Abstract
The multisubunit conserved oligomeric Golgi (COG) complex has been shown previously to be involved in Golgi function in yeast and mammalian tissue culture cells. Despite this broad conservation, several subunits, including Cog5, were not essential for growth and showed only mild effects on secretion when mutated in yeast, raising questions about what functions these COG complex subunits play in the life of the cell. Here, we show that function of the gene four way stop (fws), which encodes the Drosophila Cog5 homologue, is necessary for dramatic changes in cellular and subcellular morphology during spermatogenesis. Loss-of-function mutations in fws caused failure of cleavage furrow ingression in dividing spermatocytes and failure of cell elongation in differentiating spermatids and disrupted the formation and/or stability of the Golgi-based spermatid acroblast. Consistent with the lack of a growth defect in yeast lacking Cog5, animals lacking fws function were viable, although males were sterile. Fws protein localized to Golgi structures throughout spermatogenesis. We propose that Fws may directly or indirectly facilitate efficient vesicle traffic through the Golgi to support rapid and extensive increases in cell surface area during spermatocyte cytokinesis and polarized elongation of differentiating spermatids. Our study suggests that Drosophila spermatogenesis can be an effective sensitized genetic system to uncover in vivo functions for proteins involved in Golgi architecture and/or vesicle transport.
INTRODUCTION
Vesicular transport of proteins and lipids throughout the cell requires that vesicles be able to recognize and fuse to specific membrane compartments. Several large protein complexes have been proposed to help target vesicles to a particular membrane domain. The exocyst (sec6/8 complex), composed of eight proteins, is thought to help recruit Golgi-derived vesicles to the plasma membrane at the final step of the secretory pathway (Hsu et al., 1996; TerBush et al., 1996; Kee et al., 1997). The multisubunit TRAPP I and Vps52/53/54 complexes are believed to serve analogous functions during ER-to-Golgi and endosome-to-Golgi trafficking, respectively (Conibear and Stevens, 2000; Sacher et al., 2001). The eight-component mammalian conserved oligomeric Golgi (COG) complex and the homologous Sec34/35 complex in yeast have been proposed to assist in Golgi vesicle targeting (Cao et al., 1998; VanRheenen et al., 1998; Walter et al., 1998; VanRheenen et al., 1999; Sacher et al., 2001; Whyte and Munro, 2001; Ungar et al., 2002). The COG complex can stimulate intra-Golgi trafficking in vitro (Walter et al., 1998), and subunits of the Sec34/35 complex have been shown to be required for ER-to-Golgi trafficking, retrograde transport through the Golgi, and trafficking between the Golgi and endosomes (Spelbrink and Nothwehr, 1999; VanRheenen et al., 1999; Whyte and Munro, 2001). However, a direct role of the COG complex in vesicle trafficking has not yet been demonstrated. Indeed, Chinese hamster ovary (CHO) cells mutant for either the Cog1 or Cog2 subunits did not show profound defects in membrane protein transport through the secretory pathway but did cause pleiotropic defects in membrane protein glycosylation (Kingsley et al., 1986; Reddy and Krieger, 1989). One possibility is that the COG complex function facilitates retrograde transport through the Golgi to maintain proper compartmentalization of enzymes that mediate a number of Golgi-based functions, including glycosylation and efficient anterograde traffic.
Recent studies in yeast and mammalian cells suggest that the COG complex (previously called the Sec34/35 complex in yeast) may be composed of two classes of subunits (Whyte and Munro, 2001; Ram et al., 2002; Ungar et al., 2002), initially defined and distinguished in Saccharomyces cerevisiae by the degree to which they were essential for viability and subcellular morphology. Deletions of any of the yeast SEC34, SEC35, SEC36, or SEC38 genes caused severe growth defects, and mutations in at least two of these (SEC35 and SEC36) caused visible defects in internal membrane organization (Whyte and Munro, 2001). In contrast, deletions of genes encoding the yeast homologues of Cog5, Cog6, Cog8, or one other yeast COG complex subunit did not cause strong defects in growth or internal membrane organization (Whyte and Munro, 2001; Ram et al., 2002). Subsequent studies showed that mutations in two of the growth-essential subunits disrupted the yeast Sec34/35 complex, whereas mutation of any one of the four subunits not essential for growth only mildly affected migration of the complex in a gel filtration column, suggesting that the functional differences between the subunit classes may reflect their physical arrangement (Ram et al., 2002). The growth-essential subunits were proposed to form a core complex to which the nonessential subunits were peripherally associated (Ram et al., 2002). Intriguingly, the mammalian COG complex appeared as a bilobed structure when visualized by deep-etch electron microscopy (EM), raising the possibility that the two classes of subunits may be physically separated in distinct lobes of the COG complex (Ungar et al., 2002).
Although the yeast homologues of Cog5, Cog6, and Cog8 and the proposed functional counterpart of Cog7 in yeast (Ungar et al., 2002) were not essential for viability, deletions of any one of them seemed to disrupt normal Golgi function. The yeast mutants displayed defects in v-SNARE recycling and some reduction in glycosylation and secretion, suggesting that the genes are required for efficient intra-Golgi traffic (Ram et al., 2002). However, the roles of these proteins and their mammalian homologues at the cellular level have not been determined. In mammalian cells, loss of function of either Cog1 or Cog2 (proposed functional counterparts of yeast SEC 36 and SEC35) disrupted Golgi morphology and several aspects of protein glycosylation but did not block traffic of membrane proteins to the cell surface (Kingsley et al., 1986; Reddy and Krieger, 1989; Ungar et al., 2002). Surprisingly, CHO cells mutant for either Cog1 or Cog2 were viable, unlike yeast cells mutant for SEC36 or SEC35. To better understand the function of the two types of COG subunits in vivo, it may be necessary to examine requirements for their function in other cell types with different demands for Golgi function, as can be found in a developing multicellular organism.
Here, we demonstrate that four way stop (fws), a gene essential for spermatogenesis, encodes the Drosophila homologue of the Cog5 protein. Functional Fws was required for cleavage furrow ingression during cytokinesis in dividing spermatocytes and for the extensive polarized cell growth that accompanies spermatid elongation. The Fws protein localized to Golgi membrane throughout spermatogenesis and mutations in the fws gene disrupted the architecture of the Golgi-derived spermatid acroblast. The subunits of Drosophila COG, including Fws, may facilitate efficient vesicle trafficking through the Golgi, possibly by mediating proper retrograde movement of Golgi enzymes between Golgi stacks to maintain the correct molecular composition of the different layers. Efficient Golgi vesicle trafficking and/or Golgi function may in turn be required for rapid and extensive increase in cell-surface area during spermatocyte cytokinesis and spermatid elongation. Our results provide the first evidence that the COG complex facilitates dramatic changes in cell shape and introduce Drosophila spermatogenesis as an effective sensitized genetic system to uncover in vivo requirements for proteins involved in Golgi function and/or membrane vesicle transport.
MATERIALS AND METHODS
Fly Strains and Husbandry
Flies were raised on standard cornmeal molasses agar at 25°C (Ashburner, 1990). Visible markers and balancer chromosomes are described in FlyBase except where otherwise noted. Oregon-R was used as the wild-type strain. fwsz-0161 and fwsz-1201 were provided by C. Zuker from his collection of ethyl methane sulfonate (EMS)-generated, viable lines. The viable Zuker lines were screened for male sterility by B. Wakimoto and D. Lindsley. The fws alleles were identified in a screen of the male steriles for mutations that disrupt cytokinesis conducted by examining squashed testes by phase-contrast microscopy. Details of the cytokinesis screen will be described elsewhere (Giansanti et al., in preparation). Stocks carrying Df(2L)qua1374 were provided by L. Cooley. Df(2L)71E was generated by the imprecise excision (Zhang and Spradling, 1993) of the P-element associated with dl1313 (gift of S. Govind). Transgenic flies carrying GFP-fws were generated by inserting GFP with BstEII linkers into a BstEII site created upstream of the start codon of genomic fws (introduced with QuikChange XL Site-Directed Mutagenesis kit from Stratagene, La Jolla, CA), cloning the tagged gene as a 5.7-kilobase (kb) HindIII-SalI fragment into pCaSpeR 4, and introducing the construct into flies by P-element–mediated germ-line transformation (Rubin and Spradling, 1982, 1983). Female fertility tests were conducted by crossing fwsz-0161/fwsz-1201 females to fwsz-0161/Cyo or fwsz-1201/Cyo males, collecting at least 1000 eggs, and counting the number that had hatched after 48 h at 25°C.
Microscopy and Immunofluorescence
Live testes were squashed and examined by phase-contrast light microscopy as described by Regan and Fuller (1990). In live squashes, DNA was visualized with 10 μg/ml Hoechst 33342 dye. To visualize α-tubulin with F-actin, GFP-Fws with Lava Lamp, GFP-Fws with α-mannosidase, or endogenous Fws, cells were squashed and then fixed with 4% formaldehyde as described by Gunsalus et al. (1995). In all cases, GFP-Fws was visualized directly by exciting the green fluorescent protein (GFP) moiety, even in fixed material. To visualize anillin with γ-tubulin, Lava Lamp alone, or α-mannosidase alone, cells were fixed with 3.7% formaldehyde and acetic acid before squashing as described by Giansanti et al. (1999). F-actin was stained with rhodamine-labeled phalloidin (Molecular Probes, Eugene, OR) diluted 1:10. Monoclonal antibodies were used to stain α-tubulin (Amersham Biosciences, Arlington Heights, IL; diluted 1:50) and γ-tubulin (Sigma, St. Louis, MO; clone GTU-88, diluted 1:100). Polyclonal antibodies were used to stain anillin (gift of C. Fields; diluted 1:20), α-mannosidase II (gift of D. Roberts; diluted 1:1000), Lava Lamp (gift of J. Sisson and B. Sullivan; diluted 1:50), and Fws (diluted 1:2000). Secondary antibodies [fluorescein-conjugated sheep anti-mouse F(ab′)2 and rhodamine-conjugated goat anti-rabbit, Boehringer Mannheim, Indianapolis, IN] were used at a dilution of 1:50 or 1:100. Samples were mounted in Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). Images were captured using a Photometrics cooled CCD camera connected to a Zeiss Axiophot microscope. Gray-scale digital images were collected separately using the IP Lab Spectrum software and then converted to Photoshop format, pseudocolored, and merged.
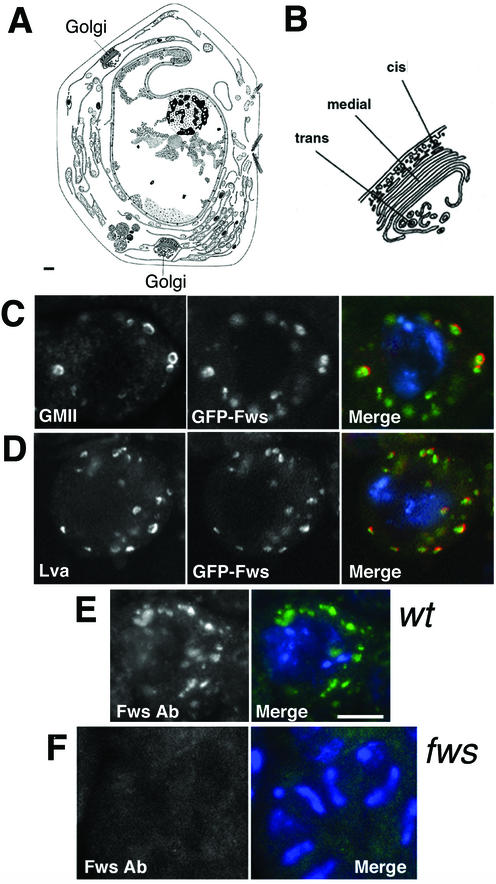
Fws antibodies were generated by injecting rabbits (at Covance Research Products, Inc.) with a gel-excised polypeptide containing the first 341 aa of Fws (expressed using the Invitrogen pBad/Topo Thiofusion system) (Invitrogen, San Diego, CA). Staining of Golgi structures in wild-type spermatocytes and spermatids was not observed in cells from fws mutant males. The antisera also stained the mitochondrial derivative in spermatids. However, because this staining still remained in fws mutant cells, the mitochondrial stain did not represent the fws protein.
Cloning of fws
The fws locus was mapped by deficiency complementation. The break points of Df(2L)71E were determined by recovering and sequencing the DNA flanking the remaining P-element. To identify testis transcripts, genomic DNA from the phage 17B5 that covered most of the region (gift of B. Suter) was used to generate probes for Northern blots containing testis RNA isolated with the Invitrogen MicroFastTrack 2.0 kit. A 7-kb SalI genomic rescue fragment containing CG6549 was cloned into pCaSpeR 4 and introduced into flies by P-element–mediated germ-line transformation. The premature stop codons in CG6549 in the fwsz-0161 and fwsz-1201 alleles were identified by amplifying the genomic region containing CG6549 by PCR and sequencing from the bulk PCR products pooled from several independent reactions. Homologues of fws were identified by tBLASTn sequence analysis and compared with the Clustal W and Boxshade programs.
RESULTS
Wild-Type Function of the four way stop Gene Is Required for Cleavage Furrow Ingression during Spermatocyte Cytokinesis and for Spermatid Elongation
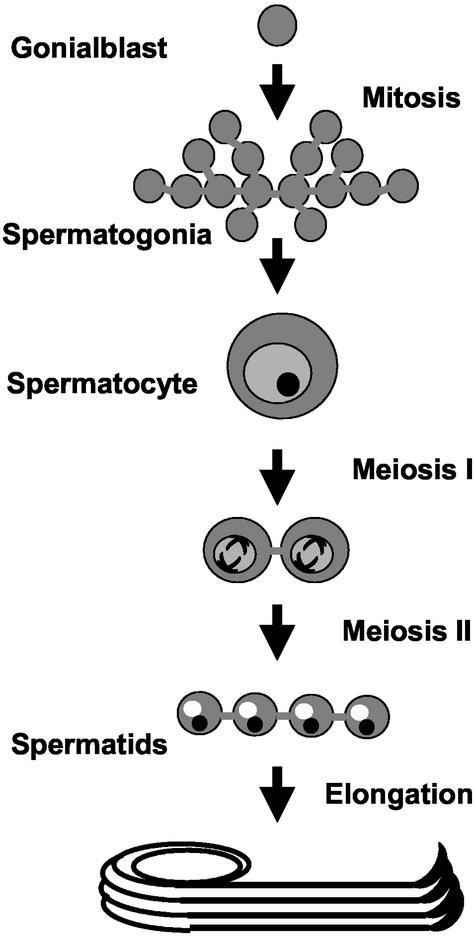
Two male sterile alleles of fws were identified in a screen for viable mutations that disrupt spermatogenesis (see MATERIALS AND METHODS). In wild-type Drosophila testes, cysts of 16 interconnected primary spermatocytes are produced from a single founding gonialblast cell by four rounds of mitosis. The 16 spermatocytes undergo two meiotic divisions in synchrony, creating a cyst of 64 haploid spermatids that subsequently elongate (Figure 1). In both gonial mitoses and meiotic divisions, cytokinesis is incomplete, and the daughter cells remain connected by ring canals (for review, see Fuller, 1993). The spermatids differentiate as interconnected cells within the cyst and become individualized at the final stages of spermatogenesis.
Figure 1.
Stages of spermatogenesis. A single gonialblast cell undergoes four rounds of mitosis to give rise to 16 spermatogonia, which then initiate differentiation by becoming spermatocytes. Each spermatocyte undergoes two meiotic divisions, giving rise to four round spermatids, which subsequently elongate.
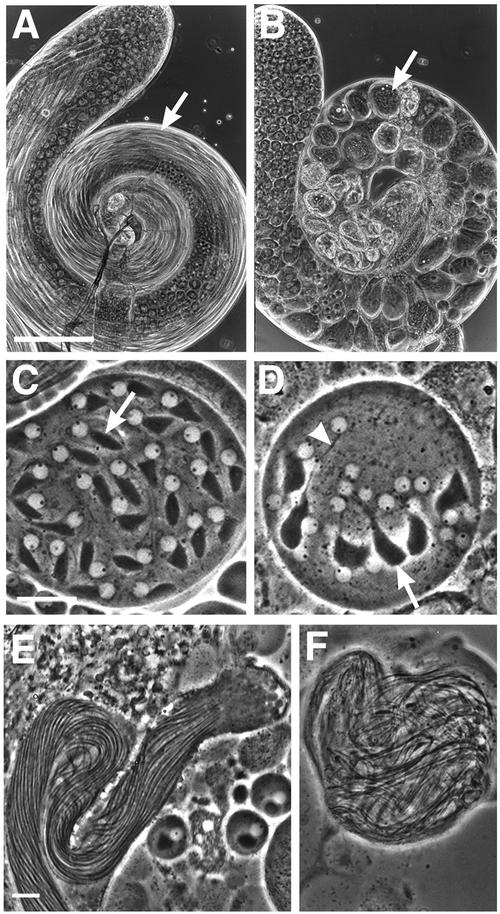
Early round spermatids in fws mutant males exhibited a multinucleate phenotype indicative of failure in spermatogonial and/or spermatocyte cytokinesis. Wild-type early round spermatids viewed by phase-contrast light microscopy (Figure 2A) have a single light nucleus (arrow) and a single dark mitochondrial derivative (arrowhead), which are approximately equal in size. Spermatids from fws mutants commonly had multiple nuclei of normal size associated with an abnormally large mitochondrial derivative (Figure 2, B and C), indicating normal chromosome segregation followed by failure of cytokinesis during meiosis (Lifschytz and Meyer, 1977). Many spermatids in fws mutant animals had two or four nuclei (Figure 2B), indicating failure of cytokinesis in one or two meiotic divisions, respectively. Some spermatids in fws mutant animals had more than four nuclei (Figure 2C), most likely caused by failure of cytokinesis during spermatogonial mitoses as well as during meiosis. Occasionally, spermatids with three nuclei were observed (Figure 2B), most likely caused by failure of cytokinesis during meiosis I, followed by the pinching off of only one daughter cell during meiosis II. The severity of defects in cytokinesis varied from cyst to cyst. For example, 100% of the spermatids counted in one fwsz-0161 mutant cyst had four or more nuclei, whereas in a different cyst from the same testis, only 29% of the spermatids contained four or more nuclei. Similar ranges of spermatid phenotypes were observed in animals homozygous for each of the two fws alleles and for various combinations of the alleles and chromosomal deficiencies that uncovered the fws locus (R.M. Farkas, unpublished observations).
Figure 2.
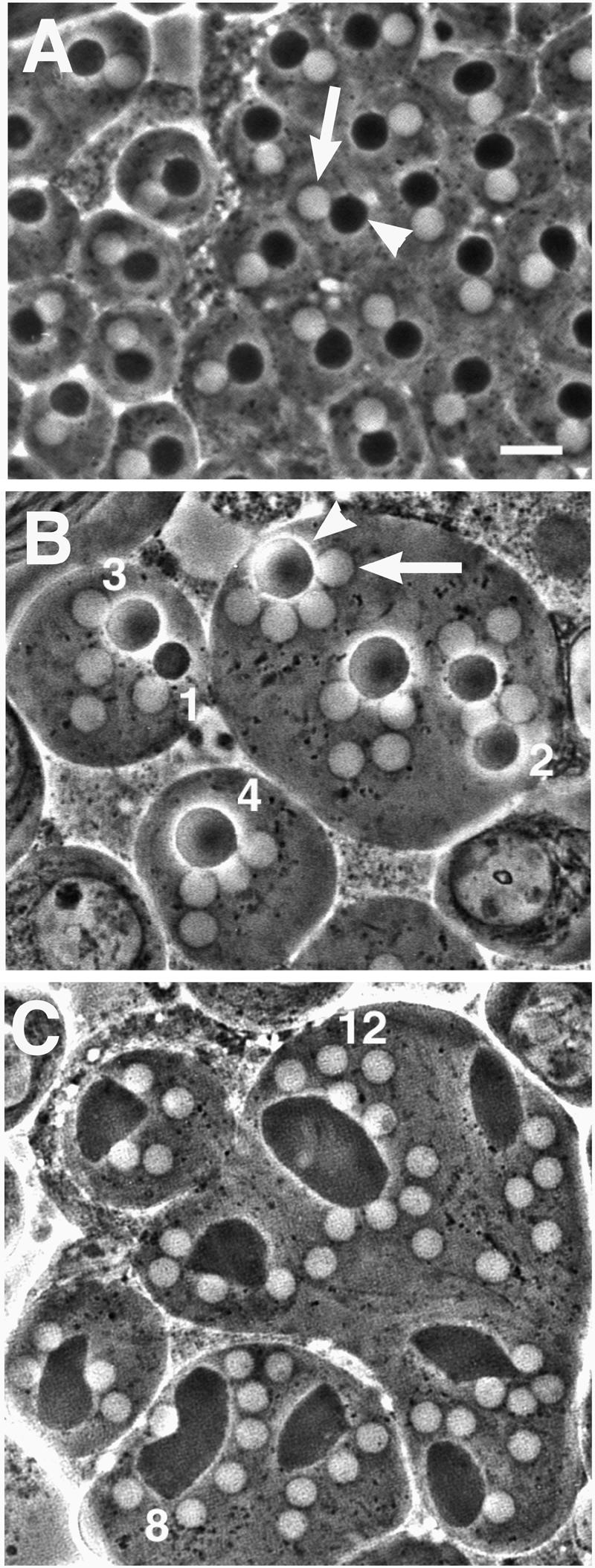
Abnormal spermatids in fws testes. Spermatids viewed by phase-contrast microscopy. (A) Wild-type spermatids contain a single light nucleus (arrow) and dark mitochondrial derivative (arrowhead) approximately equal in size. (B and C) Spermatids from fwsz-0161 mutants contain large mitochondrial derivatives (arrowhead) associated with multiple, normal-sized nuclei (arrow). The number of nuclei associated with select mitochondrial derivatives are noted. Bar, 10 μm.
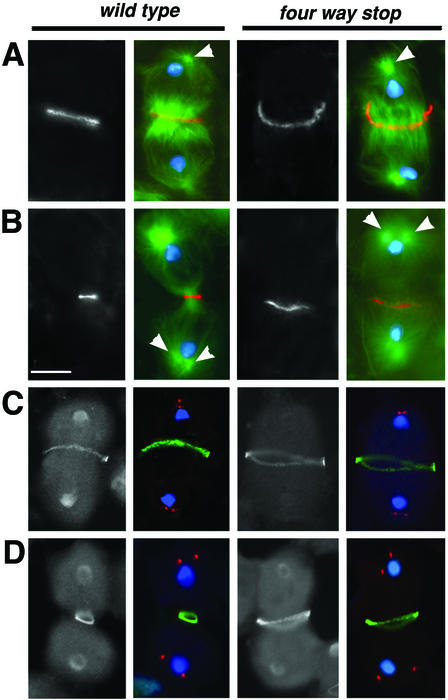
To pinpoint the specific stage of cytokinesis that requires fws function, we compared wild-type and fws- mutant spermatocytes at various stages of cytokinesis, using markers for F-actin and microtubules. Wild-type spermatocytes in early telophase I (Figure 3A) exhibit an equatorial F-actin ring, a single microtubule aster at each pole of the cell (arrowhead), and a central spindle, a robust array of microtubules with plus-ends that overlap near the center of the cell. As a wild-type spermatocyte progresses through telophase, the two centrosomes at each spindle pole begin to separate and rotate around the nuclei along with the microtubule asters they nucleate, the actin ring constricts, and the central spindle microtubules pinch together at the center of the cell (Figure 3B). Aster rotation can occur even in the absence of contractile ring and central spindle constriction and so can be used to distinguish early telophase from late telophase (R.M. Farkas and M.G. Giansanti, unpublished observations).
Figure 3.
Defects in furrow ingression in fws spermatocytes. (A and B) Spermatocytes in telophase stained with phalloidin to visualize F-actin (red), anti-α-tubulin antibodies to visualize microtubules (green), and DAPI to visualize the DNA (blue). Panels in black and white show F-actin staining only. fwsz-0161 cells shown in right panels. (A) Early telophase. (B) Mid to late telophase. Progression through telophase can be assessed by the positions of the microtubule asters (arrowheads). (C and D) Spermatocytes stained with DAPI to visualize DNA (blue), antibodies against γ-tubulin to mark the center of the asters (red), and antibodies against anillin (green) to mark the leading edge of the cleavage furrow. Panels in black and white show anillin staining only. fwsz-0161/Df(2L)qua1374 cells shown in right panels. (C) Late anaphase. (D) Mid to late telophase. Bar, 10 μm.
Spermatocytes mutant for fws appeared normal in early telophase but showed defects in contractile ring constriction in mid to late telophase. Ninety-five percent of fwsz-0161/fwsz-0161 (n = 39) and 100% of fwsz-0161/Df(2L)qua1374 (n = 44) mutant cells in early telophase exhibited a normal F-actin ring and central spindle (Figure 3A). By the time the microtubule asters had visibly separated (arrowheads), the F-actin ring appeared almost fully constricted in wild-type spermatocytes but was barely constricted and/or broken in 85% of the 34 fwsz-0161/fwsz-0161 mid-late telophase cells examined and in 92% of the 38 fwsz-0161/Df(2L)qua1374 cells examined (Figure 3B). The central spindle also appeared less dense in fws late telophase cells compared with wild type.
To confirm that the defects in actin ring constriction were accompanied by a failure in cleavage furrow ingression, we compared anillin localization in wild-type and fws-mutant cells. Anillin, an actin-binding protein that colocalizes with F-actin throughout telophase, has a pleckstrin-homology domain and is believed to associate closely with the plasma membrane as the cleavage furrow ingresses (Field and Alberts, 1995; Oegema et al., 2000). The anillin ring formed normally in fws anaphase spermatocytes (Figure 3C) but appeared barely constricted in all 22 fwsz-0161/Df(2L)qua1374 telophase cells examined (Figure 3D), suggesting that fws is required for plasma membrane ingression during cytokinesis.
Testes from fws mutant males also exhibited striking abnormalities in spermatid elongation. In wild-type testes, spermatids extend to a final length of 1.8 mm. Each cyst of 64 interconnected spermatids develops as a syncytium, which begins as a sphere and eventually elongates into a rope-like structure that extends through the entire length of the testis (Figure 4A, arrow). Within each spermatid, the microtubule-based axoneme increases in length, and the mitochondrial derivative (Figure 4C, arrows) extends along the flagellum. The axonemes and closely associated mitochondrial derivatives of 64 spermatids aligned in parallel gives the elongated cyst a striated appearance (Figure 4E).
Figure 4.
Spermatid elongation defects in fws testes, viewed by phase-contrast microscopy. (A) Wild-type testis filled with cysts of elongated spermatids (arrow). (B) fwsz-0161/Df(2L)qua1374 testis filled with abnormal ovoid cysts (arrow). (C) Wild-type spermatids in early stages of elongation exhibit an oblong mitochondrial derivative (arrow). (D) fwsz-0161 spermatids initiate elongation of the axoneme (arrowhead) and mitochondrial derivative (arrow). (E) Cyst of wild-type spermatids in advanced stages of elongation. The parallel elongated axonemes and mitochondrial derivatives give the cyst a striated appearance. (F) Cyst of fwsz-0161 spermatids. A and B, Bar, 100 μm; C–F, bar, 10 μm (comparable pictures of wild-type and fws are at the same magnification).
In fws mutant testes, spermatid cysts remained ovoid (Figure 4B, arrow). Although axonemes and mitochondrial derivatives initiated elongation (Figure 4D) and seemed to have elongated substantially in later-stage spermatid cysts (Figure 4F), each cyst as a whole did not elongate effectively. Axonemes and mitochondrial derivatives became so tangled in late-stage spermatid cysts that it was impossible to determine whether these subcellular structures achieved full length. The defect in spermatid elongation was unlikely to be a consequence of the failure of cytokinesis, because mutations in many other genes that disrupt cytokinesis in spermatocytes do not disrupt spermatid elongation (R.M. Farkas, unpublished observations).
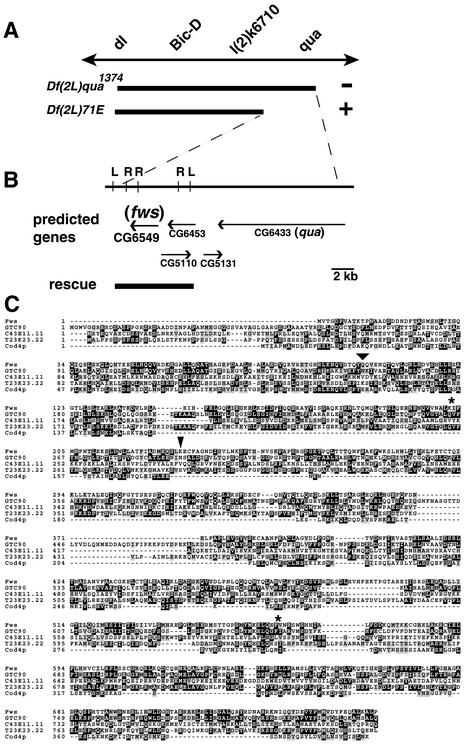
Mutations in the four way stop Locus Disrupt the Drosophila Homologue of Cog5
Mutations associated with two viable but male-sterile alleles of fws were localized to an 18-kb region on the basis of complementation tests with a series of chromosomal deletions (Figure 5A). A 7-kb SalI fragment of genomic DNA containing the entire reading frame of the predicted gene CG6549 (Figure 5B) rescued the cytokinesis and elongation phenotypes when crossed into a fws mutant background. Identification of CG6549 as fws was confirmed by sequence analysis of the mutant alleles (see below).
Figure 5.
fws encodes the Drosophila homologue of Cog5. (A) Genetic map of region containing fws. Bars show chromosomal deficiencies that complement (+) and fail to complement (−) the fws phenotypes. (B) Physical map defined by break points of the deficiencies, with restriction enzyme sites for SalI (L) and EcoRI (R). Predicted genes are based on data from the Berkeley Drosophila Genome Project website. (C) Sequence alignment of Drosophila melanogaster Fws (top line) with Homo sapiens GTC-90/Cog5 (second line), C. elegans predicted protein C43E11.11 (third line), Arabidopsis thaliana predicted protein T23K23.22 (fourth line), and S. cerevisiae Cod4p (bottom line). Amino acids that are identical in at least two species are shaded in black, and similar amino acids are shaded in gray. The positions of the stop codons in fwsz-0161 and fwsz-1201 are designated with a wide arrowhead and narrow arrowhead, respectively. Positions of introns are marked with asterisks.
The fws gene is predicted to encode a 751-aa protein with ∼25% identity and ∼50% similarity to human Cog5, its S. cerevisiae homologue, and proteins predicted from the Arabidopsis and Caenorhabditis elegans genomes (Figure 5C) (Walter et al., 1998). No other sequences encoding predicted proteins with similarity to Fws were identified by tBLASTn analysis of the Drosophila genome. Sequence analysis of genomic DNA amplified from fws mutant animals demonstrated that both EMS-induced fws alleles carried early nonsense mutations in the protein-coding region of the Drosophila Cog5 homologue (Figure 5C).
The fws gene encodes a 2.4-kb transcript detected in adult males, females, and embryos by Northern blot analysis (R.M. Farkas, unpublished data). No alternative forms of the transcript were observed. Although fws mRNA was present in females and in embryos, the fws mutations did not dramatically affect female fertility or embryogenesis: when mutant females were crossed to wild-type or fws/+ males, 75–85% of the eggs laid hatched. A similar hatching rate was observed among wild-type flies. The fws/+ and fws/fws progeny of the mutant females developed into adults with normal external morphology.
Four Way Stop Protein Localizes to Golgi in Spermatocytes and Differentiating Spermatids
To determine whether the Fws protein localized to Golgi, consistent with Fws being the fly homologue of Cog5, we engineered transgenic flies that expressed a GFP-Fws fusion protein under the control of the fws promoter from a fragment of genomic DNA containing the fws locus (see MATERIALS AND METHODS). When crossed into a fws homozygous mutant background, the GFP-fws construct rescued the male sterility and the defects in cytokinesis and spermatid elongation caused by the fws mutations. One or two copies of the GFP-fws construct caused no discernible phenotype in a wild-type background. The subcellular localization of the GFP-Fws fusion protein was determined by direct fluorescence from the GFP moiety in both live squashes and fixed cells.
In wild-type spermatocytes, the GFP-Fws fusion protein localized to multiple spherical structures that seemed to be Golgi stacks (Figure 6). The morphology of the Fws-containing structures and their positions in spermatocyte cells were consistent with existing transmission EM data, which showed multiple clusters of Golgi cisternae scattered throughout the spermatocyte cytoplasm (Figure 6, A and B) (Tates, 1971). In addition, the Fws-containing structures were recognized by antibodies against two different Drosophila Golgi proteins, α-mannosidase II (GMII) (Rabouille et al., 1999) and Lava lamp (Lva) (Sisson et al., 2000). The GFP-Fws protein did not colocalize precisely with GMII or Lva in fixed cells. Lva and GMII staining often appeared as a ring or crescent surrounding GFP-Fws (Figure 6, C and D, arrows), suggesting that Lva and GMII may reside mostly in cis cisternae or around the edges of cisternae, whereas Fws protein appeared to localize throughout the Golgi. The GFP-Fws showed similar patterns in live and fixed cells (R.M. Farkas, unpublished data). Polyclonal antibodies raised against Fws protein labeled the same characteristic subcellular structures as GFP-Fws in spermatocytes from wild-type males that lacked the GFP-fws construct (Figure 6E). Golgi staining was not seen in fws homozygous mutant cells incubated with the Fws antibody (Figure 6F), indicating that the Golgi staining was caused by the Fws protein.
Figure 6.
Localization of Fws to the Golgi apparatus in spermatocytes. (A) Diagram of a thin section of a Drosophila spermatocyte based on transmission electron microscopy (printed with permission of A. D. Tates) (see also Fuller, 1993). Bar, 1 μm. (B) Membrane organization of the Golgi apparatus. (C and D) Wild-type spermatocytes expressing GFP-Fws (green), stained with antibodies against GMII (C) or Lva (D) to visualize the Golgi membranes (red) and DAPI to visualize the DNA (blue). (E) Wild-type spermatocyte stained with antibodies against Fws (green) and DAPI to visualize the DNA (blue). (F) Antibody control: Two fws mutant spermatocytes stained with antibodies against Fws (green) and DAPI to visualize the DNA (blue). C–E, Bar, 10 μm.
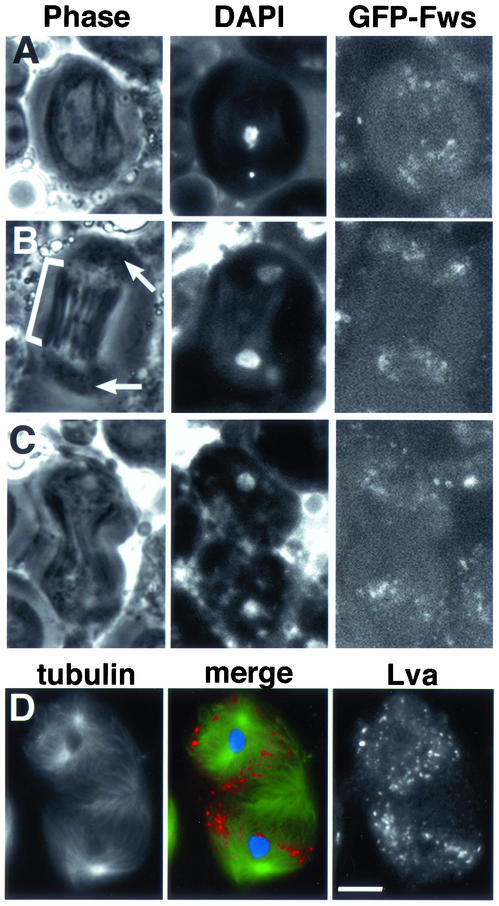
Because the fws mutations caused defects in cytokinesis during male meiosis and in spermatid elongation, we examined the subcellular localization of fws protein during these stages of spermatogenesis. The structures containing GFP-Fws seemed to fragment as cells entered meiotic division. By metaphase of meiosis I, Fws protein and Golgi markers appeared in small puncta in the cytoplasm and remained so throughout cytokinesis (Figure 7). In live metaphase and anaphase cells, spots of GFP-Fws protein appeared in the polar regions of the cell (Figure 7, A and B), excluded from the region of the central spindle (bracket) and astral membranes (arrow). In live cells in mid telophase, dots of GFP-Fws appeared scattered through the cytoplasm but were generally excluded from the region of the central spindle (Figure 7C). By the end of telophase, when dividing spermatocytes were highly constricted, dots of GFP-Fws filled most of the two nascent daughter cells in an even distribution but still seemed to be generally excluded from the midbody region. Similarly, in fixed cells, the Golgi marker Lva localized in dots that were dispersed throughout the cytoplasm but excluded from the region of the midbody at late telophase (Figure 7D).
Figure 7.
Fws localization in wild-type spermatocytes undergoing meiosis I. Live spermatocytes expressing GFP-Fws and stained with Hoechst dye to visualize DNA in (A) metaphase, (B) early telophase, and (C) mid telophase. (B) Astral membranes are marked by arrows, and central spindle is marked by bracket. (D) Fixed spermatocyte in telophase stained with Lva antibodies to visualize Golgi membrane (red), anti-α-tubulin antibodies to visualize microtubules (green), and DAPI to visualize the DNA (blue). Black and white panels show anti-tubulin and anti-Lva staining, respectively. Bar, 10 μm.
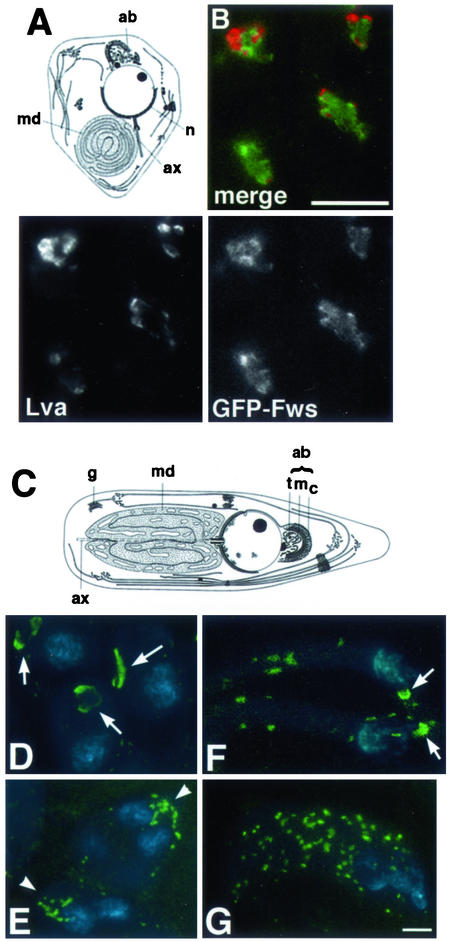
In wild-type early round spermatids, GFP-Fws protein localized with Lva and GMII to the acroblast, a cone-shaped structure composed of Golgi cisternae positioned adjacent to the spermatid nucleus (Figure 8A) (Tates, 1971). Antibodies against Lva (Figure 8, B and D) and GMII (R.M. Farkas, unpublished data) stained the edges and tips of the acroblast, whereas GFP-Fws appeared in a triangular pattern, filling the center of the cone and overlapping with Lva (Figure 8B) and GMII at the triangle periphery. Endogenous Fws protein also seemed to localize to the acroblast, because antibodies against Fws stained a triangular structure of the appropriate size and position in wild-type cells but not in fws cells (R.M. Farkas, unpublished data). GFP-Fws continued to colocalize with Lva and GMII in elongating spermatids, appearing in puncta distributed along the spermatid flagella (R.M. Farkas, unpublished data).
Figure 8.
Role of fws in acroblast formation. (A) Diagram of wild-type round spermatid based on transmission electron microscopy (printed with permission of A. D. Tates) (see also Fuller, 1993). Major structures include the Golgi-derived acroblast (ab), nucleus (n), mitochondrial derivative (md), and axoneme (ax). (B) Four wild-type early round spermatids expressing GFP-Fws (green) stained with Lva antibodies to visualize the Golgi (red). Bar, 10 μm. (C) Diagram of a spermatid initiating elongation based on transmission electron microscopy (printed with permission of A. D. Tates) (see also Fuller, 1993). Major structures include the Golgi-derived acroblast (ab), mitochondrial derivative (md), nucleus (n), Golgi stacks (g), and axoneme (ax). (D–G) Spermatids fixed and stained with DAPI to visualize DNA (blue) and Lva antibodies to visualize the Golgi (green). (D) Three wild-type early round spermatids with acroblasts (arrows) associated with their nuclei. (E) Two fwsz-0161/Df(2L)qua1374 early round spermatids, each with two nuclei, displaying abnormal acroblast morphology (arrowheads). (F) Two wild-type elongating spermatids with compact acroblasts (arrows) associated with the nuclei and several Golgi-based punctate structures along the tails. (G) A single fwsz-0161/Df(2L)qua1374 spermatid containing four nuclei at a stage comparable to cells in (F). Note the lack of acroblasts and the unusually large number of punctate Golgi structures along the tails. Bar, 10 μm.
Normal Assembly of the Golgi-Based Acroblast in Haploid Spermatids Requires Functional Fws Protein
In early round spermatids from fws mutant males, structures labeled with the Golgi marker Lva appeared in puncta clustered in an arched pattern at the position normally occupied by the acroblast (Figure 8E). However, in the absence of fws function, these punctate Golgi structures did not form the continuous curved sheet characteristic of wild-type acroblasts (Figure 8, C and D) (Tates, 1971). Later-stage fws spermatids with slightly elongated mitochondrial derivatives lacked any semblance of acroblasts, and the number of Lva-stained spots scattered along the fws spermatid flagella seemed higher than in wild-type cells (Figure 8, A, F, and G). No gross defects in Golgi morphology were detected in cells at earlier stages in fws testes on the basis of Lva and GMII immunostaining (R.M. Farkas, unpublished data).
DISCUSSION
The fws gene encodes the Drosophila homologue of S. cerevisiae Cod4p and mammalian Cog5, which are subunits of large Golgi-associated protein complexes (now called the COG complex) that have been implicated in Golgi architecture and function, including glycosylation and vesicle transport (Kingsley et al., 1986; Reddy and Krieger, 1989; Walter et al., 1998; Chatterton et al., 1999; Whyte and Munro, 2001; Ram et al., 2002; Ungar et al., 2002). Although several subunits of the yeast complex were found to be essential for growth and for normal subcellular morphology, deletions of the yeast Cog5 homologue had no effects on either, making the function of Cog5 in the complex unclear (Whyte and Munro, 2001). We have shown that fws is necessary during Drosophila spermatogenesis, in spermatocytes for normal ingression of the cleavage furrow during cytokinesis in the meiotic divisions, and in differentiating spermatids for the dramatic cellular elongation that accompanies growth of the sperm tail. In addition, wild-type function of fws is required in early spermatids for normal assembly or maintenance of the large-scale architecture of the Golgi-derived spermatid acroblast, which apparently normally forms from Golgi particles dispersed throughout the cell during the meiotic divisions. Our results suggest that Fws/Cog5 may be required for maximally efficient Golgi function, with the effects at the cellular and subcellular levels of loss of function of the protein most apparent only in a sensitized system that may place great demands on COG complex function and on normal Golgi architecture, function, and/or membrane trafficking.
A requirement for Fws in Golgi vesicle transport could explain each of the phenotypes observed in the fws mutant testes. In fws spermatids, Golgi membrane seemed fragmented rather than fused into the characteristic large, arched acroblast cisternae. In wild-type testes, the acroblast cisternae form after meiosis II by the aggregation and fusion of Golgi vesicles that had been dispersed during cell division (Tates, 1971; this study). The defects observed in acroblast structure in fws mutants could reflect a failure to efficiently fuse Golgi membrane into cisternae after meiosis. It is also possible that the defects were in acroblast stability rather than acroblast formation. In the absence of functional Fws, the equilibrium of anterograde and retrograde Golgi transport may alter, ultimately destabilizing nascent acroblast cisternae. Consistent with our observations that Drosophila COG may help establish or maintain Golgi architecture, mutations in mammalian Cog1 or Cog2 affect Golgi function and were found to distort Golgi cisternae in mutant CHO cells viewed by EM (Ungar et al., 2002). Defects in membrane traffic through the Golgi could be direct effects of lack of fws or indirect results of abnormal Golgi architecture caused by inefficient retrograde traffic to maintain proper Golgi compartments in the mutant. Interestingly, null mutants in a gene encoding a Golgi-associated protein, GOPC, in mice also resulted in male sterility and failure of vesicle fusion to form acrosomes in early round spermatids (Yao et al., 2002), similar to the acrosomal defect observed in spermatids from Drosophila males mutant for fws.
Defects in Golgi trafficking and/or architecture could cause the spermatid elongation and cytokinesis phenotypes observed in fws mutants. Spermatid elongation requires an increase in cell-surface area, which may be achieved by the delivery of Golgi-derived membrane to the plasma membrane. Studies in several systems have suggested that Golgi-derived vesicles are crucial for cleavage furrow ingression during cytokinesis. In dividing Xenopus zygotes and in Drosophila embryos undergoing cellularization (a specialized form of cytokinesis), Golgi-based vesicles fuse to the plasma membrane to form the furrow walls (Bluemink and de Laat, 1973; Byers and Armstrong, 1986; Aimar, 1997). Although we did not observe defects in spermatocyte Golgi architecture at the level of light microscopy, it is possible that there were subtle defects in intra-Golgi trafficking significant enough to disrupt spermatocyte cytokinesis. An alternative possibility is that particular glycoproteins modified in the Golgi must be delivered to the cell surface or to the vicinity of the contractile ring to initiate or mediate constriction of the cleavage furrow during cytokinesis in spermatocytes. Indeed, mutations in the mammalian Cog1 or Cog2 subunits were originally identified because of pleiotropic defects in glycosylation of cell-surface proteins that led to instability of the LDL receptor (Kingsley et al., 1986).
The activity of Fws did not seem to be generally required for cell division. Although fws mRNA was expressed in female flies and in embryos, the fws alleles examined did not have striking effects on viability, female fertility, or embryogenesis. It is possible that the truncated Fws proteins encoded by the two mutant alleles analyzed retained some degree of function sufficient for these processes. However, the viability of apparently null mutant fws flies is entirely consistent with the viability of yeast cells mutant for the S. cerevisiae homologue of Cog5. Fws may be functionally redundant with other proteins or pathways involved in Golgi function expressed in cells outside of the testis. However, we favor the possibility that Fws, and by inference certain subunits of the COG complex, may not be absolutely required for Golgi trafficking or architecture but may raise the efficiency of these processes. The phenotype of loss-of-function mutations in either the Cog1 or Cog2 subunits of the COG complex in CHO cells is consistent with this possibility (Kingsley et al., 1986; Reddy and Krieger, 1989).
Different cell types and different cellular processes may require different rates of exocytosis or levels of Golgi function. Because spermatocytes are relatively large (∼20 μm in diameter) and undergo cleavage furrow ingression in ∼20 minutes (Tates, 1971; R.M. Farkas, unpublished observations), they may require particularly rapid and extensive membrane addition during cytokinesis. Because of the rapid rate of cleavage furrow ingression in primary spermatocytes, new membrane added may need to come from preexisting stores, either from Golgi vesicles or by the recruitment and recycling although the Golgi of endosomal vesicles. In either case, the demands of such rapid membrane mobilization may make Fws function essential for cleavage furrow ingression during spermatocyte cytokinesis. Similarly, the dramatic expansion in cell surface that accompanies the 100-fold increase in cell length during Drosophila spermatid elongation may demand a level of efficiency that requires Fws function. Perhaps because of its distinctively large size, assembly and/or stability of the acroblast may also require more efficient membrane trafficking, more effective retrograde movement, or more effective Golgi–Golgi vesicle fusion than other Golgi-based structures. Thus, the events of male gamete differentiation in Drosophila may place high demands on Golgi function, making Drosophila spermatogenesis an effective sensitized system for genetic and functional analysis of pathways involved in Golgi architecture and function and/or membrane vesicle trafficking in vivo. Consistent with this possibility, hypomorphic mutations in the syntaxin 5 gene of Drosophila were found to cause defects in spermatocyte cytokinesis and spermatid elongation in a parallel study (Xu et al., 2002).
ACKNOWLEDGMENTS
We are grateful to Matt Fish for injections of the rescue and GFP constructs and to C. Zucker, B. Wakimoto, D. Lindsley, L. Cooley, S. Govind, B. Suter, D. Roberts, B. Sullivan, and J. Sisson for providing fly stocks and reagents. We are indebted to A.D. Tates for his beautiful EM cytology work, which aided in our interpretation of immunofluorescence data, and to Amy Kiger for providing the picture of the wild-type testis. We thank the members of the Fuller and Gatti laboratories, especially Carmen Robinett, as well as Suzanne Pfeffer, Gerry Waters, and Hugh Pelham for helpful discussions in the preparation of this manuscript, and an anonymous reviewer for useful critical comments. This work was supported by National Institutes of Health (NIH) training grant 2T52GM07790 to R.M.F. and NIH grant 1R01GM62276 to M.T.F.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0343. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0343.
REFERENCES
- Aimar C. Formation of new plasma membrane during the first cleavage cycle in the egg of Xenopus laevis: an immunocytological study. Dev Growth Differ. 1997;39:693–704. doi: 10.1046/j.1440-169x.1997.t01-5-00005.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- Bluemink JG, de Laat SW. New membrane formation during cytokinesis in normal and cytochalasin B-treated eggs of Xenopus laevis. I. Electron microscope observations. J Cell Biol. 1973;59:89–108. doi: 10.1083/jcb.59.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers TJ, Armstrong PB. Membrane protein redistribution during Xenopus first cleavage. J Cell Biol. 1986;102:2176–2184. doi: 10.1083/jcb.102.6.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Hirsch D, Schwartz JJ, Bickel PE, Rosenberg RD, Lodish HF, Krieger M. Expression cloning of LDLB, a gene essential for normal Golgi function and assembly of the ldlCp complex. Proc Natl Acad Sci USA. 1999;96:915–920. doi: 10.1073/pnas.96.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 71–147. [Google Scholar]
- Giansanti MG, Bonaccorsi S, Gatti M. The role of anillin in meiotic cytokinesis of Drosophila males. J Cell Sci. 1999;112:2323–2334. doi: 10.1242/jcs.112.14.2323. [DOI] [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Ting AE, Hazuka CD, Davanger S, Kenny JW, Kee Y, Scheller RH. The mammalian brain rsec6/8 complex. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- Kee Y, Yoo JS, Hazuka CD, Peterson KE, Hsu SC, Scheller RH. Subunit structure of the mammalian exocyst complex. Proc Natl Acad Sci USA. 1997;94:14438–14443. doi: 10.1073/pnas.94.26.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM, Kozarsky KF, Segal M, Krieger M. Three types of low density lipoprotein receptor-deficient mutant have pleiotropic defects in the synthesis of N-linked, O-linked, and lipid-linked carbohydrate chains. J Cell Biol. 1986;102:1576–1585. doi: 10.1083/jcb.102.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Meyer GF. Characterization of male meiotic-sterile mutations in the genetic control of meiotic divisions and gametogenesis. Chromosoma. 1977;64:371–392. doi: 10.1007/BF00294944. [DOI] [PubMed] [Google Scholar]
- Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kuntz DA, Lockyer A, Watson R, Signorelli T, Rose DR, van den Heuvel M, Roberts DB. The Drosophila GMII gene encodes a Golgi alpha-mannosidase II. J Cell Sci. 1999;112:3319–3330. doi: 10.1242/jcs.112.19.3319. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Li B, Kaiser CA. Identification of sec36p, sec37p, and sec38p: components of yeast complex that contains sec34p and sec35p. Mol Biol Cell. 2002;13:1484–1500. doi: 10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Krieger M. Isolation and characterization of an extragenic suppressor of the low-density lipoprotein receptor-deficient phenotype of a Chinese hamster ovary cell mutant. Mol Cell Biol. 1989;9:4799–4806. doi: 10.1128/mcb.9.11.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan CL, Fuller MT. Interacting genes that affect microtubule function in Drosophila melanogaster: two classes of mutation revert the failure to complement between haync2 and mutations in tubulin genes. Genetics. 1990;125:77–90. doi: 10.1093/genetics/125.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Vectors for P element-mediated transformation in Drosophila. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Field C, Ventura R, Royou A, Sullivan W. Lava lamp, a novel peripheral Golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol. 2000;151:905–918. doi: 10.1083/jcb.151.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink RG, Nothwehr SF. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol Biol Cell. 1999;10:4263–4281. doi: 10.1091/mbc.10.12.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tates AD. Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. Ph.D. Thesis. Leiden, Netherlands: Rijksuniversiteit de Leiden; 1971. [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter DM, Paul KS, Waters MG. Purification and characterization of a novel 13 S hetero-oligomeric protein complex that stimulates in vitro Golgi transport. J Biol Chem. 1998;273:29565–29576. doi: 10.1074/jbc.273.45.29565. [DOI] [PubMed] [Google Scholar]
- Whyte JRC, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Xu, H., Brill, J.A., Hsien, J., Boulianne, G.L., and Trimble, W.S. Syntaxin 5 is required for cytokinesis and spermatid differentiation in Drosophila. Dev. Biol. 251, 294–306. [DOI] [PubMed]
- Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci USA. 2002;99:11211–11216. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Spradling AC. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]