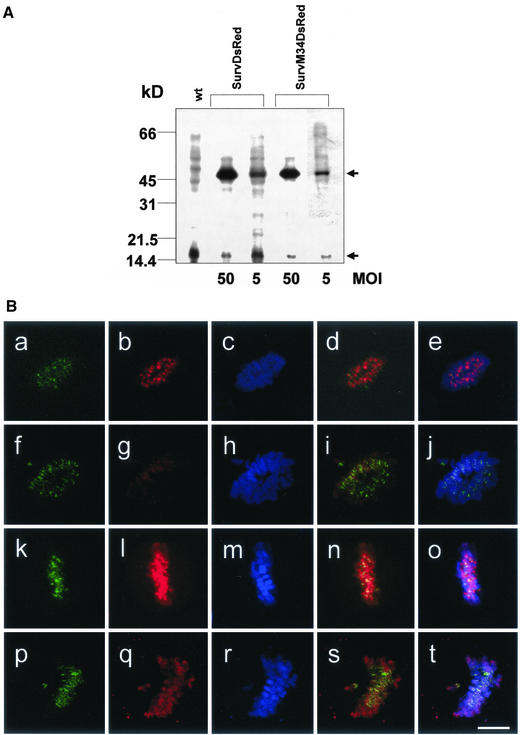
To determine the expression levels of the ectopic proteins, we performed immunoblot analysis of nonsynchronized HeLa cells with high and low expression of survivinDsRed and survivinM34DsRed, respectively. As expected, the immunoblots showed that HeLa cells transduced with 50 MOI of pczCFG5.1survDsRed and pcz-CFG5.1survM34DsRed, respectively, expressed more transgenic protein than the cells transduced with 5 MOI of the appropriate virus (Figure 8A). The expression level of 50 MOI-survDsRed was in the same range as the expression level of 50 MOI-survM34DsRed. In each cell line the transgenic protein expression was compared with the endogenous protein level. Expression of survivinDsRed and survivinM34DsRed was increased by a factor of 10.8 and 12.7, respectively, compared with the endogenous survivin in cells transduced with a high MOI. The survivinDsRed protein level of cell lines transduced with a low MOI was 2.3-fold (survivin-DsRed) and 4.1-fold (survivinM34DsRed) higher than the endogenous survivin protein level.
Figure 8.
(A) Survivin expression levels of HeLa cell lines transduced with 5 MOI and 50 MOI of survivinDsRed or survivinM34DsRed. Detection of survivin in HeLa cell lysates by Western blot analysis by using the anti-survivin mAb 9B1D9. Concomitant detection of endogenous survivin (bottom arrow) and retrovirally expressed survivin fusion proteins (top arrow) in permanent HeLa cell lines transduced with different multiplicity of infections. Molecular weight markers are indicated on the left. (B) Phosphorylation-defective survivinM34DsRed is not targeted to kinetochores. Confocal laser scanning microscopy analysis of HeLa cell lines stably transduced with 5 MOI pcz-CFG5.1-survivinDsRed (a–e) or with 5 MOI of the p34cdc2 phosphorylation-defective pcz-CFG5.1-survivinM34DsRed (f–j), with 50 MOI of pcz-CFG5.1-survivinDsRed (k–o) or with 50 MOI pcz-CFG5.1-survivinM34DsRed (p–t). a, f, k, and p, detection of the chromosomal passenger protein Aurora B-kinase by an anti-AIM-1/Aurora B-kinase antibody in indirect FITC-immunofluorescence. b, g, l, and q, provirally expressed survivinDsRed or survivinM34DsRed. c, h, m, and r, TO-PRO-3 staining of DNA. d, i, n, and s, merge of Aurora B-kinase and survivinDsRed or survivinM34DsRed signals. e, j, o, and t, triple merge with TO-PRO-3 dye. Bar, 5 μm.
Previously, it has been reported that mutants of the passenger proteins INCENP and Aurora-B-kinase that fail to bind to kinetochores cause defective cytokinesis (Mackay et al., 1998; Tatsuka et al., 1998). Therefore, we were interested to find out whether survivinM34DsRed is capable to localize at kinetochores. To this end, we analyzed HeLa cells with low and high expression of the survivinM34DsRed or the nonmutated survivinDsRed, respectively. In addition, the chromosomal passenger protein Aurora B-kinase was detected by a mAb and DNA was stained by TO-PRO-3 dye. SurvivinDsRed was clearly targeted to the centromeres in cells transduced with a low MOI (Figure 8B, a–e), whereas the survivinM34DsRed failed to accumulate at the kinetochores when transduced with 5 MOI (Figure 8B, f–j) but was detected in midbodies (our unpublished data). It even failed to bind to the chromosomes. In cells transduced with 50 MOI, both survivin fusion proteins were able to bind to chromosomes (Figure 8B, k–t). However, in contrast to wild-type survivinDsRed the mutated survivinDsRed failed to or did only slightly accumulate at kinetochores as was evidenced by merging the three fluorescence signals (Figure 8B, d, i, n, and s, as well as e, j, o, and t).
DISCUSSION
Overexpression of 142 aa survivin fused to DsRed at low and high levels allowed us to continuously chase the still equivocal localization of survivin during the cell cycle and to correlate it with coexpressed EGFP-labeled α-tubulin. In addition, we obviated problems arising from the human survivin 165 aa and 137 aa isoforms (Mahotka et al. 1999), which eventually exhibit localizations and functions different from originally described survivin.
In accordance with previously published data on survivin localization (Skoufias et al., 2000; Uren et al., 2000; Rodriguez et al., 2002), survivinDsRed was found to be excluded from the nucleus during interphase. It was, however, detected in centrosomes during this phase of the cell cycle (Figures 1, D and E, 2A). In prometaphase it strongly bound to chromosomes and became concentrated at the kinetochores in metaphase (Figures 2A and 3). The accumulation of survivin at kinetochores resembled the dynamics of INCENP and Aurora-B-kinase (Ainsztein et al., 1998; Adams et al., 2000), which are recruited together with survivin to the centromere to build a so-called chromosomal passenger complex (Wheatley et al., 2001). At telophase, the survivinDsRed was colocalized with the ends of the microtubuli of the central spindle, which during cytokinesis was condensed to the midbody by the actin-myosin contractile ring.
When HeLa cells were transduced with a high MOI, survivinDsRed was not only localized at kinetochores but also bound to the chromosomal arms from prophase to anaphase. This suggests that survivin in superabundance cannot completely be targeted to the kinetochores by complexing with INCENP or other relevant proteins. We demonstrated by a chromosomal binding assay that survivinDsRed bound to the chromosomes, whereas DsRed tetramers failed to bind. Therefore, interaction of the survivinDsRed with the chromosomes was apparently mediated by its survivin proportion. Furthermore, our own data demonstrate that the subcellular localization of endogenous survivin and a survivinEGFP fusion protein expressed in HeLa cells is not different from that of survivinDsRed during mitosis. From this it is concluded that the tetrameric structure of survivinDsRed is not pivotal for the localization of this fusion protein. In contrast to reports where the association of survivin with the mitotic spindle was shown by staining with an anti-survivin antibody (Li et al., 1998, 1999; Fortugno et al., 2002), we were not able to detect the endogenous survivin or the overexpressed survivin fusion proteins at this localization. One might speculate whether binding to the mitotic spindle is a characteristic of one of the survivin isoforms. This question could be answered by further analysis of survivin 165 aa and 137 aa isoforms fused to living color proteins.
Live cell cinematography revealed that survivin leaves the kinetochores at beginning anaphase and freely diffuses into the cytoplasm. At telophase, reassembly of survivin at the ends of the polar spindles was immediately followed by cell cleavage. Targeting of survivin to the telophase disk suggests that it is involved in cytokinesis. The redistribution of survivin could delay cytokinesis and thus provide a safeguard for the successful segregation of the chromatides. During anaphase we did not observe a direct transmission of survivinDsRed to the central spindle as it was recently described for INCENP (Wheatley et al., 2001b).
The phenotype created by the high-level expression of the phosphorylation-defective survivinM34DsRed showed a similar cytokinesis defect as did cells transfected with mutated INCENP (Mackay et al., 1998) or with an inactive Aurora-B-kinase (Tatsuka et al., 1998). First analysis of HeLa cells transduced with 50 MOI of survivinM34DsRed revealed an increased multinuclearity of adherent cells. Furthermore, detached multinucleated cells accounting for ∼17% of the whole cultured cell population were detected in the cell culture supernatant. This indicates that a defective cytokinesis occurred during proliferation of the cells. The elevated incidence of multinuclearity is reflected by the impaired cell proliferation observed in HeLa cells with a strong expression of the mutant survivin (Figure 5d).
When the effects of phosphorylation-defective survivin were analyzed in more detail, the transmission of the survivinM34DsRed to the centromeres seemed to be impaired but it was still able to reassemble at the telophase disk (Figure 6B). Membrane blebbing and fusion events in the vicinity of the developing midbody were seen at onset of cytokinesis in every cell analyzed by time-lapse video imaging (n = 11). This could be responsible for the increased incidence of multinuclearity. Yet, the majority of cells was able to complete cytokinesis despite the blebbing and fusion events. We believe that the membrane blebbing is due to the inability of survivinM34DsRed to cooperate with proteins involved in cell cleavage. Several studies of mammalian cells revealed the role of a kinesin motor protein (i.e., CHO1/MKLP1, Zen-4; Kaitna et al., 2000; Powers et al., 1998;) as part of the chromosomal passenger complex at the ends of polar microtubuli, demarcating the cleavage furrow. It is proposed that this kinesin bundles the central spindle and midbody matrix before the establishment of the contractile actin/myosin ring. It might well be that mutant survivin interferes with the correct recruitment of the Zen-4 kinesin and that this results in the observed phenotype.
We found a prolonged metaphase in cells transduced with a high MOI of survivinDsRed, suggesting an extended period of sister chromatid cohesion. However, we did not find differences in the duration of metaphase in cells expressing survivinDsRed compared with cells expressing the phosporylation-defective survivinDsRed. Overexpression of mutant survivin apparently did not influence mitotic progression but led to an aberrant cell cleavage, indicating that phosphorylation of survivin is essential for cytokinesis. A recent study showed that the p34cdc2-cyclinB1 was able to phosporylate survivin at Thr34 (O'Connor et al., 2000). The p34cdc2 has been detected on the mitotic spindle, which is, however, remote from the kinetochore-localized survivin. As an alternative we propose Aurora-B-kinase, which is detected at the same localization as survivin, to be a candidate kinase to phosphorylate survivin. This hypothesis is supported by the fact, that overexpression of a kinase-inactive Aurora-B-kinase caused a multinuclearity phenotype that resembles the phenotype observed in cells transduced with a high MOI of phosphorylation-defective survivin.
In contrast to a recent study (O'Connor et al., 2000), we did not detect a profound increase of apoptosis by introducing the phosphorylation-defective survivin. It has been hypothesized that loss of phosphorylation on Thr34 results in dissociation of survivin–caspase complexes, thus facilitating induction of apoptosis.
At variance with this view is our finding that transduction with even 50 MOI survivinM34DsRed particles yielded permanent cell lines and caused only a slight increase (3%) of apoptosis. We suggest that the observed cytokinesis defects of these cells may contribute to the slightly increased apoptosis. At a critical grade of multinuclearity or aneuploidy errors in chromosomal segregation may finally result in apoptosis. At present, we do not know whether the differing apoptosis-phenotypes caused by overexpressed mutant survivin reflect technical differences.
That the tetrameric structure of survivinM34DsRed was responsible for the failure to cause apoptosis was rendered unlikely because recently performed experiments revealed that also phosphorylation-defective survivinM34EGFP and survivinM34 linked to EGFP via an internal ribosomal entry site did not cause excessive apoptosis in HeLa cells (Temme and Rieber, unpublished data). In line with our data, depletion of survivin by using antibody injection or small interfering RNA in vertebrate cells did not result in pronounced apoptosis, but led to a gradual increase in multinucleation (Kallio et al., 2001; Wheatley, Carvalho, and Earnshaw, personal communication).
Survivin has originally been described as an inhibitor of apoptosis protein. It was hypothesized that survivin could link mitosis to apoptosis, thus providing a death switch for cells undergoing defective mitosis. We were interested to see how cell lines overexpressing the chimeric survivinDsRed or mutant survivinM34DsRed cope with proapoptotic reagents. Furthermore, we tried to determine whether the IAP function of overexpressed survivin was linked to mitosis. In contrast to recent studies (Li et al., 1998, 1999) cells overexpressing survivinDsRed were protected against the microtubule-destroying drugs colchicin and nocodazole. Because survivinDsRed was not localized at the mitotic spindle, these data do not support the view that the antiapoptotic effect of survivin depends on its association with the mitotic spindle. Interestingly, survivinDsRed was also capable to diminish apoptosis induced by the DNA-damaging reagents cisplatin and doxorubicine. At variance with recent reports (O'Connor et al., 2000; Grossman et al., 2001), overexpressed survivinM34DsRed also had a protective effect against proapoptotic reagents compared with HeLa control cells transduced with DsRed. Thus, survivin lacking p34cdc2 phosphorylation apparently is not a dominant negative effector of apoptosis. Furthermore, overexpressed survivinDsRed and survivinM34DsRed were able to partially inhibit the apoptosis-inducing effect of C2 ceramide, suggesting that survivin can inhibit the apoptotic cascade after mitochondrial damage presumably by complexing caspases. Therefore, we conclude that the IAP function of survivin is not necessarily linked to the cell cycle.
In summary, this study confirms survivin as a chromosomal passenger protein that is released from kinetochores at beginning of anaphase and reassembles at the telophase disk. Furthermore, evidence is provided that phosphorylation of survivin represents a crucial step for the execution of cytokinesis. We suggest that mutant survivin destabilizes the organization of the chromosomal passenger complex at the cleavage furrow, resulting in abnormal cleavage of cells.
ACKNOWLEDGMENTS
We thank S. Heinicke, S. Schwind, and B. Utess for excellent technical assistance and Dr. T. Ott and Prof. Dr. K. Willecke (Institute for Genetics, University Bonn, Germany) for providing the HeLa wild-type cells. This work was supported by a grant of the Ministry of Environment and Agriculture (Az. 56-8811.61/71), the State of Saxony, Germany (to A.T and E.P.R.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0182. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0182.
REFERENCES
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC. INCENP binds the Aurora-related kinase AIRK2, and is required to target it to chromosomes, the central spindle, and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Ainsztein AM, Kandel-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;7:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DP, Plescia J, Altieri DC, Chen J, Rosenberg SH, Zhang H, Ng SC. Survivin does not inhibit caspase-3 activity. Blood. 2000;96:4002–4003. [PubMed] [Google Scholar]
- Berberich-Siebelt F, Klein-Hessling S, Santner-Nannan B, Hepping N, Lindemann D, Schimpl A, Berberich I, Serfling E. C/EBPβ enhances IL-4 but impairs IL-2, and IFN-g induction in T cells. Eur J Immunol. 2000;30:2576–2585. doi: 10.1002/1521-4141(200009)30:9<2576::AID-IMMU2576>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Ann Biochem. 1976;772:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Conn CW, Hennigan RF, Dai W, Sanchez Y, Stambrook PJ. Incomplete cytokinesis, and induction of apoptosis by overexpression of the mammalian polo-like kinase, Plk3. Cancer Res. 2000;60:6826–6831. [PubMed] [Google Scholar]
- Conway EM, Pollefeyt S, Cornelissen J, DeBaere I, Steiner-Mosonyi M, Ong K, Baens M, Collen D, Schuh AC. Three differentially expressed survivin cDNA variants encode proteins with distinct antiapoptotic functions. Blood. 2000;95:1435–1442. [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins – suppressors of apoptosis. Genes Dev. 1999;13:1253–1262. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh A, Asanuma K, Moriai R, Yamada M, Koyanagi Y, Sato T, Yagihasi A, Nakamura M, Kobayashi D, Watanabe N. Expression of survivin mRNA in CD34-positive cells. Clin Chim Acta. 2001;306:149–151. doi: 10.1016/s0009-8981(01)00416-8. [DOI] [PubMed] [Google Scholar]
- Fortugno P, Wall NR, Giodini A, O'Connor DS, Plescia J, Padgett KM, Tognin S, Marchisio PC, Altieri DC. Survivin exists in immunochemically distinct subcellular pools, and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci USA. 2001;98:635–640. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J Biol Chem. 1997;272:24154–24158. doi: 10.1074/jbc.272.39.24154. [DOI] [PubMed] [Google Scholar]
- Hofer A, Nagel F, Wonka F, Krinke HE, Golfert F, Funk RH. A new perfusion cell chamber system for determination of heat shock effects by means of video-enhanced microscopy. Med Biol Eng Comput. 1999;37:667–669. doi: 10.1007/BF02513364. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. INCENP, and an Aurora-like kinase form a complex essential for chromosome segregation, and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Stover ML, Liu P, Kalajzic Z, Rowe DW, Lichtler AC. Use of VSV-G pseudotyped retroviral vectors to target murine osteoprogenitor cells. Virology. 2001;284:37–45. doi: 10.1006/viro.2001.0903. [DOI] [PubMed] [Google Scholar]
- Kallio MJ, Nieminen M, Eriksson JE. Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation, and mitotic exit. FASEB J. 2001;15:2721–2723. doi: 10.1096/fj.01-0280fje. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T. Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proc Natl Acad Sci USA. 1999;96:1457–1462. doi: 10.1073/pnas.96.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R, Yamakawa H, Utsunomiya H, Ito K, Sato S, Yajima A. Expression of survivin, and Bcl-2 in the normal human endometrium. Mol Hum Reprod. 2000;6:529–534. doi: 10.1093/molehr/6.6.529. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–686. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, Villa A, Marchisio PC, Altieri DC. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Mackay AM, Ainsztein AM, Eckley DM, Earnshaw WC. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J Cell Biol. 1998;140:991–1002. doi: 10.1083/jcb.140.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097–6102. [PubMed] [Google Scholar]
- Mesri M, Wall RW, Li J, Kim RW, Altieri DC. Cancer gene therapy using a survivin mutant adenovirus. J Clin Invest. 2001;108:981–990. doi: 10.1172/JCI12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore SW, Chen J, Jakob C, Zakula D, Matayoshi ED, Wu W, Zhang H, Li F, Ng SC, Altieri DC. Crystal structure, and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol Cell. 2000;6:173–182. [PubMed] [Google Scholar]
- O'Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Bossinger O, Rose D, Strome S, Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Span SW, Ferreira CG, Kruyt FA, Giaccone G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein survivin. Exp Cell Res. 2002;275:44–53. doi: 10.1006/excr.2002.5492. [DOI] [PubMed] [Google Scholar]
- Schmitz M, Diestelkoetter P, Weigle B, Schmachtenberg F, Stevanovic S, Ockert D, Rammensee HG, Rieber EP. Generation of survivin-specific CD8+ T effector cells by dendritic cells pulsed with protein or selected peptides. Cancer Res. 2000;60:4845–4849. [PubMed] [Google Scholar]
- Shi Y. Survivin structure: crystal unclear. Comment in Nat Struct Biol. 2000;8:620–623. doi: 10.1038/77904. [DOI] [PubMed] [Google Scholar]
- Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3, and -7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Mollinari C, Lacroix FB, Margolis RL. Human survivin is a kinetochore-associated passenger protein. J Cell Biol. 2000;25:1575–1582. doi: 10.1083/jcb.151.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuka M, Katayama H, Ota T, Tanaka T, Odashima S, Suzuki F, Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–4816. [PubMed] [Google Scholar]
- Uren AG, Coulson EJ, Vaux DL. Conservation of baculovirus inhibitor of apoptosis repeat proteins BIRps in viruses, nematodes and yeast. Trends Biochem Sci. 1998;23:59–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin, and the inner centromere protein INCENP show similar cell-cycle localization, and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Huang H, Dutil E, Kaiser A, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:620–623. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres, and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Kandels-Lewis SE, Adams RR, Ainsztein AM, Earnshaw WC. INCENP binds directly to tubulin, and requires dynamic microtubules to target to the cleavage furrow. Exp Cell Res. 2001b;262:122–127. doi: 10.1006/excr.2000.5088. [DOI] [PubMed] [Google Scholar]
- Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;113:4363–4371. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]