Abstract
Protein phosphatase 1 (PP1) is a ubiquitous serine/threonine phosphatase that regulates many cellular processes, including cell division. When transiently expressed as fluorescent protein (FP) fusions, the three PP1 isoforms, α, β/δ, and γ1, are active phosphatases with distinct localization patterns. We report here the establishment and characterization of HeLa cell lines stably expressing either FP-PP1γ or FP alone. Time-lapse imaging reveals dynamic targeting of FP-PP1γ to specific sites throughout the cell cycle, contrasting with the diffuse pattern observed for FP alone. FP-PP1γ shows a nucleolar accumulation during interphase. On entry into mitosis, it localizes initially at kinetochores, where it exchanges rapidly with the diffuse cytoplasmic pool. A dramatic relocalization of PP1 to the chromosome-containing regions occurs at the transition from early to late anaphase, and by telophase FP-PP1γ also accumulates at the cleavage furrow and midbody. The changing spatio-temporal distribution of PP1γ revealed using the stable PP1 cell lines implicates it in multiple processes, including nucleolar function, the regulation of chromosome segregation and cytokinesis.
INTRODUCTION
Reversible protein phosphorylation is the major general mechanism that regulates most physiological processes in eukaryotic cells. Protein phosphatase I (PP1) is involved in a wide range of cellular processes and is believed to derive both its intracellular localization and its substrate specificity from proteins with which it associates, termed “targeting” subunits (see Cohen, 2002 for review).
Analysis of the subnuclear targeting of PP1 is complicated by the fact that it is expressed in mammalian cells as three closely related isoforms, α, β/δ, and γ1, which are encoded by separate genes (Sasaki et al., 1990; Barker et al., 1993; Barker et al., 1994). These isoforms are more than 89% identical in amino acid sequence, yet show distinct subcellular localization patterns when analyzed by antibody staining of fixed cells (Andreassen et al., 1998). Although all three isoforms are found in the nucleus in interphase cells, only PP1γ shows a strong accumulation in the nucleolus, suggesting a role in the regulation of one or more nucleolar processes. Evidence also points to PP1 as a major counteracting phosphatase to several cell cycle–regulated kinases (Fernandez et al., 1992; Berndt et al., 1997), with antibody staining also suggesting distinct patterns for the isoforms during mitosis (Andreassen et al., 1998; Zeitlin et al., 2001).
We were interested in examining the organization of the specific isoforms throughout the cell cycle but found that PP1 isoform-specific antibodies from three different commercial sources, while giving clean signals on Western blots for both endogenous and expressed PP1, gave weak and irreproducible signals for immunostaining. An alternative approach to analyzing the localization of PP1 isoforms in vivo is to express each isoform fused to a fluorescent protein (FP) tag, which avoids problems of antibody cross-reactivity and fixation effects while also permitting imaging of protein in live cells. A critical issue, however, is whether the presence of the FP-tag influences PP1 function or localization. We have already demonstrated that the respective FP-PP1 fusion proteins are active phosphatases, with distinct localization patterns that can be disrupted by overexpression of a PP1 targeting subunit (Trinkle-Mulcahy et al., 2001). On transient transfection of expression plasmids, FP-PP1α is found mainly in a diffuse nucleoplasmic pool and largely excluded from the nucleolus, whereas FP-PP1γ accumulates predominantly within the nucleolus. FP-PP1β is found in both the nucleoplasm and the nucleolus but does not appear to accumulate within the nucleoli to the same extent as FP-PP1γ (Trinkle-Mulcahy et al., 2001). We were particularly interested in PP1γ because of its nucleolar accumulation, which is consistent with its identification in our recent proteomics study of purified nucleoli (Andersen et al., 2002). Having the ability to purify nucleoli allows us to analyze tagged PP1γ in this structure both by microscopy and by biochemistry.
Two different HeLa cell lines stably expressing FP-PP1γ were established and characterized, as was a cell line expressing FP alone. Fluorescence time-lapse imaging reveals that localization of FP-PP1γ is dynamic throughout the cell cycle, with specific patterns implicating it in the regulation of nucleolar function, chromosome segregation, and cytokinesis.
MATERIALS AND METHODS
Establishment and Characterization of FP-PP1γ Stable HeLa Cell Lines
EGFP-PP1γ, EYFP-PP1γ, and ECFP-NIPP1 were obtained as described previously (Trinkle-Mulcahy et al., 2001). For the establishment of HeLaEGFP-PP1γ, 1 μg of EGFP-PP1γ plasmid was transfected into a 10-cm dish of HeLa cells using Effectene transfection reagent (Qiagen, Santa Clara, CA). The HeLaEGFP cell line was established in a similar manner, transfecting the EGFP-C1 plasmid (Clontech, Palo Alto, CA) alone. For the establishment of HeLaEYFP-PP1γ, 1 μg of EYFP-PP1γ plasmid and 0.1 μg of ECFP-NIPP1 plasmid were cotransfected into a 10-cm dish of HeLa cells using Effectene. After 18 h, medium containing 200 μg/ml G418 was added to select for cells that had stably incorporated the plasmid into their genomic DNA. Colonies were picked and subcloned and then expanded for biochemical and microscopic analyses. Characterization of expressed FP-PP1 by Western blotting, immunoprecipitation, and phosphatase assays was performed as previously described (Trinkle-Mulcahy et al., 2001). Cellular fractionation and large-scale purification of nucleoli was performed as described previously (Andersen et al., 2002), using 20 14-cm dishes of cells for each experiment. FACS analyses were performed using a FACScan (Becton Dickinson, Mountain View, CA).
Cell Fixation and Immunostaining
Cells were grown on glass coverslips and fixed for 10 min in 3.7% paraformaldehyde in 37°C PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9). After a 10-min permeabilization with 1% Triton X-100 in phosphate-buffered saline (PBS), cells were blocked with 1% donkey serum for 10 min and then incubated with primary antibodies for 1 h, washed, and incubated with secondary antibodies for 45 min. If required, cells were stained with DAPI (0.3 μg/ml; Sigma). After a final set of washes, cells were mounted in Vectashield media (Vector Laboratories, Burlingame, CA).
Antibodies were obtained from a variety of sources. Isoform-specific PP1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), Calbiochem (La Jolla, CA), and Oxford Biomedical Research and tested over a range of dilutions and fixation conditions for immunofluorescence. For Western blotting, all were used according to the manufacturer's recommendation. CREST (1:5000 for immunofluorescence) was a generous gift from Professor W. Earnshaw (Wellcome Trust Centre for Cell Biology, Edinburgh, Scotland). Anti-CENP F (mitosin; 1:100 for immufluorescence) and anti–aurora B kinase (AIM 1; 1:100 for immunofluorescence) were from BD Transduction Laboratories (Lexington, KY). Rhodamine-phalloidin was from Molecular Probes (Eugene, OR). Anti-PCNA was used at 1:500 for immunofluorescence. Anti–α-tubulin (DM1A; 1:10,000 for immunofluorescence) was from Sigma. Antifibrillarin (72B9; 1:10 for immunofluorescence) was a generous gift from Professor E. M. Tan. Anticoilin (204/10; Bohmann et al., 1995) was used at 1:350 for immunofluorescence. Actinomycin D (Sigma), okadaic acid (Calbiochem), Inhibitor 2 (New England Biolabs, Beverly, MA), monastrol (Tocris, Ballwin, MO), nocodazole (Sigma) and taxol (Paclitaxel, Sigma) were used as described in the text.
Isolated nucleoli were processed for transmission electron microscopy as described previously (Andersen et al., 2002). Sections were cut using a Reichart ultracut ultramicrotome and visualized in a JEOL 1200 EX TEM (Peabody, MA).
Fluorescence Microscopy and Photobleaching Experiments
For live cell microscopy, cells were grown on glass coverslips and mounted in phenol Red–free media in a closed, heated chamber (Bioptechs FCS2, Butler, PA; or Bachofer POC, Reutlingen, Germany). Live and fixed cell images were obtained on a Deltavision Restoration microscope (Applied Precision Instruments, Issaquah, WA) using a MicroMax 5 MHz cooled CCD camera (Roper Scientific, Tucson, AZ) and running SoftWoRx (Applied Precision) deconvolution and data analysis software.
Quantitation of relative fluorescence intensity in various subcellular compartments was performed on high-resolution three-dimensional (3D) data set images collected using a Zeiss (Thornwood, NY) 63× objective with a numerical aperture of 1.4. A series of 0.5-μm Z-sections was taken through each cell (40 sections for interphase cells and 80 sections for mitotic cells), ensuring that the entire cell volume was included. For each optical section, the data were collected using a binning of 2 × 2, resulting in an image pixel size of 0.212 × 0.212 μm. Utilizing the image analysis programs included in the SoftWoRx data analysis software, 2D polygons were drawn around each subcellular compartment/object of interest, in each Z-section in which they appeared. The integrated intensities of these polygons were summed to give an integrated intensity for the 3D volume corresponding to the intracellular compartment/object, after which an average background fluorescence per pixel (calculated from a region of similar size outside the cell) was subtracted. The total cellular fluorescence was also calculated in this manner, by selecting the entire cell as the region of interest over the full range of Z-sections. Dividing the total intensity value for a particular region of interest by the total intensity value for the whole cell reveals the fraction of the total fluorescence intensity in that particular subcellular compartment/object.
FRAP (fluorescence recovery after photobleaching) experiments were performed using a Zeiss LSM510 confocal microscope. After an initial image collected with the laser attenuated to 10% full power, a defined region of the cell was photobleached with the laser at full power. Subsequent images, taken at 3-s intervals with the laser attenuated to 10% full power, were obtained in order to follow the recovery of the fluorescent signal within the bleached region. Metaphase cells were fixed at the end of the experiment to show that the spindles remained intact and FP-PP1γ localization had not altered. Fluorescence quantitation was performed using the LSM510 software. A region of background fluorescence was defined outside the cell, and subtracted from both the experimental and control regions before further analysis. The relative intensity of the bleached region over time was then normalized to that of a similar unbleached region in the same cell, to account for normal photobleaching that occurs during image acquisition throughout the course of the experiment. These normalized values were presented as fractions of the starting intensity, to demonstrate both the photobleaching and the recovery from photobleaching over time. It should be noted that the kinetics of recovery of photobleached FP-PP1γ, particularly at the kinetochore, are so rapid that the fluorescence intensity shows a significant recovery before the first postbleaching image can be taken. To ensure that the photobleaching conditions used here were sufficient to bleach the pools of FP-PP1γ found at the kinetochore and nucleolus, the experiments were performed under similar conditions with fixed cells. Under these conditions, FP-PP1γ can be efficiently bleached but, as expected, cannot recover (unpublished data). The initial recovery that occurs before taking the first postbleaching image is therefore a limitation of the imaging system, based on the lag time it takes for the system to switch from the full laser power required for bleaching to the laser power (10%) at which the first postbleaching image is taken. Although this limitation precludes the accurate measurement of absolute half-times for recovery, it does allow us to compare the recovery profiles of the photobleached nucleolar and kinetochore pools of FP-PP1γ, and the kinetochore pools of FP-PP1γ after various drug treatments.
RESULTS
Characterization of Stable Cell Lines
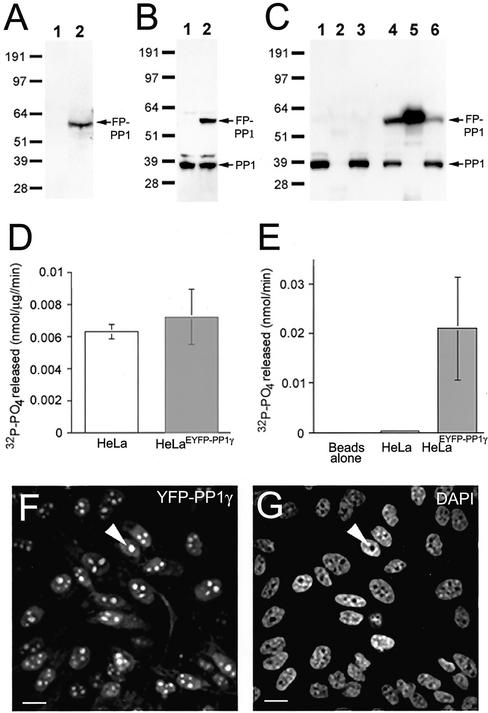
HeLaEYFP-PP1γ cells express full-length FP-PP1γ at a similar level to endogenous PP1γ (Figure 1, A and B). Importantly, total phosphatase activity in HeLaEYFP-PP1γ cells is equivalent to parental cells (Figure 1D), indicating regulation of any excess phosphatase activity. The FP tag allows recovery of >50% of the fusion protein from cell lysates by immunoprecipitation (Figure 1C, lanes 4–6). Endogenous PP1 does not coprecipitate with the fusion protein (Figure 1C, lane 5), confirming the phosphatase activity associated with the beads results from FP-PP1γ (Figure 1E). No activity is recovered using a catalytically inactive fusion protein (Trinkle-Mulcahy et al., 2001).
Figure 1.
HeLaEYFP-PP1γ cells express full-length, active fusion protein. (A) Lysates (30 μg total protein) from HeLa (1) and HeLaEYFP-PP1γ (2) cells probed on a Western blot with anti-GFP antibodies. (B) Similar lysates probed with anti-PP1 antibodies. (C) HeLa and HeLaEYFP-PP1γ cell lysates before (1 and 4) and after (3 and 6) incubation with anti-GFP antibodies coupled to protein G sepharose. Anti-PP1 antibodies detect endogenous PP1 in both lysates and the fusion protein in the HeLaEYFP-PP1γ lysate. The fusion protein was specifically immunodepleted by >50% from HeLaEYFP-PP1γ lysates (5), whereas no PP1-positive bands were pulled down from HeLa lysates (2). (D) Total in vitro phosphorylase, a phosphatase activity associated with HeLa and HeLaEYFP-PP1γ cell lysates. (E) Phosphatase activity associated with anti-GFP beads alone and with anti-GFP beads incubated with 50 μg total protein from HeLa or HeLaEYFP-PP1γ cell lysates. Data are means ± SD for n = 4. The EYFP signal in a field of HeLaEYFP-PP1γ cells is shown in F, with the DAPI-stained DNA pattern in G. A nucleolus is marked by the arrowhead. Scale bar, 10 μM.
Fluorescence imaging of HeLaEYFP-PP1γ cells reveals a diffuse cytoplasmic pool of FP-PP1γ, a more intense nucleoplasmic pool and prominent accumulation within nucleoli (Figure 1, F and G). These data are consistent with transient expression assays (Trinkle-Mulcahy et al., 2001) and antibody staining of untransfected cells (Andreassen et al., 1998). The population is homogeneous, with >95% of the cells expressing FP-PP1γ at similar levels. Quantitation of the fluorescent signal reveals that the nuclear pool (nucleoplasmic and nucleolar combined) of FP-PP1γ accounts for 31.8 ± 5.5% of the total fluorescent protein in the cell, whereas the nucleolar pool represents 5.4 ± 1.7% of the total fluorescent protein in the cell (n = 7). If these measurements are limited to the nuclear pool, the nucleolar FP-PP1γ signal is found to represent 17.1 ± 5.1% of the nuclear FP-PP1γ signal (n = 7).
Properties of Nucleolar FP-PP1γ
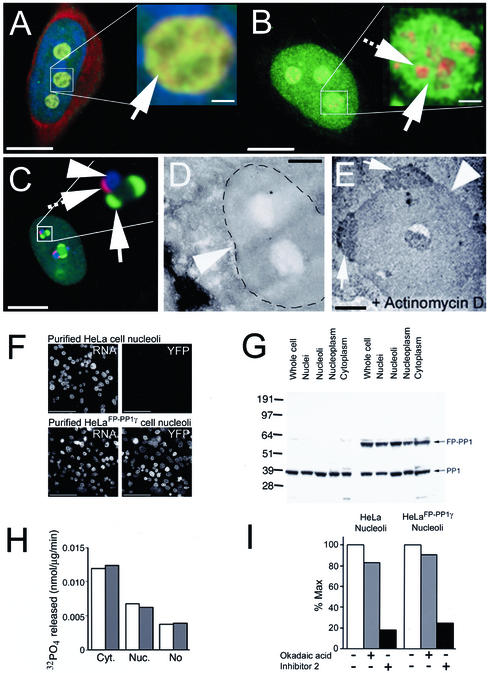
The nucleolar pool of FP-PP1γ specifically localizes to the granular compartment that contains the bulk of the rRNA (Figure 2A), remaining spatially distinct from the dense fibrillar component marked by fibrillarin (Figure 2B). Inhibition of transcriptional activity using actinomycin D causes a relocalization of FP-PP1γ to perinucleolar caps Figure 2C). These caps are distinct from those formed by fibrillarin and by p80 coilin (see Figure 2C inset; Raska et al., 1990; Carmo-Fonseca et al., 1992). Using transmission electron microscopy, with silver-conjugated antibodies to the FP domain detecting the fusion protein, perinucleolar caps containing FP-PP1γ can be shown on actinomycin d–treated nucleoli (Figure 2E). These caps do not appear on control nucleoli (Figure 2D).
Figure 2.
Purification and characterization of FP-PP1γ- enriched nucleoli from HeLaEYFP-PP1γ cells. (A) HeLaEYFP-PP1γ cell fixed and stained with DAPI for DNA (blue) and Pyronin Y for RNA (red). FP-PP1γ (green) colocalizes with the Pyronin Y–stained RNA in the granular compartment. Inset: an enlarged nucleolus, with the granular compartment indicated by an arrow. (B) HeLaEYFP-PP1γ cell fixed and stained with antifibrillarin (red) to mark the dense fibrillar component of the nucleolus. Nucleolar FP-PP1γ (green) is spatially distinct from this compartment, as shown clearly in the enlarged inset. The arrow indicates FP-PP1γ in the granular component, whereas the hashed arrow points to the dense fibrillar component labeled by antifibrillarin. (C) HeLaEYFP-PP1γ cell treated with actinomycin D. The nucleolus in the enlarged inset shows the spatial distinction between the perinucleolar caps caused by FP-PP1γ (green, arrow), fibrillarin (blue, arrowhead) and p80 coilin (red, hashed arrow). (D) Transmission electron micrograph of a nucleolus (arrowhead) from a HeLaEYFP-PP1γ cell. (E) Similar micrograph of a nucleolus (arrowhead) from an actinomycin d--treated HeLaEYFP-PP1γ cell. Perinucleolar caps containing FP-PP1γ were detected using anti-GFP antibodies and are marked by arrows. (F) Nucleoli were purified from HeLa and HeLaEYFP-PP1γ cells. rRNA is stained with Pyronin Y, and a YFP signal is only detected in the HeLaEYFP-PP1γ nucleoli. (G) Cellular fractions (30 μg total protein per lane) from HeLa (lanes 1–5) and HeLaEYFP-PP1γ (lanes 6–10) cells probed on a Western blot with anti-PP1γ antibodies. The fractions probed include whole cell lysates (1, 6), nuclei (2, 7), nucleoli (3, 8), nucleoplasm (4, 9), and cytoplasm (5, 10). (H) Total in vitro phosphorylase a phosphatase activity associated with HeLa (white bars) and HeLaEYFP-PP1γ (gray bars) cytoplasmic, nucleoplasmic, and nucleolar fractions. (I) Sensitivity of nucleolar phosphatase activity to inhibitors that distinguish between PP1 (Inhibitor 2-sensitive) and PP2A (low level okadaic acid-sensitive). Activity is expressed relative to maximal activity in the absence of inhibitors. All phosphatase assays were repeated twice (each time in duplicate) with similar results.
Nucleolar FP-PP1γ remains associated with this subnuclear body when cells are fractionated using a purification method used in a recent nucleolar proteomics project (Figure 2F; Andersen et al., 2002). Western blotting reveals that HeLaEYFP-PP1γ cells express full-length FP-PP1γ at a similar level to endogenous PP1γ throughout all the cellular fractions in which this isoform is found (Figure 2G), and that regulation of any excess phosphatase activity is maintained throughout these fractions (Figure 2H). The bulk of the total phosphatase activity in purified nucleoli from both HeLa and HeLaEYFP-PP1γ cells is sensitive to Inhibitor 2 and thus can be attributed to PP1 (Figure 2I).
Cell Cycle Progression
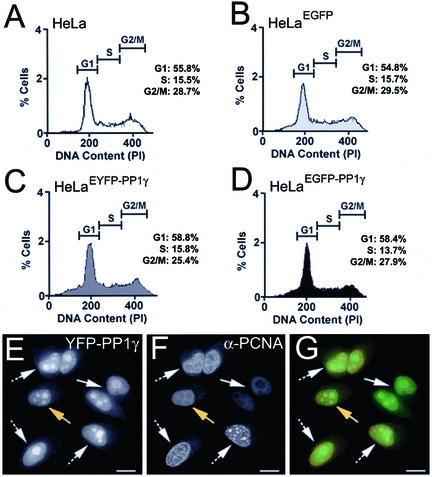
Addition of exogenous PP1 affects mammalian cell cycle progression (Fernandez et al., 1992; Berndt et al., 1997). Cell lines constitutively expressing exogenous PP1 must therefore regulate excess activity to prevent cell cycle defects. We compared cell cycle progression of HeLaEYFP-PP1γ and HeLaEGFP-PP1γ cells with that of both parental HeLa cells and HeLaEGFP cells. The growth rates of the respective cell lines are equivalent, and FACS analysis confirmed that the relative populations in G1, S, and G2/M are similar (Figure 3, A–D). The broadened shoulder observed on the G1 peak for all three stable cell lines may indicates a small increase in the number of apoptotic cells, representing ∼2–5% of the total population. When HeLaEYFP-PP1γ cells were fixed and stained with an antibody to proliferating cell nuclear antigen (PCNA; Figure 3F), a protein whose localization pattern can be used as a marker for cell cycle stages, no differences in FP-PP1γ localization and/or levels were observed between cells in G1, S, or G2 (Figure 3, E–G).
Figure 3.
Cell cycle distribution of HeLaFP-PP1γ cells. A-D: Distribution of cells in G1, S and G2/M for the parental HeLa (A), HeLaEGFP (B), HeLaEYFP-PP1γ (C), and HeLaEGFP-PP1γ (D) cell lines, as determined by FACS analysis. HeLaEYFP-PP1γ cells (E) were stained with anti-PCNA (F) to mark cell cycle stages, and the two signals are shown merged in G. Cells in G1 are indicated by an arrow, those in S-phase by a hashed arrow, and those in G2 by a yellow arrow. Scale bar, 10 μM.
Dynamic Cell Cycle Localization of FP-PP1γ
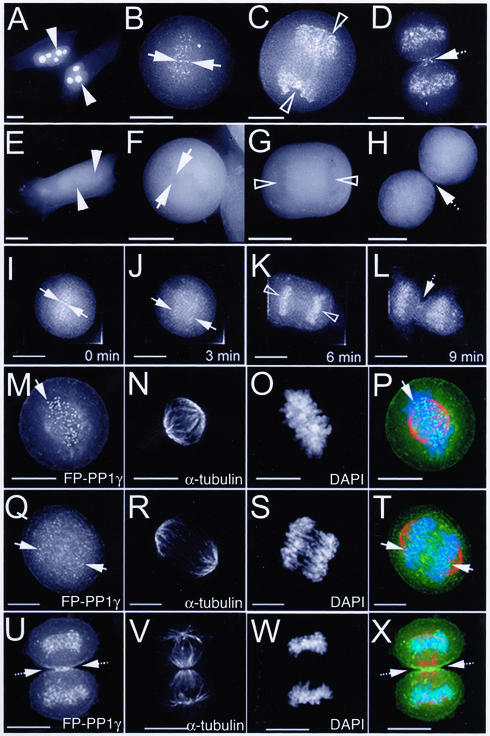
Although the typical interphase localization pattern for FP-PP1γ is maintained throughout the G1, S, and G2 stages of the cell cycle (Figure 4A), at M phase, FP-PP1γ localization changes dramatically. Bright foci appear in the region of condensed chromosomes as the nuclear envelope breaks down (Figure 4B). These foci are detectable until late anaphase, at which point a significant fraction (19.9 ± 2.2%, n = 5) of the total FP-PP1γ fluorescent signal relocates to the chromosomes (Figure 4C and J–K). At telophase, FP-PP1γ is observed both in the reforming nuclei and at the cleavage furrow and spindle midzone (Figure 4D). These patterns differ markedly from the diffuse distribution observed for GFP alone in HeLaEGFP cells (Figure 4, E–H). Fluorescence time-lapse imaging of a single HeLaEYFP-PP1γ cell reveals the short time span over which this relocalization occurs (Figure 4, I–L).
Figure 4.
Dynamic distribution of FP-PP1γ throughout cell division. Live HeLaEYFP-PP1γ (A–D) and HeLaEGFP (E–H) cells were imaged using a Deltavision restoration microscope. All images shown here are 2D maximum projections of Z-series data. In interphase cells (A and E), nucleoli are marked by arrowheads. Regions of condensed chromatin are marked by arrows in metaphase cells (B, F) and by open arrowheads in anaphase cells (C and G). The spindle midzone region is marked by a hashed arrow in telophase cells (D and H). Time-lapse imaging of a single mitotic HeLaEYFP-PP1γ cell confirms these patterns and reveals the short time period over which they occur (I–L). Mitotic HeLaEYFP-PP1γ cells were also fixed and stained with anti–α-tubulin to mark spindles (N, R, and V) and DAPI to mark chromosomes (O, S, and W). The FP-PP1γ signal (M, Q, and U) is shown for cells in metaphase (M–P), early anaphase (Q–T), and telophase (U–X). All three patterns are shown merged in P, T and X. Scale bar, 10 μM.
HeLaEYFP-PP1γ cells were fixed and stained with both DAPI and anti–α-tubulin to map the relationship between FP-PP1γ, chromatin, and spindles throughout mitosis. The foci observed in metaphase cells align on the metaphase plate at the ends of spindle fibers (Figure 4, M–P). They are detectable until early anaphase (Figure 4, Q–T), suggesting a centromere localization pattern. By telophase, FP-PP1γ is retargeted to the chromosomes, cleavage furrow and midbody (Figure 4, U–X).
Mapping Mitotic Accumulations of FP-PP1γ to Known Structures
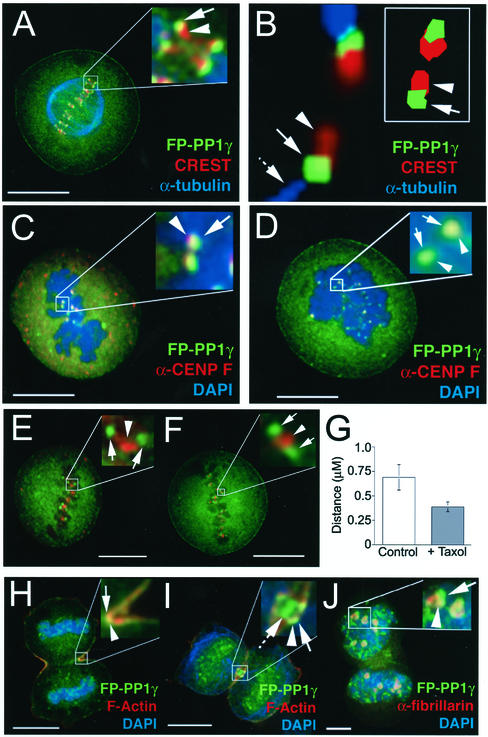
CREST, a human autoimmune sera that recognizes several centromere proteins (Earnshaw and Rothfield, 1985), labels paired foci that are spatially distinct from FP-PP1γ foci (Figure 5A). A high-resolution image of a single pair of centromeres clearly maps the FP-PP1γ foci to the space between the end of the spindle fiber and the CREST-labeled outer centromere region (Figure 5B). The 2D model (Figure 5B, inset) is based on a single section from the same region and demonstrates that there is some overlap between the FP-PP1γ signal and the CREST signal. This spatial mapping implies a kinetochore localization, which was confirmed by staining with an antibody to the kinetochore marker CENP-F (Figure 5C; Zhu et al., 1995). Disruption of spindle fibers by treatment with nocodazole does not change the kinetochore localization of FP-PP1γ (Figure 5D). Similar results were obtained using taxol, which releases tension on the kinetochore by stabilizing microtubules (Waters et al., 1998; Figure 5, E–G) and monastrol, an inhibitor of the mitotic kinesin Eg5 (unpublished data; Kapoor et al., 2000). This indicates that PP1 is a stable component of the mammalian kinetochore. Quantitation of fluorescent signal in metaphase HeLaEYFP-PP1γ cells revealed that 0.008 ± 0.003% of the total cellular FP-PP1γ signal is found at each kinetochore (n = 5), a value that does not change significantly upon treatment with either taxol or nocodazole (unpublished data). The total number of FP-PP1γ–labeled kinetochores in each cell was found to be 92.8 ± 8.0 (n = 5), which is in agreement with the number of CREST-stained centromeres observed in these cells (96.0 ± 5.6). This slight deviation from the expected number of centromeres/kinetochores, which would be 92 for a standard diploid cell, is not surprising, because HeLa cells are often aneuploid. In the case of the parental HeLa cells used to establish all of the stable cell lines; however, chromosome spreads have shown that any deviation from the diploid number of chromosomes is minimal (unpublished results).
Figure 5.
FP-PP1γ accumulates at kinetochores, cleavage furrow, and midbody. (A) HeLaEYFP-PP1γ metaphase cell stained with anti–α-tubulin to marker mitotic spindles, and with CREST, a centromere marker. FP-PP1γ foci are marked by arrows and centromeres by arrowheads. (B) High-resolution image of a pair of centromeres from a HeLaEYFP-PP1γ metaphase cell stained with anti–α-tubulin and CREST. The spatial orientation of the FP-PP1γ signal (arrowhead) with respect to the end of the spindle fiber (hashed arrow) and the centromere (arrowhead) can be clearly seen. The inset shows a 2D model of a single section through a pair of centromeres, which reveals a slight overlap between the FP-PP1γ and CREST signals. (C and D) HeLaEYFP-PP1γ metaphase cells stained with DAPI and anti-CENP F, a kinetochore marker. FP-PP1γ foci (arrow) colocalize with kinetochores (arrowhead) both in control cells (C) and in cells treated with nocodazole to disrupt microtubules (D). (E and F) HeLaEYFP-PP1γ metaphase cells stained with anti–aurora B, a markerfor the inner centromere region. FP-PP1γ foci are marked by arrows and the inner centromere region by an arrowhead. The spatial distinction between the two signals is maintained both in control cells (E) and when cells are treated with taxol to release tension on kinetochores (F). The release of tension can be quantitated as a reduction in the mean distance between paired kinetochores marked by FP-PP1γ (G; n = 57). (H) HeLaEYFP-PP1γ telophase cell stained with DAPI and rhodamine-phalloidin, to visualize F-actin. FP-PP1γ accumulates at the cleavage furrow (arrow), colocalized with F-actin (arrowhead). (I) A cell at a later stage of telophase, where the cortical accumulation of FP-PP1γ (arrow) remains colocalized with F-actin (arrowhead) in a ring-like structure at the junction between the two daughter cells, with an additional accumulation of FP-PP1γ at the midbody (hashed arrow). (J) A cell in which nucleoli are starting to reform, as indicated by an antibody to the nucleolar protein fibrillarin (arrowhead). FP-PP1γ is starting to reaccumulate in nucleoli (arrow), which is its predominant interphase localization pattern. Scale bars, 10 μM.
Antibodies to aurora B, a marker for the inner centromere region, label an area directly between paired FP-PP1γ foci in metaphase cells (Figure 5E), and the FP-PP1γ/aurora B signals remain spatially distinct throughout mitosis (unpublished data). When tension on metaphase kinetochores is released by taxol treatment (Figure 5F), FP-PP1γ and aurora B still remain spatially distinct, at least at the level of resolution available by light microscopy.
As mitosis progresses, accumulations of FP-PP1γ appear on the chromosomes as already shown and at the cleavage furrow and midbody (Figure 5, H and I). Figure 5H shows the overlap between the pool of FP-PP1γ and F-actin at the central section of the cell where the cleavage furrow forms. Cortical FP-PP1γ accumulation coincides with appearance of the cleavage furrow in anaphase. In late telophase (Figure 5I), FP-PP1γ (arrow) still overlaps cortical F-actin (arrowhead) but is also observed at the spindle midbody (hashed arrow). FP-PP1γ eventually reaccumulates in newly-formed nucleoli marked by an antibody to the nucleolar protein fibrillarin (Figure 5J).
Rapid Kinetics of FP-PP1γ Turnover
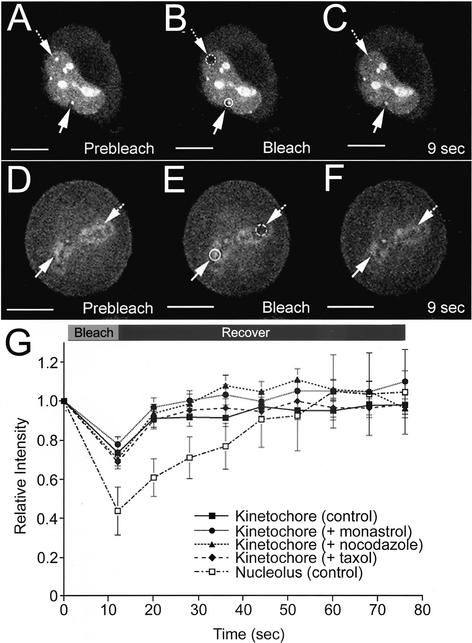
FRAP experiments were performed on live metaphase HeLaEYFP-PP1γ cells to measure exchange of FP-PP1γ at two distinct intracellular pools. Nucleolar FP-PP1γ recovers rapidly from photobleaching (Figure 6, A–C and G), as does the kinetochore pool of FP-PP1γ in metaphase cells (Figure 6, D–F and G). No significant difference in recovery kinetics was observed for FP-PP1γ at kinetochores in cells treated with nocodazole, monastrol, or taxol (Figure 6G), indicating that the rapid exchange of PP1γ at the kinetochore is independent of an intact, functional spindle mechanism.
Figure 6.
Nucleolar and kinetochore pools of FP-PP1γ show rapid turnover rates. Live HeLaEYFP-PP1γ cells were subjected to FRAP experiments in which a specific pool of EYFP-PP1γ was photobleached and recovery of fluorescent signal monitored over time. For the interphase HeLaEYFP-PP1γ cell shown in A–C, a nucleolus (hashed circle/arrow) was photobleached, whereas a nonbleached nucleolus (circle/arrow) was monitored for comparison. The signal recovered by ∼50% in 9 s (C). The experiment was repeated three times, and a plot of signal recovery over time for a bleached nucleolus normalized to a nonbleached nucleolus in the same cell is shown (G; □), with each time point presented as mean relative intensity ± SD. For the metaphase HeLaEYFP-PP1γ cell shown in D–F, a kinetochore region (hashed circle/arrow) was photobleached, whereas a nonbleached kinetochore region (circle/arrow) was monitored for comparison. The signal showed an apparent full recovery by 9 s (F). The experiment was repeated five times, and a plot of signal recovery over time for a bleached kinetochore normalized to a nonbleached kinetochore in the same cell is shown (G; ▪), with each time point presented as mean relative intensity ± SD. Similar recovery kinetics were observed for this intracellular pool when HeLaEYFP-PP1γ cells were treated with monastrol to inhibit the Eg5 mitotic kinesin (G; ●, n = 3), with nocodazole to disrupt microtubules (G; ▴, n = 4) or with taxol to release tension on kinetochores (G; ♦, n = 4). Scale bars, 10 μM.
DISCUSSION
We have established stable cell lines expressing FP-tagged PP1γ and used these cells to analyze the in vivo organization of pools of PP1γ at different stages of the cell cycle. Live cell imaging analyses have shown that the localization of PP1γ is highly dynamic and indicates specific subcellular sites and times throughout interphase and mitosis where PP1γ activity is likely to be involved.
It has proven difficult to derive the stable cell lines expressing functional FP-PP1. For example, transient transfection studies revealed that cells do not tolerate long-term expression of FP-tagged PP1. Although the control cell line, HeLaEGFP, was readily established, a HeLaEGFP-PP1γ cell line was difficult to establish and resulted in a single stable clone. Interestingly, we observed that the number of stable clones incorporating FP-PP1γ could be increased by cotransfection of low levels of an expression vector encoding the PP1 regulator NIPP1 (Nuclear Inhibitor of PP1; Van Eynde et al., 1995). No NIPP1 fusion protein was detected in any of the resulting cell clones, confirming that it had not been stably incorporated. We infer that low levels of NIPP1 were transiently expressed during the early stages of selection, thereby regulating excess PP1 activity and allowing cells to survive long enough to stably incorporate the FP-PP1γ plasmid. For comparative purposes, all experiments presented here were performed on both the FP-PP1γ cell line established in this manner (HeLaEYFP-PP1γ) and on the FP-PP1γ cell line established by transfection of the PP1 plasmid alone (HeLaEGFP-PP1γ), with equivalent results. This cotransfection approach may therefore aid the establishment of cell lines expressing other protein phosphatases or other enzymes that can be toxic if their activity is not regulated.
Expressing phosphatases as fluorescent fusion proteins provides the opportunity to study their regulation using a combined microscopy/biochemistry approach. The pattern observed for FP-PP1γ, for example, represents the cumulative pattern for a variety of PP1γ complexes present in the cell, and the fluorescent tag permits quantitation of signal levels at various intracellular sites. Live cell imaging shows the nucleolar accumulation of FP-PP1γ in interphase cells, which represents ∼5% of the total cellular FP-PP1γ signal. The significance of this accumulation is highlighted by the fact that >80% of the total phosphatase activity in purified nucleoli was found to be sensitive to Inhibitor 2 and thus attributable to PP1. This suggests a possible role for PP1 in the regulation of one or more nucleolar processes, such as rRNA transcription, rRNA maturation or ribosome subunit assembly.
The dynamic localization of PP1 throughout mitosis suggests targeting of phosphatase activity to specific sites and at specific times during cell division. Movement of FP-PP1γ to the decondensing chromatin in telophase, for example, suggests recruitment of PP1 activity for a function connected with this stage of the cell cycle. Two known regulatory events that occur at this time and location are dephosphorylation of histone H3 and nuclear lamin B. Reversible phosphorylation of histone H3 may be involved in the regulation of chromatin condensation (Cheung et al., 2000), and the chromosome targeting of FP-PP1γ in late anaphase correlates with the rapid disappearance of a Ser10 phosphoepitope on histone H3 (unpublished data). Chromatin-associated PP1 has also been shown to be recruited for nuclear lamina assembly (Steen et al., 2000). Although we observed no significant overlap between FP-PP1γ and either immunostained histones or lamin B (unpublished data), the initial retargeting occurs within a very short time span (<3 min) and these processes may only involve a small percentage of the total PP1 at this site. Therefore, the involvement of PP1γ in one or both of these regulatory pathways is a possibility.
The spatial organization of FP-PP1γ foci with respect to the aurora B kinase signal in mitotic cells is interesting in light of the fact that PP1 can either oppose or directly inhibit aurora B activity on one of its substrates, histone H3 (Francisco et al., 1994; Hsu et al., 2000; Murnion et al., 2001) and has also been suggested to oppose its activity on kinetochore proteins (Tanaka et al., 2002). Although aurora B localizes to centromeres in G2 (Zeitlin et al., 2001), PP1γ accumulation is not evident at kinetochores until prophase (unpublished data). Changes in the physical organization of the kinetochore may therefore modulate the competition between aurora B and PP1 for their substrates.
Kinetochore and spindle midzone localization has also been observed for a GFP construct of Glc7, the yeast orthologue of PP1 (Bloecher and Tatchell, 2000), placing PP1 at precise sites where and when phosphatase activity is known to be required (Sassoon et al., 1999; see Nigg, 2001 for review). Although a phosphatase has been reported not to be the likely tension-sensitive component of kinetochore phosphorylation (Nicklas et al., 1998), there is evidence for PP1 playing a role in the microtubule binding of the kinetochore complex (Sassoon et al., 1999; Bloecher and Tatchell, 2000) and in spindle stability (Tournebize et al., 1997). A role for PP1 in the regulation of cytokinesis has also been suggested (Fernandez et al., 1992; Cheng et al., 2000), which is consistent with the colocalization of FP-PP1γ and F-actin at the cleavage furrow and spindle midzone.
Significantly, PP1γ is found in the center of the midbody, where CENP-F also localizes, rather than the outer regions to which aurora B/INCENP/surviving localize. This central region is the site of membrane fusion events in cytokinesis, and PP1 has been shown to regulate the terminal step of membrane fusion in yeast (Peters et al., 1999). The midzone accumulation of PP1γ may also be related to the regulation of CENP-A by aurora B kinase, because it is disrupted by expression of dominant-negative phosphorylation site mutants of CENP-A that cause delays in the terminal stage of cytokinesis (Zeitlin et al., 2001).
The rapid exchange FP-PP1γ at both the nucleolus and the kinetochore revealed by FRAP analyses is of particular interest because it argues against a static model of PP1 targeting. A slower turnover rate, on the order of minutes or tens of minutes, would have indicated a more stable association of the phosphatase with its relevant targeting subunit and/or substrate (i.e., some sort of docking mechanism). In fact, it is clear that there is a rapid flux of PP1 through both the nucleolar pool in interphase cells and the kinetochore pool in metaphase cells, which provides the opportunity for rapid retargeting to another site if required. It remains to be determined whether PP1 is exchanging on its own or as a complex with a targeting subunit(s). The fact that the rapid rate of exchange of FP-PP1γ at the kinetochore persists after treatment with various inhibitors that destabilize the mitotic spindle complex suggests that this pool exchanges directly with the diffuse cytoplasmic pool. The reason for the rapid turnover of kinetochore-localized PP1 may be to provide a sensitive regulatory mechanism for its activity at this critical step in cell division.
In summary, the spatio-temporal mapping of stably expressed FP-PP1γ in live cells has revealed dynamic accumulations within the nucleolus in interphase cells, and at the kinetochore, chromosomes, cleavage furrow and midbody throughout mitosis. This suggests roles for PP1 in the regulation of multiple processes, including nucleolar function and the regulation of chromosome segregation and cytokinesis. Future work will focus on identification of both the relevant substrates and the targeting subunits that direct PP1 to these sites and regulate its activity.
ACKNOWLEDGMENTS
We thank Professor William Earnshaw for his generous gift of CREST autoimmune sera, Dr. Bruno Frenguelli for the use of the Zeiss LSM510, Dr. Tomoyuki Tanaka for his critical reading of the manuscript, and Anthony Leung for technical advice. L.T.-M. was supported by a Biotechnology and Biological Sciences Research Council fellowship. J.R.S. is a Wellcome Trust Career Development Fellow and is supported by Cancer Research UK. A.I.L. is a Wellcome Trust Principal Research Fellow.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0376. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0376.
REFERENCES
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Lacroix FB, Villa-Moruzzi E, Margolis RL. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J Cell Biol. 1998;141:1207–1215. doi: 10.1083/jcb.141.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker HM, Brewis ND, Street AJ, Spurr NK, Cohen PT. Three genes for protein phosphatase 1 map to different human chromosomes: sequence, expression and gene localization of protein serine/threonine phosphatase 1 beta (PPP1CB) Biochim Biophys Acta. 1994;1220:212–218. doi: 10.1016/0167-4889(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barker HM, Craig SP, Spurr NK, Cohen PT. Sequence of human protein serine/threonine phosphatase 1 gamma and localization of the gene (PPP1CC) encoding it to chromosome bands 12q24.1-q24.2. Biochim Biophys Acta. 1993;1178:228–233. doi: 10.1016/0167-4889(93)90014-g. [DOI] [PubMed] [Google Scholar]
- Berndt N, Dohadwala M, Liu CW. Constitutively active protein phosphatase 1alpha causes Rb-dependent G1 arrest in human cancer cells. Curr Biol. 1997;7:375–386. doi: 10.1016/s0960-9822(06)00185-0. [DOI] [PubMed] [Google Scholar]
- Bloecher A, Tatchell K. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Biol. 2000;149:125–140. doi: 10.1083/jcb.149.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutation analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond A I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Dean NM, Honkanen RE. Serine/threonine protein phosphatase type 1gamma1 is required for the completion of cytokinesis in human A549 lung carcinoma cells. J Biol Chem. 2000;275:1846–1854. doi: 10.1074/jbc.275.3.1846. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1–targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Brautigan DL, Lamb NJ. Protein phosphatase type 1 in mammalian cell mitosis: chromosomal localization and involvement in mitotic exit. J Cell Biol. 1992;116:1421–1430. doi: 10.1083/jcb.116.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L, Wang W, Chan CS. Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem. 2001;276:26656–26665. doi: 10.1074/jbc.M102288200. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Campbell MS, Ward SC, Gorbsky GJ. Tension-sensitive kinetochore phosphorylation in vitro. J Cell Sci. 1998;111:3189–3196. doi: 10.1242/jcs.111.21.3189. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Peters C, Andrews PD, Stark MJ, Cesaro-Tadic S, Glatz A, Podtelejnikov A, Mann M, Mayer A. Control of the terminal step of intracellular membrane fusion by protein phosphatase 1. Science. 1999;285:1084–1087. doi: 10.1126/science.285.5430.1084. [DOI] [PubMed] [Google Scholar]
- Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Shima H, Kitagawa Y, Irino S, Sugimura T, Nagao M. Identification of members of the protein phosphatase 1 gene family in the rat and enhanced expression of protein phosphatase 1 alpha gene in rat hepatocellular carcinomas. Jpn J Cancer Res. 1990;81:1272–1280. doi: 10.1111/j.1349-7006.1990.tb02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RL, Martins SB, Tasken K, Collas P. Recruitment of protein phosphatase 1 to the nuclear envelope by A-kinase anchoring protein AKAP149 is a prerequisite for nuclear lamina assembly. J Cell Biol. 2000;150:1251–1262. doi: 10.1083/jcb.150.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tournebize R, Andersen SS, Verde F, Doree M, Karsenti E, Hyman AA. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 1997;16:5537–5549. doi: 10.1093/emboj/16.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Sleeman JE, Lamond AI. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- Van Eynde A, Wera S, Beullens M, Torrekens S, Van Leuven F, Stalmans W, Bollen M. Molecular cloning of NIPP-1, a nuclear inhibitor of protein phosphatase-1, reveals homology with polypeptides involved in RNA processing. J Biol Chem. 1995;270:28068–28074. doi: 10.1074/jbc.270.47.28068. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Mancini MA, Chang KH, Liu CY, Chen CF, Shan B, Jones D, Yang-Feng TL, Lee WH. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]