Abstract
Although conserved counterparts for most proteins involved in the G2/M transition of the cell cycle have been found in all eukaryotes, a notable exception is the essential but functionally enigmatic fungal kinase NIMA. While a number of vertebrate kinases have been identified with catalytic domain homology to NIMA, none of these resemble NIMA within its extensive noncatalytic region, a region critical for NIMA function in Aspergillus nidulans. We used a bioinformatics approach to search for proteins with homology to the noncatalytic region of NIMA and identified mixed lineage kinase 3 (MLK3). MLK3 has been proposed to serve as a component in MAP kinase cascades, particularly those resulting in the activation of the c-Jun N-terminal kinase (JNK). Here we describe the first in-depth study of endogenous MLK3 and report that, like NIMA, MLK3 phosphorylation and activity are enhanced during G2/M, whereas JNK remains inactive. Coincident with the G2/M transition, a period marked by dramatic reorganization of the cytoplasmic microtubule network, endogenous MLK3 transiently disperses away from the centrosome and centrosomal-proximal sites where it is localized during interphase. Furthermore, when overexpressed, MLK3, like NIMA, localizes to the centrosomal region, induces profound disruption of cytoplasmic microtubules and a nuclear distortion phenotype that differs from mitotic chromosome condensation. Cellular depletion of MLK3 protein using siRNA technology results in an increased sensitivity to the microtubule-stabilizing agent taxol. Our studies suggest a new role for MLK3, separable from its function in the JNK pathway, that may contribute to promoting microtubule instability, a hallmark of M phase entry.
INTRODUCTION
Entry into mitosis in all eukaryotes requires the activity of Cdc2/cyclin B (Nurse, 1990). This central mitotic regulator has been characterized extensively and most of the proteins isolated genetically or biochemically as G2/M regulators impinge directly on control of Cdc2/cyclin B kinase activity. In the filamentous fungus Aspergillus nidulans, a second mitotic kinase, NIMA, is also essential for proper execution of the G2/M transition (Osmani et al., 1991). NIMA is a Ser/Thr protein kinase encoded by the nimA (never in mitosis A) gene of A. nidulans and was discovered as a temperature sensitive mutant in a screen to identify genes involved in cell cycle regulation (Osmani et al., 1988). Conditional loss of NIMA function causes arrest in G2 phase, despite the presence of fully active Cdc2/cyclin B (Osmani et al., 1991). When overexpressed in A. nidulans, NIMA induces chromatin condensation and disruption of cytoplasmic microtubules (Osmani et al., 1988). Furthermore, cells overexpressing the kinase enter a lethal pseudomitotic state from any phase of the cell cycle (Osmani et al., 1988; Krien et al., 1998; De Souza et al., 2000). This effect is dominant, occurring even without active Cdc2/cyclin B or when DNA synthesis has been prevented.
The majority of known mitotic regulators perform functions that are highly conserved through eukaryotic evolution. A number of studies suggest that there might be a NIMA-like component that plays a role in the vertebrate cell cycle. First, overexpression of wild-type NIMA in human or Xenopus laevis cells promotes apparent chromatin condensation, as is true in A. nidulans (Lu and Means, 1994; O'Connell et al., 1994; Lu and Hunter, 1995). Minimally, this indicates that NIMA targets might be conserved and suggests that NIMA-like activities operate during mitosis to modulate chromatin mechanics. Second, overexpression of a catalytically inactive variant of NIMA in both human cells and A. nidulans results in a G2 arrest (Lu and Means, 1994; Lu and Hunter, 1995), suggesting that similar NIMA-interacting elements are present during the G2/M transition in vertebrates. However, although these data argue that a NIMA-like activity is likely to be conserved across species, efforts to identify vertebrate NIMA homologues have been unsuccessful.
The only demonstrated NIMA homologue (that complements nimA mutations in A. nidulans) is the product of the nim-1 gene of Neurospora crassa (Pu et al., 1995). Not only does Nim-1 have 87% amino acid identity/similarity to NIMA in the catalytic domain, it shares 52% identity/similarity in its C-terminal extension. In other organisms, a number of NIMA-related kinases have been identified. Examples of these kinases include Fin1 in fission yeast (Krien et al., 1998), the Nrks in trypanosomes (Gale and Parsons, 1993), Kin3/NPK1 in budding yeast (Jones and Rosamond, 1990; Barton et al., 1992; Schweitzer and Philippsen, 1992), Fa2p in Chlamydomonas (Mahjoub et al., 2002), and the Neks and Stk2 in mammals (Letwin et al., 1992; Cance et al., 1993; Schultz and Nigg, 1993; Levedakou et al., 1994; Schultz et al., 1994; Rhee and Wolgemuth, 1997; Arama et al., 1998; Chen et al., 1999; Tanaka and Nigg, 1999; Kandli et al., 2000; Holland et al., 2002). Although all of these kinases have N-terminal catalytic domains with similarities in sequence to NIMA, their C-terminal extensions diverge considerably in both length and sequence.
Genetic complementation studies of nimA mutants in A. nidulans have revealed that the C-terminal extension of NIMA is essential for its in vivo function, although it is not required for its kinase activity (Pu and Osmani, 1995). In addition, the presence of NIMA's C-terminal extension is both necessary and sufficient to induce a G2 arrest after overexpression in either A. nidulans or human cells (Lu and Means, 1994; Lu and Hunter, 1995); this effect is dose dependent, suggesting that the key interaction sites are saturable. Given the biological importance of this domain to NIMA function, and the likelihood that some of NIMA's G2/M activities in A. nidulans are conserved in eukaryotic cell cycle control, it seemed likely that a mammalian NIMA-like molecule would have similarities in this region. Therefore, we conducted a database search designed to identify kinases having homology to NIMA in the C-terminal extension. This search, surprisingly, uncovered the kinase MLK3/SPRK/PTK (Ezoe et al., 1994; Gallo et al., 1994; Ing et al., 1994).
MLK3 (mixed lineage kinase 3) is a member of a family of Ser/Thr protein kinases that have the features of mitogen-activated protein kinase kinase kinases (MAPKKKs). In the adult tissues that have been examined it appears to be ubiquitously expressed (Gallo et al., 1994; Ing et al., 1994). Overexpression of MLK3 in mammalian cell lines leads to the activation of c-Jun NH2-terminal kinase (JNK; Rana et al., 1996; Teramoto et al., 1996; Tibbles et al., 1996) through apparent phosphorylation and activation of the dual specificity kinases, MAPK kinase-4 (MKK-4/SEK1) or MKK-7 (Rana et al., 1996; Whitmarsh et al., 1998). In some cell types, ectopic expression of MLK3 has also been reported to promote activation of the MAPK p38 pathway (Rana et al., 1996; Tibbles et al., 1996), the ERK pathway (Hartkamp et al., 1999), NF-κB (Hehner et al., 2000), and p70 S6 kinase (Lambert et al., 2002). Coexpression of MLK3 with GTP-bound forms of Rho family members Cdc42 or Rac has been found to increase the catalytic activity of MLK3 and to potentiate its ability to activate JNK (Teramoto et al., 1996; Bock et al., 2000). In addition, activated Cdc42, when coexpressed with MLK3, facilitates its homodimerization (Leung and Lassam, 1998), which, although not strictly necessary for MLK3 activation, promotes the autophosphorylation that is required for its activity (Vacratsis and Gallo, 2000; Leung and Lassam, 2001).
Although MLK3 is best known as an upstream activator of JNK, our observations regarding the provocative similarities between NIMA and MLK3 in their noncatalytic domains prompted us to evaluate MLK3 as a potential regulator of cell cycle events. We report here, in the first in-depth study of endogenous MLK3 protein, that this kinase does indeed have a role in the cell cycle and this aspect of its function appears to be independent of JNK. Specifically our data implicates MLK3 as a factor that promotes MT instability during prometaphase. These data provide evidence for a previously unsuspected MLK3 function in the mitotic program.
MATERIALS AND METHODS
Psi Blast Search and Alignment
Amino acid sequences of the C-terminal extension of NIMA and Fin1 were used to search the NCBI protein database using the PSI-BLAST program. Pairwise alignments were done using the Lipman-Pearson algorithm (DNASTAR software, (Lipman and Pearson, 1985; Pearson and Lipman, 1988). Structural predictions were generated by the COILS program of PredictProtein (Lupas, 1996). Nuclear localization sequences were assigned according to signature features described by Dingwall and Laskey, 1991 (Dingwall and Laskey, 1991). PEST sequences (Rogers et al., 1986) were assigned using PESTfind (www.at.embnet.org/embnet/tools/bio/PESTfind/). Similarity of the cyclin B destruction box to a region of NIMA was noted by Pu et al. (1995).
Plasmid Construction
For npGEX-MLK3, the 3′ end of the MLK3 cDNA within pGEX-MLK3 (kindly provided by Dr. J. Sivio Gutkind, National Institutes of Health, Bethesda, MD) that included ∼1 kb of contiguous nonMLK3, nonvector nucleotides was excised from the KpnI site (MLK3 nucleotide 1773) and replaced with a PCR fragment that included only MLK3 sequences and specified BamHI and EcoRI sites 3′ to the stop codon and sequenced. npGEX-MLK3K144R was generated by replacing the NcoI/KpnI fragment of npGEX-MLK3 containing the 5′ portions of MLK3 (nts 1–1773) with that from pCEFL-MLK3K144R (also supplied by Dr. Gutkind). To generate plasmids used in transient transfections, coding sequences were first inserted into a donor univector of the Univector Plasmid Fusion System (Liu et al., 1998). To create pUNI70-MLK3 and pUNI-MLK3K144R, the NcoI/BamHI fragments of npGEX-MLK3 and npGEX-MLK3K144R were ligated into NcoI/BamHI pUNI70. The NIMA sequences were cloned into pUNI70 from pETa-NIMA or pETa-NIMAK40 M using NcoI/NotI digestion to create pUNI70-NIMA and pUNI70-NIMAK40 M. All pUNI70 constructs were recombined into pHM200-HA3 using Cre recombinase as described (Liu et al., 1998). To produce the inducible MLK3 plasmid used for stable transformation, the MLK3 insert of npGEX-MLK3 was excised with BamHI and inserted into the BamHI site of pcDNA4/TO (Invitrogen, Carlsbad, CA), to generate pcDNA4/TO-MLK3, and the orientation determined by restriction endonuclease mapping. To create pET42a-MLK3:477–847 sequence encoding amino acids 477–847 of MLK3 was amplified. The digested product was inserted into NcoI/BamHI pET42a (Novagen, Madison, WI) and sequenced. pBSIISK-MLK3 used to produce MLK3 in vitro was made by inserting the BamHI fragment of npGEX-MLK3 into the BamHI site of pBSIISK (Stratagene, La Jolla, CA).
MLK3 Antisera Generation
Antibodies used for immune precipitation were generated in rabbits against GST-MLK3 477–847 encoded by plasmid pET42a-MLK3:477–847. Protein was purified from bacterial lysate using glutathione resin (AmershamPharmacia Biotech, Piscataway, NJ) according to the pET System Manual (Novagen). Purified protein was mixed with Freund's adjuvant (Sigma Chemical Co., St. Louis, MO) and injected into two rabbits twice monthly, with bleeds at the time of injection. The Ig fraction from immune and nonimmune sera was obtained by precipitation with 50% ammonium sulfate followed by resuspension and dialysis in PBS. MLK3-specific binding of sera was tested by immune precipitation of 35S-labeled MLK3, produced by in vitro transcription/translation of pBSIISK-MLK3 with T3 RNA polymerase in reticulocyte lysate (Promega, Madison, WI), mixed with similarly labeled control proteins.
Cell Culture Reagents
Cell culture media and additions obtained from Life Technologies BRL (Rockville, MD) were as follows: DMEM, MEME, MEM, McCoy's 5a, fetal bovine serum (FBS), penicillin-streptomycin (Pen-Strep), MEM sodium pyruvate, and MEM nonessential amino acids. Stock additives were as follows: tetracycline (Sigma Chemical Co.), 10 mg/ml in ethanol; nocodazole (Sigma), 7.5 mg/ml in DMSO; SB203580 (Calbiochem, San Diego, CA) 10 mM in DMSO; SP600125 (Calbiochem) 10 mM in DMSO; paclitaxel (taxol; Sigma), 10 mg/ml in DMSO; anisomycin (Sigma), 10 mg/ml in sterile water; blasticidin (Invitrogen), 5 mg/ml in sterile water, and zeocin (Invitrogen), and 100 mg/ml in sterile water and opaque tubes.
RNA Preparation
Twenty-three–nucleotide double-stranded RNAs were synthesized by Dharmacon Research (Lafayette, CO). The targeting sequence of human MLK3 (accession nos. L32976 and U07747), AAGACCCTGAAGATCACCGACTT, corresponds to the coding region 781–803 relative to the first nucleotide of the start codon. GL2 luciferase siRNA (Dharmacon Research) served as a control.
Cell Line Propagation and Transfection
HEK293 cells were grown in MEME containing 10% FBS and transiently transfected with plasmid DNA using the calcium phosphate procedure of Chen and Okayama (1987). The T-REx-293 cell line (Invitrogen), grown in DMEM (high glucose) containing 10% tetracycline-minus FBS (Hyclone, Logan, Utah), 1% Pen-Strep and 5 μg/ml blasticidin, was transformed with SspI-linearized pcDNA4/TO-MLK3 DNA. After 24 h, 200 μg/ml zeocin was included in the growth media to allow selection for stable transformants, 20 of which were subsequently cloned and expanded. Immunoblot analyses showed that all 20 clones expressed high levels of MLK3 protein after incubation with 1 μg/ml tetracycline for 24 h (unpublished data). HeLa cells maintained in MEM containing 10% FBS, 1% sodium pyruvate, 1% Pen-Strep, 1% nonessential amino acids, were synchronized by double thymidine block; cells were incubated in medium supplemented with 2 mM thymidine for 18 h, in unsupplemented medium for 8 h, and in medium supplemented with 2 mM thymidine for another 18 h and released from G1/S block into fresh unsupplemented medium at time 0 h.
For flow cytometry analysis, cells were detached from the plates by trypsinization and then fixed in ice-cold 70% ethanol. Fixed cells were pelleted and stained in a solution of 40 μg/ml propidium iodide and 80 μg/ml RNase A. Flow cytometry analysis was performed on a FACScan (Becton Dickinson Immunocytometry, San Jose, CA). SAOS2 cells were maintained in McCoy's 5a medium containing 15% FBS. For siRNA transfection, cells were seeded in six-well plates (1 × 105 cells/well) and incubated for 24 h before siRNA addition. Using Oligofectamine reagent (Invitrogen) for transfections, 258 pM siRNA was added in a volume of 0.43 ml to each well (Elbashir et al., 2001) and cells were reincubated for a minimum of 45 h before assay or further treatments.
Immunoblots, Immunoprecipitation, and Kinase Assays
Cultured cells were collected by trypsinization and washing in cold PBS. Washed cell pellets either were flash frozen in liquid N2 and stored at −80°C for later lysis or lysed directly by resuspending in lysis buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20 mM β-glycerophosphate, 5 mM EDTA, 10% glycerol, 1 mM sodium vanadate, 5 μM microcystin, 100 μM PMSF, and 100 μg/ml Pefabloc (Boehringer Mannheim, Indianapolis, IN) and sonicating on ice with 20 pulses of 1 s. For immunoblots, 20 μg lysed samples were mixed with SDS sample buffer to 1 μg/μl and separated by SDS-PAGE. Immunoblots were incubated with primary antibodies overnight at 4°C: rabbit anti-MLK3 (Santa Cruz, sc-536), 1:400; rabbit antiphospho-histone H3 (Upstate Biotechnology, Lake Placid, NY, 06–570), 1:1000; rabbit anti-JNK1 (Santa Cruz Biotechnology, Santa Cruz, CA, sc-571), 1:500; and mouse anti–HA F-7 (Santa Cruz, sc-7392), 1:400. For immune precipitation, lysed samples were cleared by centrifugation at 14,000 × g for 10 min at 4°C, supernatants were collected, and Triton X-100 was added to 0.1% for anti-JNK or anti-MLK3 immune precipitations (IPs), and NP-40 was added to 0.5% for anticyclin B1 IPs. IPs were carried out by rotating at 4°C for 4 h (MLK3) or overnight (JNK and cyclin B1) with: anti-MLK3 (anti-GST-MLK3 477–847) 2 μg immune or nonimmune sera in 100–500 μg protein; anti-JNK1 (Santa Cruz, sc-571), 10 μl in 200 μg protein; anticyclin B1 (PharMingen, San Diego, CA, no. 554177), 1 μl in 100 μg protein. Immune complexes were collected by incubation for 1 h at 4°C with protein A sepharose CL4B (Sigma, MLK3 and JNK IPs) or protein A/protein G agarose (Oncogene Research Products, Boston, MA, cyclin B1 IPs), washed three to five times with lysis buffer containing the appropriate detergent and two times with one of the after kinase assay buffers: cyclin B1 IPs, 20 mM HEPES, pH 7.5, 5 mM MgCl2, 2.5 mM MnCl2, 1 mM DTT; JNK and MLK3 IPs, 20 mM HEPES, pH 7.5, 10 mM MgCl2. Cyclin B1 kinase assays were carried out in the presence of 50 μM ATP, 0.1 μg/μl histone H1, and 0.3 μCi/μl [γ-32P]ATP (6000 Ci/mmol, Amersham Pharmacia) for 7 min at 30°C. JNK kinase assays included 100 μM ATP, 75 ng/μl GST-N-Jun, 10 μM PKI (Sigma, P0300) and 0.3 μCi/μl [γ-32P]ATP (6000 Ci/mmol) for 15 min at 30°C. MLK3 kinase assays included 100 μM ATP, 10 μM PKI, 0.1 μg/μl MBP (Sigma), and 0.3 μCi/μl [γ-32P]ATP (6000 Ci/mmol) for 35 min at 30°C. Reactions in all cases were stopped by adding equal volume 2× SDS sample buffer and heating to 90°C for 10 min.
Pin1 Binding Assays
GST or GST-Pin1 beads were prepared as described (Winkler et al., 2000). Clarified cell extract was incubated for 1 h rocking at 4°C with 1/100 volume beads equilibrated in PBS. Beads were repeatedly pelleted by brief centrifugation and resuspended in 10 volumes cold PBS + 0.5% Triton X-100. After five washes, beads were resuspended in 5 volumes SDS sample buffer for resolution by electrophoresis and immunoblot.
Phosphatase Treatment
Reactions were performed with 400 U lambda protein phosphatase (λ-PPase; New England Biolabs, Beverly, MA) per 100 μg cell extract with 1XPTase buffer and MgCl2 (supplied), with or without 50 mM EDTA as the PPase inhibitor, in total volume that was typically 1 μg/μl cell extract. Reactions were performed at 30°C for 1 h and stopped by supplementation to 50 mM EDTA and dilution fourfold in lysis buffer containing 5 μM microcystin and 20 mM β-glycerophosphate. At this stage, extract was analyzed directly by immunoblot.
Immunofluorescence Microscopy
Cells were grown on poly-d-lysine–coated coverslips and fixed by immersion in −20°C methanol for 5 min. After washes in PBS, slides were blocked and stained in 3% BSA/0.2% Tween-20/PBS. Primary antibody, mixed in blocking solution, was incubated on coverslips overnight at 4°C: rabbit anti-MLK3 (Santa Cruz, sc-536), 1:50; MLK3 blocking peptide (Santa Cruz, sc-536 P), 3:50; rabbit antiphospho-histone H3 (Upstate Biotechnology, 06–570), 5 μg/ml; mouse anti–gamma-tubulin (Sigma, T6557), 1:1000; mouse anti–alpha-tubulin (Sigma, T5168), 1:2000; mouse anti–HA F-7 (Santa Cruz, sc-7392), 1:100. Immune staining was visualized with FITC-conjugated goat anti-rabbit IgG (Caltag Laboratories, Burlingame, CA, L42001), 1:100, and biotin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, 115–065-146), 1:100 followed by Texas red–conjugated streptavidin (Jackson ImmunoResearch, 016–070-084), 5 μg/ml. Cells were mounted in ProLong Antifade (Molecular Probes, Eugene, OR) containing 0.2 μg/ml Hoechst dye 33258 to visualize DNA. For conventional microscopy, cells were examined using a Zeiss Axioskop microscope (Thornwood, NY) and images captured using a Pentamax cooled CCD camera (Princeton Instruments, Princeton, NJ), interfaced with MetaMorph software (Universal Imaging Corp., West Chester, PA). Confocal microscopy was performed using a Zeiss LSM 410 laser scanning confocal microscope. Images were processed using Adobe Photoshop (San Jose, CA).
RESULTS
MLK3 and NIMA Primary Sequences Share Several Features
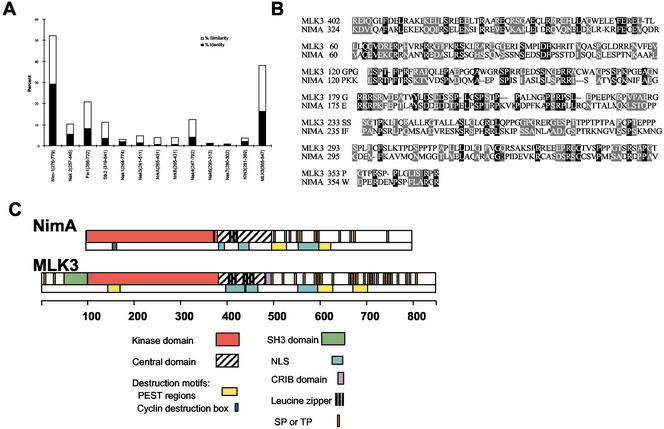
The primary amino acid sequences of the noncatalytic C-terminal extensions of NIMA and Fin1 were used to search the NCBI protein database using the PSI-BLAST program imposing relaxed parameters. MLK3 was identified and found to be 16% identical and 22% similar to NIMA in this region (aa 281–699 of NIMA), using the Lipman-Pearson algorithm (Lipman and Pearson, 1985; Pearson and Lipman, 1988; Figure 1, A and B). Only Neurospora Nim-1, among all other members of the NIMA subfamily, has higher homology in the same region (Figure 1A). In addition, the noncatalytic C-terminal extensions of two new NIMA family members, Nek8/Nercc1 (Holland et al., 2002; Roig et al., 2002) and Fa2p (Mahjoub et al., 2002) identified during the revision of this article show little homology to NIMA in this region (2% identical and 4% similar for Nek 8, and 3% identical and 6% similar for Fa2p, unpublished data).The other members of the MLK family (MLK1, MLK2, LZK, and DLK) are less similar to NIMA than MLK3 (unpublished data).
Figure 1.
MLK3 and NIMA are similar, especially in the C-terminal regions. (A) The amino acid sequence of NIMA's C-terminal extension (aa 281–699) was aligned pairwise with the corresponding regions, as indicated, of the NIMA-related kinases and with MLK3 using the Lipman-Pearson algorithm (DNASTAR software). The percent of identical and similar residues for each alignment is indicated. (B) The Lipman-Pearson alignment of NIMA (aa 281–699) with MLK3 (365–847). The N (MLK3 aa 365–401 and NIMA aa 281–324) and C (MLK3 aa 369–847 and NIMA aa 371–699) terminal sequences of these regions, although included in the alignment, were not of sufficient similarity, as determined by this program, to be aligned and so were omitted in the figure. Identical residues are indicated by black-shaded boxes, similar residues by gray-shaded boxes. (C) Schematic alignment of NIMA and MLK3 protein subsequences, drawn to scale (amino acid position).
The molecular domains of NIMA and MLK3 are depicted schematically in Figure 1C. Although the degree of primary sequence homology is only moderate, we found the similar domain arrangements of these proteins to be quite striking. Both molecules have N-terminal Ser/Thr kinase domains followed by central domains, the structures of which are predicted to be predominantly coiled-coils (COILS program, PredictProtein; www.embl-heidelberg.de/predictprotein/predictprotein.html; Lupas, 1996). These domains also contain leucine zippers and putative nuclear localization sequences. Finally, the C-terminal tails of both proteins are rich in S/T-P sites and PEST motifs. Despite the presence of several regions unique to MLK3 (an N-terminal SH3 domain and a centrally located CRIB (Cdc42/Rac Interaction Binding) domain (Burbelo et al., 1995), both the ordering and spacing all other identifiable modular domains are well conserved between NIMA and MLK3.
Endogenous MLK3 Undergoes Biochemical Alterations During the Cell Cycle
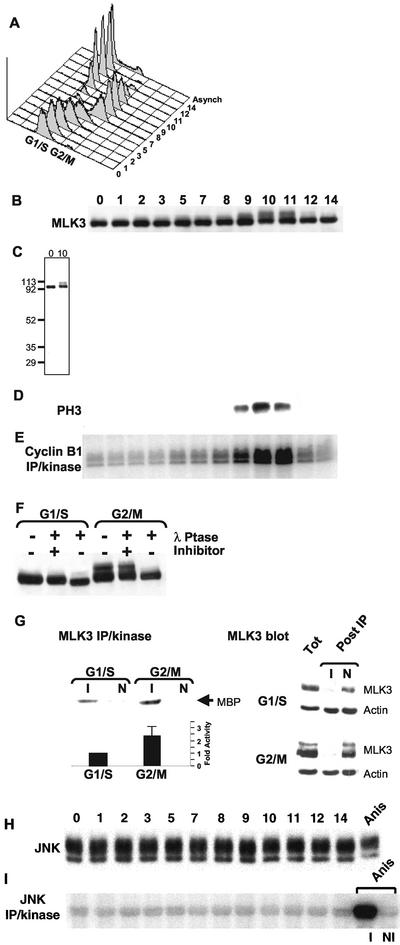
To examine the regulation of endogenous MLK3 during the cell cycle, HeLa cells were synchronized at the G1/S boundary by a double thymidine block and then collected at successive time points after release from arrest and subjected to various analyses (Figure 2). Immunoblots indicated that the levels of the 93-kDa MLK3 protein did not change appreciably during the cell cycle. However, the appearance of a higher Mr species (proportionally 20%) was found in the samples collected at 9, 10, and 11 h after release from the G1/S block (Figure 2B). No other reactive species were detectable in these samples indicating the specificity of the anti-MLK3 antibody (Figure 2C). The 9-, 10-, and 11-h periods after release from the G1/S block corresponded with G2/M of the cell cycle as judged by three criteria: 1) 4n DNA content in flow cytometric analysis (Figure 2A); 2) peak levels of phosphorylated histone H3 (Figure 2D); and 3) maximal kinase activity of Cdc2/cyclin B1 (Figure 2E). The higher Mr form of MLK3 protein disappeared after treatment with lambda phosphatase (λ Ptase, Figure 2F). Indeed, the bulk of MLK3 present in the 93-kDa bands in both G1/S and G2/M samples also shifts slightly to a higher mobility form after phosphatase treatment. Thus, it appears that MLK3 is phosphorylated at all stages of the cell cycle and that a fraction of this protein becomes hyperphosphorylated during G2/M.
Figure 2.
Cell cycle stimulation of MLK3's phosphorylation state and kinase activity in the absence of JNK activation. HeLa cells were synchronized at G1/S by a double-thymidine block (0) and then released into media lacking thymidine. Samples were collected at subsequent times for 14 h and analyzed as described. (A) Flow cytometric quantification of DNA content per cell (x-axis, propidium iodide stain) vs. cell number (y-axis). Distribution of DNA content of cells from an asynchronous culture is also shown (Asynch). Twenty micrograms of each sample was separated by SDS-PAGE and subjected to immunoblot analysis using anti-MLK3 (B) or antiphosphorylated histone H3 (D). (C) Full-length immunoblot of MLK3 from 0- and 10-h samples. Molecular weight markers are indicated on the left. (E) Anticyclin B1 immune precipitation/kinase assay analysis (100 μg/sample) using histone H1 as substrate. (F) Immunoblot of MLK3 in G1/S (0 h) and G2/M (10 h) samples that were incubated with λ-phosphatase in the presence or absence of the phosphatase inhibitor, EDTA, or were left untreated. (G, upper left panel) MLK3 kinase activity toward myelin basic protein (MBP) was determined for G1/S (0 h) and G2/M (9 h) cell extracts after immunoprecipitation with anti-MLK3 (I) or with nonimmune (N) antibodies. (G, lower left panel) Average values of MLK3 immune precipitation/kinase assays from six different synchronous cultures collected at G1/S (0 h) and G2/M (9, 10, 11 or 12 h, depending on culture). Shown is the average fold stimulation of activity from G2/M samples (2.4 times) relative to G1/S (set at 1) ± the SD (0.67). (G, right panels) Immunoblots of MLK3 and actin in G1/S and G2/M samples before (Tot) and after (Post IP) immunoprecipitation with anti-MLK3 (I) or with nonimmune (N) antibodies. (H) Immunoblot of samples from (A) reacted with anti-JNK antibodies. Twenty micrograms of a sample collected from an asynchronous HeLa culture treated with anisomycin for 20 min was also included (Anis). (I) Anti-JNK immune precipitation/kinase assay analysis (100 μg/sample) of the samples in (H) using GST-N-Jun as substrate. As part of the JNK assays, the anisomycin-treated sample also was subjected to immune precipitation with the anti-JNK antibody (I) or with a nonimmune control antibody (NI) followed by incubation in kinase assay buffer. Products of the kinase reactions (E and H) were separated by SDS-PAGE and substrate incorporation of 32P was detected by phosphorimager exposure.
To determine whether the mitotic forms of MLK3 are accompanied by a change in its kinase activity, anti-MLK3 immune precipitates, or those of nonimmune antibodies, prepared from both G1/S- and G2/M-phase extracts were incubated in a kinase assay buffer containing MBP (myelin basic protein) as substrate (Figure 2G). Quantitative analysis of 32P incorporation into MBP revealed that the MLK3 activity from the G2/M sample was enhanced ∼2-fold relative to G1/S (Figure 2G, left panels). In six times that this experiment was repeated using six different pairs of cell samples, the MLK3 kinase activity in G2/M cell extracts was stimulated an average of 2.4-fold over that in G1/S (Figure 2G, lower left panel, SD: 0.67). To compare the efficiency of MLK3 precipitation from G1/S and G2/M samples in these experiments, aliquots of the samples were collected before and after immunoprecipitation and then immunoblotted for MLK3 and actin, as a control protein (Figure 2G, right panel). Under these conditions of immunoprecipitation, MLK3 was cleared equivalently from both G1/S and G2/M samples with immune sera (>90% in both cases, unpublished data), but not with nonimmune antibodies (Figure 2G, right panel). The actin levels in both samples were not effected by precipitation with either immune or nonimmune antibodies. These results show that the activity of MLK3 kinase, which can be detected at all cell cycle stages, is enhanced during G2/M to a degree that is similar to the fold induction reported for NIMA (Ye et al., 1995).
The basal activity of MLK3 appears to require phosphorylation (Leung and Lassam, 2001) and can be stimulated by cellular conditions that also lead to the generation of hyperphosphorylated forms of this kinase, although the contribution of hyperphosphorylation to the activity increase has not been established (Bock et al., 2000). It was of interest, then, to test whether or not hyperphosphorylation of MLK3 during G2/M was responsible for the increased activity at this time. Although the buffer conditions required for λ phosphatase activity completely inactivated the kinase, even in the absence of added phosphatase, we were able to partially dephosphorylate MLK3 in G2/M samples by incubating these extracts in the absence of added phosphatase-inhibitors for 1 h at 30°C before immune precipitation and assay of kinase activity (unpublished data). Although the highest Mr species of MLK3 disappeared from these samples there was no effect of this treatment on the kinase activity (unpublished data). Thus, formation of the highest Mr species of MLK3 by hyperphosphorylation during mitosis is not required for kinase stimulation, and although we cannot completely rule out a role for phosphorylation in the activity increase, there are no data to indicate that it is a requirement.
The demonstration that the activity of MLK3 is stimulated at G2/M raised the question as to whether this event is accompanied by the activation of JNK, known to be a downstream target of MLK3 (Rana et al., 1996; Teramoto et al., 1996; Tibbles et al., 1996). Our cell cycle analysis of JNK activity shows that the levels and mobility of JNK protein (two species shown of p54 and p46) remain constant (Figure 2H), and there is no detectable JNK activation at any stage of the unperturbed cell cycle (Figure 2I). This is in contrast to the higher mobility and activated form of JNK present in lysates of an asynchronous HeLa cell culture treated for 20 min with anisomycin, an inhibitor of protein synthesis that acutely activates the JNK pathway (Figure 2, H and I, Anis). Thus, JNK does not appear to be a target of endogenous MLK3 during the cell cycle.
In summary, we have found that the levels of MLK3 protein remain relatively constant throughout the cell cycle but that a hyperphosphorylated form of the protein appears transiently during G2/M, disappearing before mitotic exit. Additionally, during G2/M the kinase activity of MLK3, measured in vitro, is elevated approximately twofold relative to its activity at G1/S. Significantly, this stimulation of MLK3 kinase activity is not accompanied by a detectable increase in JNK activity. These results suggest that MLK3 may have a cell cycle role and that this role is likely to be distinct from its function in activating the JNK pathway.
Distribution of Endogenous MLK3 Changes as a Function of the Cell Cycle
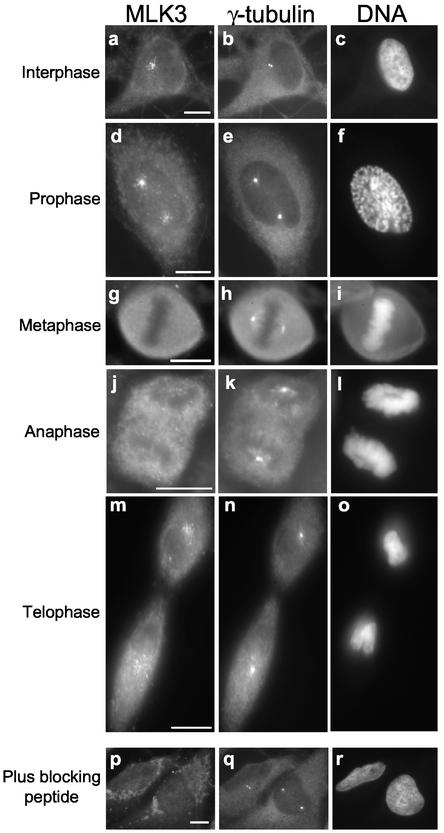
In interphase cells, characterized by a flattened morphology with decondensed chromosomes contained within a well delineated nucleus (Figure 3c), MLK3 immunostaining resulted in the appearance of numerous foci (sprinkles) that radiated as a loose gradient from a point in the cell proximal to, but outside of, the nucleus (Figure 3a). In addition, there appeared to be a uniform distribution of cytoplasmic staining (Figure 3a). When cells were costained for γ-tubulin, one or two closely spaced fluorescent dots representing the centrosome consistently appeared in the center of the MLK3 sprinkle gradient (Figure 3, compare b with a). Preincubation of the anti-MLK3 antibody with blocking peptide completely abolished sprinkle staining and most of the cytoplasmic staining, whereas γ-tubulin staining was unaffected (Figure 3, p and q). This distribution of MLK3 is independent of fixation method, because it was observed in cells after treatments with either aldehyde or organic solvent fixatives (unpublished data).
Figure 3.
MLK3 is localized near the centrosome in a cell cycle–dependent manner. Asynchronous HeLa cells growing on coverslips were fixed with methanol and then triple stained for MLK3 (left panels; a, d, g, j, m, and p), γ-tubulin (middle panels; b, e, h, k, n, and q), and DNA (right panels; c, f, i, l, o, and r). Samples p, q, and r were stained in the presence of MLK3-blocking peptide. Representative cells from different cell cycle stages are shown: interphase (a–c); prophase (d–f); metaphase (g–i); anaphase (j–l); telophase (m–o). Scale bar, 10 μM, applies to all images of a given field.
During prophase when the chromosomes had begun to condense (Figure 3f) and the centrosome had split (Figure 3e), MLK3 staining revealed two sprinkle gradients each arrayed around one of the two centrosomes (Figure 3, d and e). The majority of the more distal sprinkles lay in an area between the two centrosomes rather than outside this area. Notably, this staining of MLK3 was apparent in cells that had already recruited extra γ-tubulin to the centrosome, an event of late prophase (Figure 3e; Khodjakov and Rieder, 1999).
At a slightly later stage of mitosis, when the chromosomes were more frankly condensed and the nuclear envelope had broken down, MLK3 was uniformly distributed throughout the cell, having no apparent association with the centrosomes. Shown is a stained metaphase cell where the γ-tubulin-positive centrosomes at the spindle poles are evident (Figure 3, g–i). The change in MLK3 distribution from centrosomal-associated sprinkles in late prophase to diffuse and uniform cellular localization at prometaphase was abrupt, as judged by the lack of any detectable intermediate distributions.
During anaphase when the chromosomes had disjoined and were separating toward the two spindle poles, the globally diffuse distribution of MLK3 was maintained with no apparent association with the centrosome or any other cellular structure (Figure 3, j–l). It was only during late telophase, after cytokinesis, that the centrosome-associated sprinkle distribution of MLK3 was reestablished (Figure 3, m–o).
MLK3 Distribution Is Dependent on Microtubule Organization
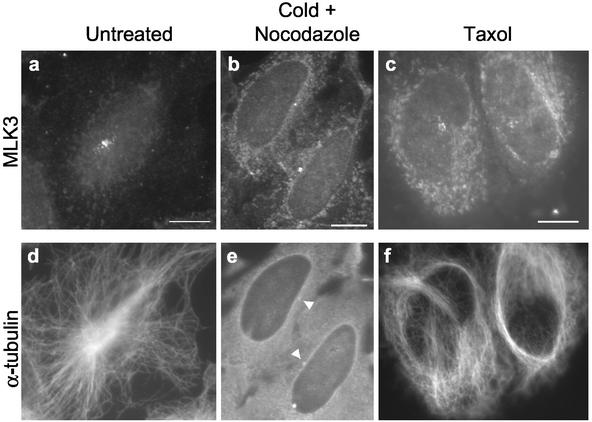
The centrosome is the primary microtubule organizing center (MTOC) of the cell, and the presence of MLK3 proximal to this organelle led us to assess the dependence of MLK3 localization on MTs. In HeLa cells that had been incubated with cold plus nocodazole and thus completely lacked cytoplasmic MTs, the number and staining intensity of MLK3-positive sprinkles were profoundly diminished (Figure 4, a and b). The few sprinkles remaining were tightly clustered around a single point that colocalized with a spot of enhanced α-tubulin staining (Figure 4e, arrowheads), suggestive of a centrosome. Costaining of similarly treated cells with MLK3 and γ-tubulin confirmed that the remaining MLK3-associated focus was at the centrosome (unpublished data). In contrast to nocodazole, taxol treatment of interphase cells both diminishes existing centrosome-nucleated MTs and induces formation of long, noncentrosome-nucleated MT bundles (DeBrabander et al., 1986). When HeLa cells were treated with taxol, the number and staining intensity of the MLK3 sprinkles were reduced relative to those found in the untreated cells (Figure 4, c and a). Furthermore, there were no additional foci of MLK3 staining in these cells despite the presence of stabilized MTs that appeared as disorganized dense bundles and loose meshworks without association with the centrosome (Figure 4f). These data indicate that the MLK3 sprinkles are associated specifically with centrosome-nucleated microtubules as well as with the centrosome itself.
Figure 4.
Localization of the centrosome-associated MLK3 sprinkles is partially dependent on microtubules. Asynchronous HeLa cells growing on coverslips were either: 1) untreated (a and d); 2) incubated in the presence of nocodazole (6 μg/ml) first on ice for 45 min and then at 37°C for 1 h (b and e); or 3) treated with taxol (5 μM) for 4 h (c and f). Cells were fixed in methanol and stained for MLK3 (a–c) and α-tubulin (d–f). Arrowheads in e denote brighter spots of α-tubulin staining indicative of centrosomes. The reduced levels of MLK3 staining after treatment with cold/nocodazole or taxol required longer photographic exposures for imaging leading to a commiserate increase in the level of cytoplasmic staining seen in b and c relative to a. Scale bars, 10 μM, applies to all images of a given field.
Ectopic Expression of MLK3 Disrupts Microtubule Organization
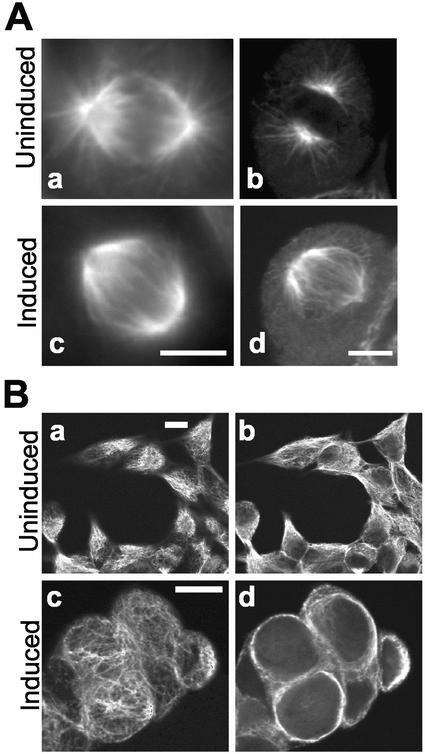
Given the presence of MLK3 at centrosome-nucleated microtubules and the known dynamic changes that these cytoskeletal structural elements undergo during G2/M when MLK3 kinase activity is maximal, we examined the effects of MLK3 ectopic expression on MTs during both interphase and M phase. We produced a stable HEK293 cell line containing an inducible wild-type MLK3 expression plasmid. After treatment of these cells with the inducing agent (tetracycline) or an equivalent fraction of vehicle alone (ethanol) for 23 h, the distribution of α-tubulin was assessed by immunofluorescence microscopy (Figure 5). Mitotic cells from uninduced cultures showed normal mitotic spindles containing both central and astral microtubules (Figure 5A, a and b). The mitotic spindles from cells of induced cultures, although similar in size to those from control cultures and possessing central microtubules, were largely devoid of astral microtubules (Figure 5A, c and d). The condensed metaphase chromosomes from these cells appeared to be normal (unpublished data), and no increase in mitotic index was observed (4% of the cells in both induced and uninduced populations were phospho-histone H3-positive; unpublished data). The MT effects in interphase cells overexpressing MLK3 were also dramatic. Although interphase cells from control cultures presented a flattened morphology and displayed cytoplasmic as well as peripheral MT networks (Figure 5B, a and b), induced cells were rounded-up and, in optical sections of the cell center, showed a stunning absence of cytoplasmic MTs (Figure 5Bd). In these cells, there was a narrow layer of α-tubulin staining on the cell periphery, which, upon adjustment of the focal section, appeared to be a meshwork of noncentrosome-nucleated MTs (Figure 5Bc). Although JNK was activated in induced cells, coincubation with the specific JNK inhibitor, SP600125, (Bennett et al., 2001) or with the p38 MAPK inhibitor, SB203580, which also partially inhibits JNK (Clerk and Sugden, 1998), had no effects on the phenotype of cells induced to express MLK3 at high levels (unpublished data). These results indicate that overexpression of MLK3 in these human cells causes disruption in microtubule organization in both interphase and mitotic cells. Similar MT effects have been reported for NIMA overexpression both in A. nidulans as well as heterologous systems (Osmani et al., 1988, 1994).
Figure 5.
MLK3 overexpression disrupts microtubule organization. HEK293 cells containing a stable, inducible, wild-type MLK3 expression plasmid grown on coverslips were incubated with tetracycline (1 μg/ml; Ac and d, Bc and d; Induced) or vehicle (ethanol) alone (0.1%; Aa and b, Ba and b; Uninduced) for 23 h. Cells were then fixed in methanol and stained for α-tubulin. (A) metaphase cells. To detect maximal immunofluorescence signal in the astral area, images were collected using conventional microscopy (Aa and c). For clarity, confocal images (antibody and d) were also collected. (B) interphase cells. Shown are representative single fields of both uninduced (Ba and b) and induced (Bc and d) cells from which confocal optical slices were captured at two different depths. Ba and c are images focused close to the topmost cellular peripheries, whereas Bb and d are images focused 2–3 μM more internally, toward the cell centers. Scale bars, 10 μM, applies to image pairs Aa and c, Ab and d and to all images of a given field in B.
Comparison of NIMA Properties with Those of MLK3
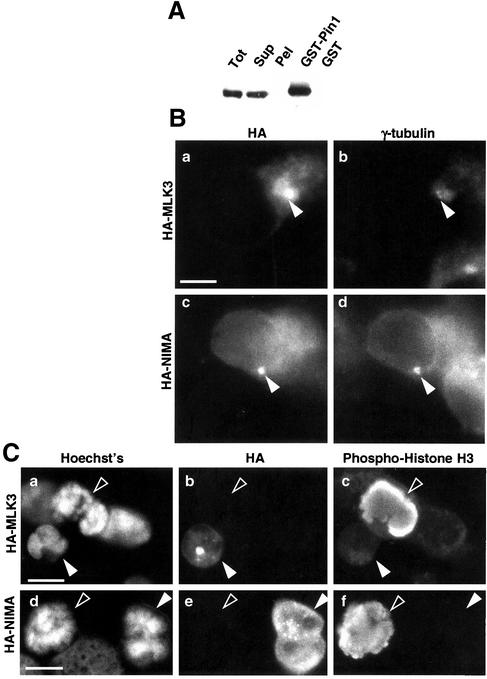
NIMA was the original bait used to identify the prolyl isomerase, Pin1, in yeast two-hybrid screens (Lu et al., 1996; Crenshaw et al., 1998). To determine whether MLK3 is also a Pin1-binding protein, pull-down assays were performed using GST-Pin1 protein immobilized to glutathione-sepharose beads. Endogenous MLK3 is readily bound from cell extracts (Figure 6A) as assessed by immunoblot. The bound MLK3 has a slightly reduced gel mobility compared with that in the lysate, suggestive of posttranslational modification-selective Pin1 binding.
Figure 6.
Comparison of MLK3 properties with those of NIMA. (A) Homogenates made from HEK293 cells (Tot) were separated by centrifugation into a soluble (Sup) and insoluble fraction (Pel). The soluble fraction was incubated with either GST-Pin1 beads or GST-beads, which were then washed extensively before the addition of SDS sample buffer. Homogenate fractions as well as bead-bound material were subjected to immunoblot analysis using MLK3-specific antibodies. The bead-bound material shown (GST-Pin1 and GST) was derived from ∼10 times the volume of soluble extract shown in the Sup sample. (B) HEK293 cells were transfected with HA-tagged MLK3 (a and b) or NIMA (c and d) expression constructs for 9 h, fixed with methanol, and stained for HA (a and c) and γ-tubulin (b and d). The γ-tubulin–staining centrosomes are indicated by the closed arrowheads. (C) HEK293 cells were transfected with HA-tagged MLK3 (a–c) or NIMA (d–f) expression constructs for 33 h, fixed with methanol, and stained for DNA (a and d), HA (b and e), and phospho-histone H3 (c and f). Transfected cells are indicated by closed arrowheads, and untransfected prometaphase cells are indicated by open arrowheads. Scale bar, 10 μM.
Although the ectopic expression of NIMA in human cells triggers disruption of microtubules (O'Connell et al., 1994), the distribution of NIMA in these transfected cells has not been determined. To examine the localization of NIMA in human cells after transfection and to compare with that of MLK3 in cells treated similarly, HA epitope-tagged NIMA and MLK3 were introduced separately into HEK293 cells by transient transfection. HA staining in both HA-NIMA– and HA-MLK3–transfected cells resulted in the appearance of one or more spots that localized closely to the γ-tubulin–reactive centrosomes (Figure 6B). In addition, a more general cytoplasmic distribution of both HA-MLK3 and HA-NIMA was detected, particularly in cells expressing higher levels of these transgenes. These results indicate that the distributions of HA-NIMA and HA-MLK3 after transfection are similar to each other and, importantly, are similar to that detected for endogenous MLK3.
A second effect of NIMA expression in human cells is a nuclear alteration that has been interpreted to be mitotic chromatin condensation as assessed by visual inspection after DNA staining (O'Connell et al., 1994; Lu and Hunter, 1995). However, because reagents for detecting specific markers of mitotic chromatin condensation were not available at the time of those studies, NIMA-induced nuclear effects in human cells have not been established unambiguously as mitotic chromatin condensation. Thus, we examined the nuclear effects of MLK3 overexpression and compared them in parallel with those of NIMA by transiently transfecting HEK293 cells with either HA epitope-tagged NIMA or MLK3. In both HA-NIMA and HA-MLK3-transfections, those cells expressing high levels of the transgene products (judged by HA staining) invariably contained nuclei which, when visually inspected after DNA staining, bore marked similarity to condensing chromosomes during mitosis (Figure 6C, compare open to closed arrowheads in a and d). However, in costaining experiments, the nuclei from these transfected cells were unreactive with an antibody for phosphorylated histone H3, a marker of mitotic chromosome condensation, whereas nearby nontransfected prophase cells reacted intensely with this antibody (Figure 6C, c and f). These data reveal that ectopic expression of NIMA or MLK3 in human cells triggers very similar nuclear distortion phenotypes but in neither case are these effects due to mitotic chromosome condensation.
MLK3 Depletion Leads to Increased Taxol Sensitivity
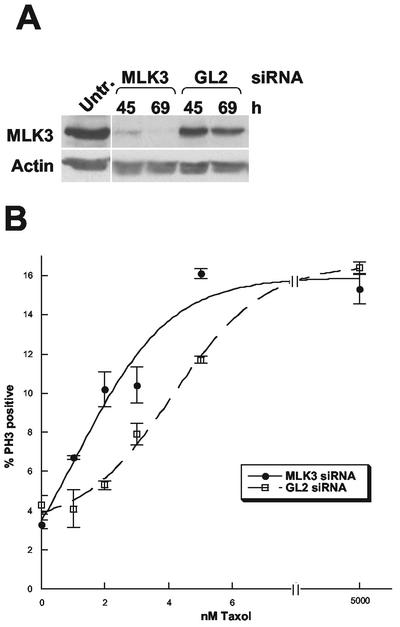
To better understand the functions of MLK3 in normally dividing, unstressed cells and to more fully evaluate its cell cycle role, we analyzed the effects of depleting this protein specifically using siRNA. To deplete MLK3, siRNA duplexes were synthesized that were directed against the coding region 781–803 of MLK3 relative to the first nucleotide of the start codon. These duplexes were used to transfect SAOS2 cells, a human cell line that, unlike HeLa cells, have intact cell cycle checkpoints during prophase, the time of MLK3 stimulation (Jha et al., 1994; Scolnick and Halazonetis, 2000). Transfections were carried out in parallel using a nonspecific RNA duplex of firefly luciferase from Photinus pyralis (GL2) at the same final concentration as the MLK3-specific siRNA (Elbashir et al., 2001). Cells were collected at 45 and 69 h after transfection, counted, and prepared for flow cytometry, immunoblot analysis, and immunocytochemistry. In these experiments MLK3 was efficiently depleted by the specific siRNA but not the GL2 control duplex, whereas actin levels were unaffected in either case (Figure 7A). There was no significant difference in the number of live cells present after transfection with either MLK3- or GL2-specific siRNAs at either 45 or 69 h (unpublished data) indicating that loss of MLK3 within this time period is not toxic. In addition, flow cytometry analysis showed no apparent differences in the cell cycle profiles of the two types of transfected samples when compared with each other or with that of untreated cells (unpublished data). Inspection of cells that were fixed and prepared for immunofluorescence microscopy using DAPI as well as antibodies against the mitotic marker phospho-histone H3, revealed no noticeable alteration of nuclear morphology in interphase cells and no significant difference in the mitotic index nor the percentage of mitotic cells in prophase, metaphase, telophase, and anaphase between the MLK3-depleted and nondepleted cells (unpublished data). These results indicate that MLK3 depletion does not affect nuclear morphology and, unlike the loss of NIMA function in Aspergillus, neither leads to a cell cycle arrest nor affects the passage through the stages of M phase
Figure 7.
siRNA depletion of MLK3 increases cell sensitivity to taxol. (A) Immunoblot of untreated SAOS2 cells (Untr.) and those transfected with MLK3 siRNA duplex or GL2 luciferase siRNA duplex (nonspecific siRNA control) and then incubated for 45 and 69 h. The top portion of the immunoblot was probed with anti-MLK3 antibody and the bottom portion with antiactin antibody to check for equal loading of total protein. (B) SAOS2 cells were transfected with MLK3 siRNA or GL2 luciferase siRNA for 54 h before the addition of taxol at increasing concentrations. After a further 16-h incubation, cells were fixed and prepared for immunofluorescence microscopy using antiphospho-histone H3 antibodies. The individual cells from three fields of each sample (125–530 cells/field) were examined under low power and the percentage of phospho-histone H3-positive cells determined, the average of which is plotted ± the SD.
The apparent relationship of MLK3 and MT organization led us to examine two issues regarding its function. The first issue was whether or not this kinase contributes to the mitotic stress checkpoint that delays entry into metaphase in response to microtubule poisons (Jha et al., 1994; Scolnick and Halazonetis, 2000). The presence of this checkpoint been demonstrated in SAOS2 cells and the exact assay parameters for determining its presence or absence in this cell type have been established (Scolnick and Halazonetis, 2000). Thus, when treated with 5 μM taxol for 16 h, these cells have a mitotic index of only 15–16% unless the mitotic stress checkpoint is compromised. In this case, where a mitotic delay is absent, the mitotic index increases to around 30% at this same 16-h time point.
Second, because MLK3 is able to disrupt microtubule organization when overexpressed and may normally carry out a similar function during G2/M when its activity is stimulated and when disassembly of the cytoplasmic MT network occurs, we evaluated whether its depletion might lead to a subtle increase in the stability of this network. Such a phenotype might not be readily apparent under normal growth conditions but could be observed by an increase in the cellular sensitivity to microtubule-stabilizing agents.
To resolve these two issues, SAOS2 cells were transfected in parallel with MLK3 and GL2-specific siRNAs for 54 h, to allow MLK3 depletion, followed by the addition of the microtubule-stabilizing agent taxol at 5 μM to determine the presence or absence of the mitotic stress checkpoint and at increasing low concentrations (1–5 nM) to determine taxol sensitivity. After a further 16-h incubation, cells were fixed and prepared for immunofluorescence microscopy using antiphospho-histone H3 antibodies. After the 5 μM taxol treatment, the mitotic index of MLK3 siRNA-transfected cells was similar to that of cells transfected with GL2 siRNA (15.3 and 16.4% phospho-histone H3 positive, respectively) and similar to that reported for SAOS2 cells in which the mitotic stress checkpoint is intact (Figure 7B). These results indicate that MLK3 does not contribute to this checkpoint.
Because a mitotic arrest is triggered by taxol-induced stabilization of microtubules, an increased sensitivity to taxol would result in an increased mitotic index after treatments with low taxol concentrations. Although the mitotic indices of MLK3 siRNA-transfected cells were similar to those of cells transfected with GL2 siRNA in the absence of taxol (3.3 and 4.3% phospho-histone H3 positive, respectively) or at high (5 μM) taxol concentrations, they were significantly higher at low (1–5 nM) concentrations of this agent (Figure 7B). The data indicate an approximate twofold decrease in the EC50 of taxol for the MLK3 siRNA transfectants compared with the GL2 siRNA transfectants. Thus, MLK3 depletion, although neither toxic to cells nor essential for passage through the cell cycle, increases cellular sensitivity to taxol at low concentrations, suggesting that its absence results in a partial stabilization of the MT network.
DISCUSSION
MLK3 as a NIMA-like Kinase with Diverse Roles
There are remarkable similarities between MLK3 and NIMA suggestive of functional relatedness. However, although both kinases appear to act during G2/M of the cell cycle, within their autologous systems (human and Aspergillus) the requirement of each is clearly different. Loss of NIMA function in Aspergillus brought about either by use of temperature-sensitive nimA alleles (Oakley and Morris, 1983; Bergen et al., 1984; Osmani et al., 1987) or expression of kinase-inactive versions of NIMA (Lu and Means, 1994; Lu and Hunter, 1995) triggers a cell cycle arrest at G2, whereas loss of MLK3 in human cells does not affect cell cycle progression. In spite of this essential difference, the similarities between these two kinases are striking. As proteins they are markedly alike both in their molecular domain arrangements and amino acid sequences, as well as their respective Pin1-binding properties. Within their autologous systems (human and Aspergillus), the kinase activities of endogenous MLK3 and NIMA are stimulated approximately twofold during M phase (Ye et al., 1995). Furthermore, both proteins appear to associate with MTs and MTOCs, however in different cell cycle stages, and, when ectopically expressed, to trigger loss of cytoplasmic microtubules (Osmani et al., 1988; O'Connell et al., 1994).
In addition to these similarities to NIMA, MLK3 also has differences both in its domain organization and biological roles. Relative to domain organization, MLK3 has unique SH3 and CRIB motifs, both of which imply signaling functions that are lacking in NIMA. The SH3 domain of MLK3 has been shown to participate in autoinhibition (Zhang and Gallo, 2001) and, although signaling molecules that overcome this SH3-mediated autoinhibition of MLK3 have yet to be identified, one potential candidate is hematopoietic progenitor protein kinase-1, a MAPKKK kinase that can activate the JNK pathway. This kinase can also interact with the SH3 domain of MLK3 and can phosphorylate MLK3 in vitro, although effects on MLK3 activity have not been observed (Kiefer et al., 1996). The CRIB domain of MLK3 has been shown to bind activated Cdc42 and to increase MLK3 catalytic activity in vivo but not in vitro (Burbelo et al., 1995; Teramoto et al., 1996; Bock et al., 2000). Additionally, coexpression of activated Cdc42 with MLK3 results in a change in the in vivo phosphorylation pattern of MLK3, suggesting that an additional component or cellular environment is required for MLK3 activation by Cdc42 (Bock et al., 2000). Interestingly, functional interactions with small GTPases have been demonstrated as well for another NIMA-like kinase, Nercc1 (Roig et al., 2002). Relative to biological roles, MLK3 has been implicated to be involved in stress response pathways, whereas NIMA has not. Although existing data, which are mainly derived from overexpression studies, suggest that the SH3 and CRIB domains influence the functions of MLK3 in these stress signaling pathways, specific roles, particularly for endogenous MLK3, are far from delineated. Finally, regarding the mitotic functions of MLK3, the domains unique to MLK3 remain to be explored.
Both MLK3 and NIMA trigger nuclear effects after ectopic expression. Within their respective cells of origin NIMA in Aspergillus has been reported to trigger bona fide mitotic chromatin condensation replete with phosphorylated histone H3 (De Souza et al., 2000), whereas MLK3 in human cells induces the formation of distorted nuclei that are visually similar to mitotic condensing chromosomes but that lack phosphorylated histone H3. The nuclear effects of NIMA when expressed in human cells have been widely interpreted as mitotic chromatin condensation (O'Connell et al., 1994; Lu and Hunter, 1995). However, now that phosphorylated histone H3 is commonly used as a marker of mitotic condensed chromatin, we show that these NIMA-induced nuclear effects in human cells are actually identical to those triggered by MLK3 and are not relevant to mitotic chromosome condensation. A similar nuclear compaction differing from mitotic chromosome condensation is also observed after ectopic expression of Fin1p, the NIMA-like kinase in fission yeast (Krien et al., 2002). Abnormal nuclear morphologies have also been noted after the expression of nuclear variants of Nercc1, a recently identified NIMA-like kinase that, like NIMA and MLK3, is activated and appears to function during G2/M (Holland et al., 2002; Roig et al., 2002). However, kinase-inactive Nercc1 variants also are able to induce these abnormal nuclear morphologies, whereas the nuclear distortion induced by NIMA or MLK3 does not occur after expression of kinase-inactive versions of these proteins (unpublished data). Thus, it is likely that the mechanisms by which these Nercc1-induced abnormal nuclei form are different than those leading to the nuclear effects triggered by NIMA or MLK3 ectopic expression.
The available data indicate that MLK3 and NIMA may act identically in several ways after ectopic expression in human cells. Not only do both trigger a nuclear distortion that is not mitotic chromatin condensation, but also they both localize near the centrosome, similar to endogenous MLK3, and they both appear to cause MT disruption (O'Connell et al., 1994). Thus, although MLK3 and NIMA are not full orthologues, they may carry out related functions during G2/M, which affect the microtubule network at this stage. Of the NIMA-like proteins examined, only MLK3 causes a MT disruption similar to that induced by NIMA, when ectopically expressed. Although the cellular response to taxol suggests that the microtubule network may be subtly stabilized in the absence of MLK3, the microtubule effects in the absence of NIMA function have not been examined. That some NIMA-like kinases, and perhaps NIMA itself, may have roles in microtubule disruption during G2/M is supported by a recent finding demonstrating that a microtubule-severing protein in Chlamydomonas, Fa2p, is a NIMA-like kinase (Mahjoub et al., 2002).
An unexpected finding of our studies is the increase in cellular sensitivity to taxol induced by MLK3 depletion. To date, the only known cellular targets that physically interact with taxol are microtubules (for review, see Wang et al., 2000). It seems probable then that MLK3 (or lack of MLK3) affects this interaction, most likely through effects on proximal microtubules. Regardless of mechanism, the ability to deplete MLK3 function, whether through siRNA or use of specific inhibitors, might have important therapeutic implications. Taxol is one of the broadest-spectrum anticancer agents currently used in the treatment of patients. Because of its serious side effects (neurotoxicity and cardiotoxicity) the identification of adjuvant treatments that improve its therapeutic index are of enormous clinical importance. The specific targeting or depletion of MLK3, which is in itself nontoxic, might provide such a taxol-potentiating antitumor treatment.
Localization of MLK3: Microtubule Tethering
MLK3's subcellular localization is similar to that described for the MT motor-associated protein, PCM-1 (pericentriolar material-1;Balczon et al., 1994). This 228-kDa protein can move along MTs in a dynein-dependent manner (Kubo et al., 1999), accounting for its concentration near the minus ends of MTs at the centrosome. Like that of MLK3, PCM-1's distribution is both dependent on the presence of MTs and dynamic during the cell cycle, appearing as numerous foci localized near the centrosomal region during interphase that become dispersed during late G2 throughout M phase (Balczon et al., 1994; Kubo et al., 1999). We do not know yet if PCM-1 colocalizes with MLK3 because of a lack of compatible antibody reagents to allow costaining. PCM-1 has been suggested to be a scaffold protein (Balczon et al., 1994; Kubo et al., 1999), raising the possibility that MLK3 may be part of a motor-containing signaling module nucleated by PCM-1 or a similar scaffold protein.
There is precedent for MLK3's involvement in MT-associated signaling modules. MLK3 is one of the kinases found to be associated with the JIPs (JNK interacting proteins), which are putative scaffolds that assemble the JNK signaling modules (Whitmarsh et al., 1998; Yasuda et al., 1999; Davis, 2000; Kelkar et al., 2000; for review, see Davis, 2000). These modules are thought to restrict JNK activity to particular regions of the cell (Whitmarsh and Davis, 1998). Recently, the JIPs have been recognized as cargo for conventional kinesin (Bowman et al., 2000; Verhey et al., 2001), which moves on MTs toward the plus ends. Interaction of scaffolds with motor proteins provides the means for dynamic spatial regulation of signaling pathways, an attractive mechanism to explain the substrate discrimination observed when the same signaling intermediates transfer signals to distinct sets of substrates depending on the type or context of their stimulation.
Intriguingly, evidence supporting a model of MLK family member activation in which JIP tethers the MLK kinase in a monomeric, unphosphorylated state until its dissociation, which allows leucine-zipper–dependent dimerization, phosphorylation, and activation has recently been published (Nihalani et al., 2001). The data described in the present study are consistent with a similar mechanism whereby MLK3 is bound to a JIP-like scaffold protein during interphase that tethers it to MTs in the region of the centrosome and facilitates its release during prophase, at which time it is hyperphosphorylated, catalytically stimulated, and presumably active toward proximal targets.
Although phosphorylation appears to be required for basal activity of MLK3 (Leung and Lassam, 2001) and a role for further phosphorylation in the stimulation of activity that occurs at G2/M cannot be ruled out, the MLK3 hyperphosphorylation, which occurs at this stage to produce an upshifted protein on SDS-PAGE, is not required for this increased activity. Such electrophoretic shifts due to hyperphosphorylation with no coincident change in activity have been seen as well for the NIMA-like kinase Nercc1/Nek8 (Roig et al., 2002). Rather than activity enhancement, the hyperphosphorylation of MLK3 at G2/M may be important instead for its dispersal from the centrosome-proximal microtubules, which occurs at this time.
In addition to the distinct, MT-associated and centrosome-proximal sites of localization, much of the MLK3 protein is diffusely cytoplasmic. A similar cytoplasmic distribution has been seen for the “core” centrosome proteins γ-tubulin, centrin, and Nek2, only a fraction (10–20%) of which are tightly associated with the centrosome (Moudjou et al., 1996; Paoletti et al., 1996; Fry et al., 1998). Although it is possible that these proteins each have cytoplasmic functions distinct from their roles at the centrosome, none have been uncovered thus far. Analogously, we expect that it is the MT-associated and centrosome-proximal population of MLK3, perhaps localized by association with a JIP-like scaffold protein, that becomes stimulated during G2/M and thus, is able to act on proximal substrates effecting MT dynamics. The modest fold-change in kinase activity at G2/M measured for bulk MLK3 protein may reflect this localization-specific activation.
Potential Cell Cycle Targets of MLK3
Our study clearly establishes that MLK3 is activated during G2/M but that the substrate is unlikely to lie within the JNK pathway. Although cell cycle activation of JNK has been noted in other studies, the timing and duration of this activation is not coincident with that of MLK3 stimulation. In one study, a transient increase in total JNK activity at the start of S phase was reported (Patel et al., 1998). Recently, in a second study, a fraction of JNK was found to localize to the centrosomes and, using phospho-specific anti-JNK antibodies that recognize only the active form, this centrosome-associated fraction was found to be activated from the beginning of S phase through anaphase (MacCorkle-Chosnek et al., 2001). Although we did not assay JNK activity immediately after release from the G1 block so would have missed a transient activation, we did not detect any increase in total JNK activity throughout the rest of the cell cycle including G2/M when the total activity of MLK3 was stimulated. Although it is possible that a low, undetectable level of JNK was stimulated during this time, it seems most probable to us that this pathway was not activated in response to the MLK3 stimulation.
What then might the ultimate target of MLK3 activation during the cell cycle be? As cells enter prometaphase, the time of MLK3 activation and dissemination, there is a sudden loss of the cytoplasmic microtubule network; an abrupt increase in MT instability, known to be promoted by phosphorylation events, results in a conversion from the long MTs of interphase to many short MTs radiating from the centrosome (Belmont et al., 1990; Zhai and Borisy, 1994; Zhai et al., 1996). Then, as the spindle forms, the MTs within the central spindle are stabilized, whereas cytoplasmic and astral MTs continue to be unstable. These changes in MT dynamics during the cell cycle are affected by changes in balance between MT-stabilizing and -destabilizing activities (for recent reviews, see Cassimeris, 1999; Andersen, 2000). A number of MT-associated proteins (MAPs) stabilize MTs by distinct mechanisms, but commonly they are negatively regulated by cell cycle–dependent phosphorylation, which occurs during M phase. In contrast, the destabilizing accessory proteins that have been identified act constitutively, for the most part, throughout the cell cycle. Thus, the MT remodeling that occurs during M phase has been attributed to reduced activity (and increased phosphorylation) of stabilizing MAPS. It is possible that MLK3 phosphorylates and thereby inhibits a MAP or MAPs at G2/M. When we used MAP-rich tubulin preparations as substrates in assays of MLK3 kinase activity, no obvious candidate targets emerged (our unpublished data), but the relevant target may not be detectable or present in these fractions.
A reduction in the length of astral microtubules comparable to the phenotype seen after ectopic expression of MLK3 has been described for Drosophila mutants of the mitotic kinase, Aurora A (Giet et al., 2002). Astral MT dynamics are also known to be influenced by the centrosomal protein and Aurora A substrate, DTACC (Gergely et al., 2000; Giet et al., 2002) and the DTACC-associated MAP, the MSPS/XMAP215 protein (Cullen et al., 1999). The equivalent of these proteins in human cells may be candidate substrates or be indirectly influenced by activated MLK3 during M phase.
Other possible targets for MLK3 could be as yet unidentified MT-destabilizing proteins that, unlike those described, are activated during M phase. One of the known MT-destabilizing proteins is XKCM1, a member of the kinesin superfamily of MT motors (Walczak et al., 1996), the destabilizing activity of which is counteracted by XMAP215 (Vasquez et al., 1994; Tournebize et al., 2000). It is noteworthy that another kinesin family member, KIF3X, has been shown to interact with the C-terminal region of MLK3 in a yeast two-hybrid assay (Nagata et al., 1998). An association of MLK3 and KIF3X in vivo has not been demonstrated, and neither the localization nor functional properties of KIF3X are known. Nevertheless, the possibility that MLK3 affects MT motor protein function is intriguing. Perhaps the dramatic distortion of interphase nuclei also seen in cells overexpressing MLK3 (or NIMA) is the result of dis-regulated motor activity along nuclear-envelope–tethered MTs.
MLK3 is present in a wide spectrum of tissues and cell types including those that are nondividing. It is possible that this kinase affects MT organization during remodeling of the cytoskeleton in all cells. In dividing cells this would occur during M phase, and in nondividing cells it would take place during processes such as outgrowth and migration. Whether all of the diverse roles of MLK3 are enacted in all the cell types where it is present or a subset of these roles are carried out only in specific cell types has not been examined.
The novel and unexpected findings of our study are that the JNK pathway kinase, MLK3 shares a number of molecular and functional similarities with the fungal kinase NIMA and, as is true for NIMA, has a cell cycle role during G2/M. This role is independent of JNK activation and appears likely to be involved in the microtubule alterations that are a hallmark of M phase entry.
ACKNOWLEDGMENTS
We are especially indebted to our colleague Sally Kornbluth for many enthusiastic and insightful discussions. We thank her and Danny Lew for critical reading of the manuscript. We are grateful to Christopher Holley, James Joseph, Patrick Fields, and Timothy Fields for helpful discussions and suggestions. For excellent technical assistance, we thank Charles Mena and especially Elizabeth MacDougall. This work was supported by National Institutes of Health grant CA-82845, DOD grant DAMD17–98-1–8077, and North Carolina Biotechnology Center grant 9905-ARG-0028.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–02–0115. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–02–0115.
REFERENCES
- Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. doi: 10.1016/s0962-8924(00)01786-4. [DOI] [PubMed] [Google Scholar]
- Arama E, Yanai A, Kilfin G, Bernstein A, Motro B. Murine NIMA-related kinases are expressed in patterns suggesting distinct functions in gametogenesis and a role in the nervous system. Oncogene. 1998;16:1813–1823. doi: 10.1038/sj.onc.1201710. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AB, Davies CJ, Hutchison CA, Kaback DB. Cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the FUN52 gene, whose product has homology to protein kinases. Gene. 1992;117:137–140. doi: 10.1016/0378-1119(92)90502-g. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen LG, Upshall A, Morris NR. S-phase, G2, and nuclear division mutants of Aspergillus nidulans. J Bacteriol. 1984;159:114–119. doi: 10.1128/jb.159.1.114-119.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock BC, Vacratsis PO, Qamirani E, Gallo KA. Cdc42-induced activation of the mixed-lineage kinase SPRK in vivo. Requirement of the Cdc42/Rac interactive binding motif and changes in phosphorylation. J Biol Chem. 2000;275:14231–14241. doi: 10.1074/jbc.275.19.14231. [DOI] [PubMed] [Google Scholar]
- Bowman AB, Kamal A, Ritchings BW, Philp AV, McGrail M, Gindhart JG, Goldstein LS. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- Cance WG, Craven RJ, Weiner TM, Liu ET. Novel protein kinases expressed in human breast cancer. Int J Cancer. 1993;54:571–577. doi: 10.1002/ijc.2910540409. [DOI] [PubMed] [Google Scholar]
- Cassimeris L. Accessory protein regulation of microtubule dynamics throughout the cell cycle. Curr Opin Cell Biol. 1999;11:134–141. doi: 10.1016/s0955-0674(99)80017-9. [DOI] [PubMed] [Google Scholar]
- Chen A, Yanai A, Arama E, Kilfin G, Motro B. NIMA-related kinases: isolation and characterization of murine nek3 and nek4 cDNAs, and chromosomal localization of nek1, nek2 and nek3. Gene. 1999;234:127–137. doi: 10.1016/s0378-1119(99)00165-1. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A, Sugden PH. The p38-MAPK inhibitor, SB203580, inhibits cardiac stress-activated protein kinases/c-Jun N-terminal kinases (SAPKs/JNKs) FEBS Lett. 1998;426:93–96. doi: 10.1016/s0014-5793(98)00324-x. [DOI] [PubMed] [Google Scholar]
- Crenshaw DG, Yang J, Means AR, Kornbluth S. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 1998;17:1315–1327. doi: 10.1093/emboj/17.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell. 2000;102:293–302. doi: 10.1016/s0092-8674(00)00035-0. [DOI] [PubMed] [Google Scholar]
- DeBrabander M, Geuens G, Nuydens R, Willebrords R, Aerts F, DeMey J, McIntosh JR. Microtubule dynamics during the cell cycle: the effects of taxol and nocodazole on the microtubule system of PtK2 cells at different stages of the mitotic cycle. Int Rev Cytol. 1986;101:215–275. doi: 10.1016/s0074-7696(08)60250-8. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Ezoe K, Lee ST, Strunk KM, Spritz RA. PTK1, a novel protein kinase required for proliferation of human melanocytes. Oncogene. 1994;9:935–938. [PubMed] [Google Scholar]
- Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Jr, Parsons M. A Trypanosoma brucei gene family encoding protein kinases with catalytic domains structurally related to Nek1 and NIMA. Mol Biochem Parasitol. 1993;59:111–121. doi: 10.1016/0166-6851(93)90012-m. [DOI] [PubMed] [Google Scholar]
- Gallo KA, Mark MR, Scadden DT, Wang Z, Gu Q, Godowski PJ. Identification and characterization of SPRK, a novel src-homology 3 domain-containing proline-rich kinase with serine/threonine kinase activity. J Biol Chem. 1994;269:15092–15100. [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkamp J, Troppmair J, Rapp UR. The JNK/SAPK activator mixed lineage kinase 3 (MLK3) transforms NIH 3T3 cells in a MEK-dependent fashion. Cancer Res. 1999;59:2195–2202. [PubMed] [Google Scholar]
- Hehner SP, Hofmann TG, Ushmorov A, Dienz O, Wing-Lan Leung I, Lassam N, Scheidereit C, Droge W, Schmitz ML. Mixed-lineage kinase 3 delivers CD3/CD28-derived signals into the IkappaB kinase complex. Mol Cell Biol. 2000;20:2556–2568. doi: 10.1128/mcb.20.7.2556-2568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PM, Milne A, Garka K, Johnson RS, Willis C, Sims JE, Rauch CT, Bird TA, Virca GD. Purification, cloning, and characterization of Nek8, a novel NIMA- related kinase, and its candidate substrate Bicd2. J Biol Chem. 2002;277:16229–16240. doi: 10.1074/jbc.M108662200. [DOI] [PubMed] [Google Scholar]
- Ing YL, Leung IW, Heng HH, Tsui LC, Lassam NJ. MLK-3: identification of a widely-expressed protein kinase bearing an SH3 domain and a leucine zipper-basic region domain. Oncogene. 1994;9:1745–1750. [PubMed] [Google Scholar]
- Jha MN, Bamburg JR, Bedford JS. Cell cycle arrest by Colcemid differs in human normal and tumor cells. Cancer Res. 1994;54:5011–5015. [PubMed] [Google Scholar]
- Jones DG, Rosamond J. Isolation of a novel protein kinase-encoding gene from yeast by oligodeoxyribonucleotide probing. Gene. 1990;90:87–92. doi: 10.1016/0378-1119(90)90442-t. [DOI] [PubMed] [Google Scholar]
- Kandli M, Feige E, Chen A, Kilfin G, Motro B. Isolation and characterization of two evolutionarily conserved murine kinases (Nek6 and nek7) related to the fungal mitotic regulator, NIMA. Genomics. 2000;68:187–196. doi: 10.1006/geno.2000.6293. [DOI] [PubMed] [Google Scholar]
- Kelkar N, Gupta S, Dickens M, Davis RJ. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol Cell Biol. 2000;20:1030–1043. doi: 10.1128/mcb.20.3.1030-1043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Tibbles LA, Anafi M, Janssen A, Zanke BW, Lassam N, Pawson T, Woodgett JR, Iscove NN. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 1996;15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- Krien MJ, Bugg SJ, Palatsides M, Asouline G, Morimyo M, O'Connell MJ. A NIMA homologue promotes chromatin condensation in fission yeast. J Cell Sci. 1998;111:967–976. doi: 10.1242/jcs.111.7.967. [DOI] [PubMed] [Google Scholar]
- Krien MJ, West RR, John UP, Koniaras K, McIntosh JR, O'Connell MJ. The fission yeast NIMA kinase Fin1p is required for spindle function and nuclear envelope integrity. EMBO J. 2002;21:1713–1722. doi: 10.1093/emboj/21.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JM, Karnoub AE, Graves LM, Campbell SL, Der CJ. Role of MLK3-mediated Activation of p70 S6 Kinase in Rac1 Transformation. J Biol Chem. 2002;277:4770–7. doi: 10.1074/jbc.M109379200. [DOI] [PubMed] [Google Scholar]
- Letwin K, Mizzen L, Motro B, Ben-David Y, Bernstein A, Pawson T. A mammalian dual specificity protein kinase, Nek1, is related to the NIMA cell cycle regulator and highly expressed in meiotic germ cells. EMBO J. 1992;11:3521–3531. doi: 10.1002/j.1460-2075.1992.tb05435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung IW, Lassam N. Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. J Biol Chem. 1998;273:32408–32415. doi: 10.1074/jbc.273.49.32408. [DOI] [PubMed] [Google Scholar]
- Leung IW, Lassam N. The kinase activation loop is the key to mixed lineage kinase-3 activation via both autophosphorylation and hematopoietic progenitor kinase 1 phosphorylation. J Biol Chem. 2001;276:1961–1967. doi: 10.1074/jbc.M004092200. [DOI] [PubMed] [Google Scholar]
- Levedakou EN, et al. Two novel human serine/threonine kinases with homologies to the cell cycle regulating Xenopus MO15, and NIMA kinases: cloning and characterization of their expression pattern. Oncogene. 1994;9:1977–1988. [PubMed] [Google Scholar]
- Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hunter T. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell. 1995;81:413–424. doi: 10.1016/0092-8674(95)90394-1. [DOI] [PubMed] [Google Scholar]
- Lu KP, Means AR. Expression of the noncatalytic domain of the NIMA kinase causes a G2 arrest in Aspergillus nidulans. EMBO J. 1994;13:2103–2113. doi: 10.1002/j.1460-2075.1994.tb06486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- MacCorkle-Chosnek RA, VanHooser A, Goodrich DW, Brinkley BR, Tan TH. Cell cycle regulation of c-Jun N-terminal kinase activity at the centrosomes. Biochem Biophys Res Commun. 2001;289:173–180. doi: 10.1006/bbrc.2001.5948. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Montpetit B, Zhao L, Finst RJ, Goh B, Kim AC, Quarmby LM. The FA2 gene of Chlamydomonas encodes a NIMA family kinase with roles in cell cycle progression and microtubule severing during deflagellation. J Cell Sci. 2002;115:1759–1768. doi: 10.1242/jcs.115.8.1759. [DOI] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Nagata K, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihalani D, Meyer D, Pajni S, Holzman LB. Mixed lineage kinase-dependent JNK activation is governed by interactions of scaffold protein JIP with MAPK module components. EMBO J. 2001;20:3447–3458. doi: 10.1093/emboj/20.13.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- O'Connell MJ, Norbury C, Nurse P. Premature chromatin condensation upon accumulation of NIMA. EMBO J. 1994;13:4926–4937. doi: 10.1002/j.1460-2075.1994.tb06820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J Cell Biol. 1983;96:1155–1158. doi: 10.1083/jcb.96.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, McGuire SL, Osmani SA. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell. 1991;67:283–291. doi: 10.1016/0092-8674(91)90180-7. [DOI] [PubMed] [Google Scholar]
- Osmani SA, May GS, Morris NR. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol. 1987;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Pu RT, Morris NR. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Patel R, Bartosch B, Blank JL. p21WAF1 is dynamically associated with JNK in human T-lymphocytes during cell cycle progression. J Cell Sci. 1998;111:2247–55. doi: 10.1242/jcs.111.15.2247. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu RT, Osmani SA. Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J. 1995;14:995–1003. doi: 10.1002/j.1460-2075.1995.tb07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu RT, Xu G, Wu L, Vierula J, O'Donnell K, Ye XS, Osmani SA. Isolation of a functional homolog of the cell cycle-specific NIMA protein kinase of Aspergillus nidulans and functional analysis of conserved residues. J Biol Chem. 1995;270:18110–18116. doi: 10.1074/jbc.270.30.18110. [DOI] [PubMed] [Google Scholar]
- Rana A, Gallo K, Godowski P, Hirai S, Ohno S, Zon L, Kyriakis JM, Avruch J. The mixed lineage kinase SPRK phosphorylates and activates the stress- activated protein kinase activator, SEK-1. J Biol Chem. 1996;271:19025–19028. doi: 10.1074/jbc.271.32.19025. [DOI] [PubMed] [Google Scholar]
- Rhee K, Wolgemuth DJ. The NIMA-related kinase 2, Nek2, is expressed in specific stages of the meiotic cell cycle and associates with meiotic chromosomes. Development. 1997;124:2167–2166. doi: 10.1242/dev.124.11.2167. [DOI] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Roig J, Mikhailov A, Belham C, Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SJ, Fry AM, Sutterlin C, Ried T, Nigg EA. Cell cycle-dependent expression of Nek2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 1994;5:625–635. [PubMed] [Google Scholar]
- Schultz SJ, Nigg EA. Identification of 21 novel human protein kinases, including 3 members of a family related to the cell cycle regulator nimA of Aspergillus nidulans. Cell Growth Differ. 1993;4:821–930. [PubMed] [Google Scholar]
- Schweitzer B, Philippsen P. NPK1, a nonessential protein kinase gene in Saccharomyces cerevisiae with similarity to Aspergillus nidulans nimA. Mol Gen Genet. 1992;234:164–167. doi: 10.1007/BF00272358. [DOI] [PubMed] [Google Scholar]
- Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nigg EA. Cloning and characterization of the murine Nek3 protein kinase, a novel member of the NIMA family of putative cell cycle regulators. J Biol Chem. 1999;274:13491–13497. doi: 10.1074/jbc.274.19.13491. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Coso OA, Miyata H, Igishi T, Miki T, Gutkind JS. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c- Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J Biol Chem. 1996;271:27225–8. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]
- Tibbles LA, Ing YL, Kiefer F, Chan J, Iscove N, Woodgett JR, Lassam NJ. MLK-3 activates the SAPK/JNK and p38/RK pathways via SEK1 and MKK3/6. EMBO J. 1996;15:7026–7035. [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Vacratsis PO, Gallo KA. Zipper-mediated oligomerization of the mixed lineage kinase SPRK/MLK-3 is not required for its activation by the GTPase cdc 42 but is necessary for its activation of the JNK pathway. Monomeric SPRK L410P does not catalyze the activating phosphorylation of Thr258 of murine mitogen-activated protein kinase kinase 4. J Biol Chem. 2000;275:27893–27900. doi: 10.1074/jbc.M002858200. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wang TH, Wang HS, Soong YK. Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer. 2000;88:2619–2628. doi: 10.1002/1097-0142(20000601)88:11<2619::aid-cncr26>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–4. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- Winkler KE, Swenson KI, Kornbluth S, Means AR. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science. 2000;287:1644–1647. doi: 10.1126/science.287.5458.1644. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Whitmarsh AJ, Cavanagh J, Sharma M, Davis RJ. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XS, Xu G, Pu RT, Fincher RR, McGuire SL, Osmani AH, Osmani SA. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO Journal. 1995;14:986–994. doi: 10.1002/j.1460-2075.1995.tb07079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Borisy GG. Quantitative determination of the proportion of microtubule polymer present during the mitosis-interphase transition. J Cell Sci. 1994;107:881–890. doi: 10.1242/jcs.107.4.881. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gallo KA. Autoinhibition of mixed lineage kinase 3 through its Src homology 3 domain. J Biol Chem. 2001;276:45598–45603. doi: 10.1074/jbc.M107176200. [DOI] [PubMed] [Google Scholar]