Abstract
Centrin, an EF hand Ca2+ binding protein, has been cloned in Tetrahymena thermophila. It is a 167 amino acid protein of 19.4 kDa with a unique N-terminal region, coded by a single gene containing an 85-base pair intron. It has > 80% homology to other centrins and high homology to Tetrahymena EF hand proteins calmodulin, TCBP23, and TCBP25. Specific cellular localizations of the closely related Tetrahymena EF hand proteins are different from centrin. Centrin is localized to basal bodies, cortical fibers in oral apparatus and ciliary rootlets, the apical filament ring and to inner arm (14S) dynein (IAD) along the ciliary axoneme. The function of centrin in Ca2+ control of IAD activity was explored using in vitro microtubule (MT) motility assays. Ca2+ or the Ca2+-mimicking peptide CALP1, which binds EF hand proteins in the absence of Ca2+, increased MT sliding velocity. Antibodies to centrin abrogated this increase. This is the first demonstration of a specific centrin function associated with axonemal dynein. It suggests that centrin is a key regulatory protein for Tetrahymena axonemal Ca2+ responses, including ciliary reversal or chemotaxis.
INTRODUCTION
Centrin, an EF hand Ca2+-binding protein, first identified in unicellular green algae and cloned in Chlamydomonas (Salisbury et al., 1984; Huang et al. 1988), is highly conserved and has been characterized in a variety of eukaryotes (Salisbury, 1995). Centrin is an integral part of microtubule-organizing centers, such as centrioles and basal bodies, and of the filamentous structures associated with these regions (Salisbury et al., 1984) including flagellar roots. Centrin is also part of the family of proteins that make up contractile stalks and fibers in protozoa such as Stentor or Vorticella (Routledge, 1978; Maciejewski et al., 1999). It may be part of the microtubule-severing apparatus at the base of the cilium (Sanders and Salisbury, 1989) and, most significantly for this study, it is found as a part of some of the inner dynein arms of Chlamydomonas axonemes (LeDizet and Piperno, 1995). Here we report on the cloning and characterization of the centrin structural gene, localization of its associated protein, and function of centrin in the control of inner arm dynein (IAD) in Tetrahymena thermophila. Because of the wide interest in the genomics and proteomics of Tetrahymena (Asai and Forney, 2000), technologies exist for studies on the functional significance of proteins such as centrin in this organism, and we have begun to exploit these advantages.
Using degenerate oligonucleotides generated against highly conserved N-terminal and internal peptide regions, we amplified a genomic DNA fragment containing ∼65% of the coding region of the Tetrahymena centrin gene by PCR. Using RACE techniques, we successfully cloned the entire length of both the cDNA and corresponding genomic sequence. Southern blotting revealed that unlike Paramecium (Madeddu et al., 1996), only a single centrin gene is present in T. thermophila. Analysis of the amino acid sequence derived from the cDNA indicated that Tetrahymena centrin is a 167-amino acid protein of 19.4 kDa calculated molecular weight, which includes four EF hand motifs and shows >80% homology to a majority of other centrin molecules. The protein also has high homology to other cloned Tetrahymena Ca2+-binding EF hand proteins including calmodulin (CaM; Maihle and Satir, 1980), TCBP23, and TCBP25 (Takemasa et al., 1989, 1990). Because all four Ca2+-binding EF hand proteins are present in a single cell, we undertook to localize centrin with respect to these other proteins both in the cell body and in the axoneme, so as to help delineate the function of centrin in Ca2+ responses, in particular with regard to the cilium. Cloning allowed us to define a unique N-terminal of Tetrahymena centrin and to generate a peptide antibody against this sequence for use in such studies. Localization studies using this and other centrin antibodies indicated that centrin was found along the ciliary axoneme and confirmed localization to the Tetrahymena IAD and not with 22S outer arm dynein (OAD).
Ca2+ controls important cellular events, including ciliary beat, in Tetrahymena. Electrophysiological (Onimaru et al., 1980) and behavioral (Leick et al., 1994) assays indicate that like Paramecium, Tetrahymena undergoes ciliary reversal. Although a Ca2+-based action potential and depolarization produce reversal and the electrical characteristics and their behavioral correlates are identical to those in Paramecium, the changes in beat form and the molecular details of Ca2+ interaction with the ciliary axoneme are not well understood for this organism. In permeabilized Tetrahymena swimming stops and beat form appears abnormal when the cells are treated with Ca2+ concentrations greater than 10−7 M (Goodenough, 1983). Presumably Ca2+ interacts directly with one or more axonemal Ca2+-binding proteins to influence dynein arm behavior, switching of doublet activity, and beat form changes. Because in Chlamydomonas ciliary beat form has been shown to primarily be regulated by IADs (Brokaw and Kamiya, 1987), we attempted to demonstrate a link between Ca2+ binding to centrin and IAD mechanoactivity. Studies using in vitro microtubule (MT) translocation by IADs were undertaken to clarify the role of centrin in IAD function that could lead to a change in beat form. The results suggest a model whereby Ca2+ binds directly to the EF hand regions of IAD associated centrin, causing an increase in IAD-generated sliding velocity. Therefore, in Tetrahymena axonemes centrin acts as a key transducer molecule, independent of phosphorylation, controlling ciliary beat by changing IAD function in order to initiate a signal transduction cascade leading to chemotaxis or backwards swimming. This is the first demonstration of a specific centrin function associated with axonemal dynein.
MATERIALS AND METHODS
Growth of Cells and Preparation of Cell Fractions
T. thermophila SB255 were grown at 2l-28°C to early or midstationary phase in complex growth medium (cf. Gorovsky, 1973) on a rotary shaker. Harvested cells were washed twice in 10 mM Tris, pH 7.2. Whole cell lysates were made by mixing the cell pellet with an equal volume SDS sample buffer and left in the freezer at −20°C. Cilia were isolated from the harvested cells by dibucaine deciliation, as previously described (Satir et al., 1976). Axonemes were prepared from isolated cilia by 1% Triton X-100 treatment for 2 h on ice, centrifuged at 20,000 × g,. and finally washed and resuspended in axoneme buffer (20 mM K acetate, 5 mM MgSO4, 0.5 mM EDTA, 30 mM HEPES, pH 7.6). As described previously (cf. Larsen et al., 1991), crude dynein was extracted from the axonemes using 0.6 M KCl in axoneme buffer on ice for 2 h and fractionated into IAD (14S) and 22S dynein fractions on a sucrose gradient (5–30%), followed by an assessment of ATPase activity and heavy-chain composition of each fraction.
Reagents
Authentic isolated Chlamydomonas centrin, mAb 20H5 against bacterially expressed Chlamydomonas centrin and polyclonal MC1 rabbit antibody against mouse centrin were generous gifts from J.L. Salisbury (Mayo Clinic, Rochester, MN). Antibody to Tetrahymena centrin N-terminal region (TcN antibody) was produced in chick by AnaSpec (San Jose, CA) and affinity purified. CaM and tubulin antibodies, purified CaM and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) were obtained from Sigma (St. Louis, MO). Antibodies to TCBP23 and -25 (TCBP 23, -25 antibodies) were kindly provided by Y. Watanabe (University of Tsukuba, Japan). The Ca2+-mimicking peptide, CALP1, was kindly provided by J.E. Blalock (University of Alabama, Birmingham, AL; Villain et al., 2000).
Electrophoresis of Proteins and Immunoblotting
SDS-PAGE was performed using 12% polyacrylamide and 0.1% SDS. On completion of migration, the proteins were electrophoretically transferred to a PVDF Immobilon P (Millipore, Bedford, MA) or a nitrocellulose membrane (Schleicher & Schüll, Dassel, Germany) in transfer buffer (Towbin et al., 1979) at 250 mA for 3 h. Protein transfer was evaluated by staining the membrane in 1% Ponceau S red prepared in 0.5% acetic acid. The membranes were then blocked by saturation with milk buffer (2% nonfat dry milk in 1× TBST [0.01 M Tris-HCl, pH 7.4, 0.l5 M NaCl, and 0.05% Tween 20]) for at least 30 min before overnight incubation of membranes in primary antibody (1:1000 dilution) at 4°C. The membranes were washed three times for 5 min each in 1× TBST and then incubated with a secondary antibody coupled to alkaline phosphatase (Sigma) at room temperature for 1 h. The membranes were again washed three times for 5 min each in 1× TBST and then incubated with a BCIP/NBT solution (Kirkegaard and Perry Labs, Gaithersburg, MD). The reactions were stopped by washing in water and air-drying the membranes.
Immunofluorescence and Immunogold Localizations
Immunolabeling of permeabilized Tetrahymena or isolated axonemes was carried out as previously described for Paramecium by Cohen and Beisson (1988). Cells starved in 10 mM Tris-HCl buffer (pH 7.2) or axonemes were mounted onto poly-l-lysine–coated microscope slides. After permeabilization for 2 min in buffer A (60 mM Pipes, 25 mM HEPES, 10 mM EGTA, 2 mM MgC12, 1% Triton X-100, pH 6.9; Schliwa and van Blerkom, 1981), preparations were fixed in 2% paraformaldehyde (freshly prepared) in buffer A for 30 min, washed three times in buffer A, and then briefly incubated in buffer B (10 mM Tris-HC1, pH 7.4, 0.15 M NaCl, 0.01% Tween-20, 3% bovine serum albumin, 5 mM CaCl2). Buffer B was used in all subsequent steps: 1° antibody (30 min), three washes 5 min each, incubation in FITC- or Cy3-labeled 2° antibody (15 min), and three final washes 5 min each. Preparations were mounted in mounting media (1× TBS, 70% glycerol, 2% n-propylgallate). The respective antibody dilutions used were 1:1000 for 1° antibodies and 1:500 for 2° antibodies (Jackson Laboratory, Bar Harbor, ME).
Fluorescence microscopy was performed using a Scanalytics EPR deconvolution system (Scanalytics Inc., Fairfax, VA; Femino et al., 1998) on an Olympus AX70 microscope. Cell reconstructions were done using Scion Image (Scion Corp., Frederick, MD) or Vox Blast (Vaytec Inc., Fairfield, IA)
Immunogold localization was performed by settling axoneme preparations onto formvar cast nickel grids coated with poly-l-lysine. Axonemal sliding was induced by floating grids on drops of axoneme buffer containing 0.7 mM ATP for 2 min. Preparations were fixed in 0.3% glutaraldehyde in axoneme buffer, rinsed in axoneme buffer, and washed with wash buffer (axoneme buffer, 0.05% Tween 20). After 2o antibody incubation, the preparation was thoroughly washed before a quick rinse in nanopure water. The preparations were negative-stained with 2% aqueous uranyl acetate for 90 s before being viewed on a JEOL (Peabody, MA) 100CXII operated at 80 kV.
PCR Amplification and Nucleotide Sequence Analysis
T. thermophila SB255 genomic DNA was isolated from cells as described by Gaertig et al. (1993). PCR amplification was accomplished by using partially degenerate oligonucleotide primers designed on the basis of highly conserved regions derived from the consensus sequence of an alignment of 15 different centrin sequences found in National Center for Biotechnology Information (NCBI) GenBank using GCG software (Wisconsin Package Version 10.2, Genetics Computer Group [GCG], Madison, WI). The nucleotide sequences, incorporating either a PstI (sense primers) or an AvrII (antisense primers) restriction enzyme site were as follows: N-term sense 1: GCGCTGCAGTTRTTYGAYACYGAYGG; N-term sense 2: GCGCTGCAGCTYTTYGAYACYGAYGG; C-term antisense 1: GACCCTAGGRATCATTTCTTRYAAYTC; C-term antisense 2: GACCCTAGGRATCATTTCTTRRAGYTC. PCR reactions for degenerate primers (50 μl) contained 1 μM of each primer, ∼50 ng genomic DNA, 0.2 mM dNTPs, 1× PCR buffer and 2 U Taq DNA polymerase (Fisher Scientific, Pittsburgh, PA). Reactions were performed as follows in a Geneamp PCR System 2400 (Perkin Elmer-Cetus, Applied Biosystems Division, Foster City, CA): 94°C for 45 s, 42°C for 1 min, 72°C for 45 s for four cycles, followed by a higher stringency sequence 94°C for 45 s, 55°C for 45 s, 72°C for 45 s for 26 cycles and 72°C for 10 min and 4°C hold. PCR conditions for nondegenerate primers were essentially the same, with the following changes: 0.2 μM of each primer; reaction sequence 94°C for 45 s, 52°C for 45 s, and 72°C for 45 s for 30 cycles. The amplification products were separated on a 1% agarose gel, isolated, and recovered using a PCR Wizard Prep kit (Promega, Madison, WI). DNA sequencing was performed by the Albert Einstein College of Medicine DNA Sequencing Facility using an ABI 377 automated sequencer (Perkin Elmer-Cetus). Sequence data were analyzed by Sequencher software (Gene Codes Corp., Ann Arbor, MI).
RACE Techniques for Generating Full-length Centrin Sequence
Total RNA was isolated from Tetrahymena strain SB255 in logarithmic growth following the protocol of Chomczynski and Sacchi (1987). Using total RNA as a template, poly-A mRNA was amplified for the production of a cDNA pool using a 3′ RACE kit (Life Technologies, Rockville, MD), following the manufacturer's protocol. Using a centrin-specific 3′ sense primer and a poly-dT containing anchor primer, the 3′ end of centrin cDNA was amplified from the original cDNA pool. Sequencing of the PCR product revealed the 3′ end of the coding region of the gene, including the poly-A tail of the mRNA. A centrin-specific 5′ antisense primer was used to generate oligo-dC–tailed single-strand cDNA from the original cDNA pool using a 5′ RACE kit (Life Technologies). The 5′ end of centrin cDNA was amplified from the oligo-dC tailed single-strand cDNA using a nested centrin-specific 5′ antisense primer and an oligo-dG–containing anchor primer. Sequencing the PCR product revealed the 5′ end of the coding region of the gene.
Southern Blot
Genomic DNA (∼10 μg) was digested with selected restriction endonucleases purchased from New England Biolabs (Beverly, MA) as either single or double digests. The digested DNA was electrophoretrically separated on a 1% agarose gel and transferred to nitrocellulose following the protocol in Maniatis et al. (1982). The blot was prehybridized in hybridization buffer for 2 h at 58°C. A radioactive probe generated from a nick-translation reaction of the full-length genomic PCR product was hybridized overnight for 16 h at 58°C at 107 cpm/ml in hybridization buffer. The blot was washed twice at room temperature in 2× SSC, 0.1% SDS and then washed at 55°C in 1× SSC, 0.1% SDS until the background count was less then 600 cpm. The blot was wrapped in plastic wrap and placed on Kodak BioMax MR Film for 16 h (Eastman-Kodak, Rochester, NY). It was developed using a Konica SRX-101A automatic film developer (Konica Medical Imaging, Wayne, NJ).
In Vitro Motility Assays for Centrin Function
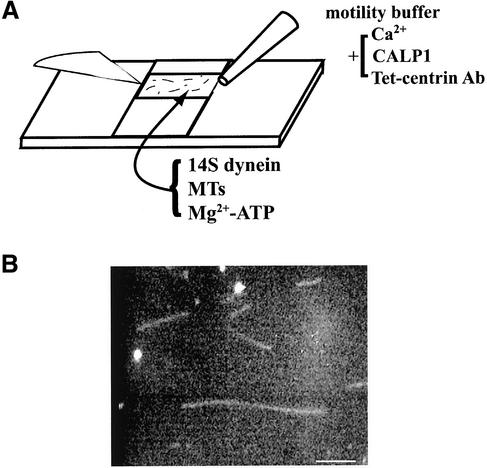
These assays were modified from Hamasaki et al. (1995) and Wada et al. (2000). Briefly, motility chambers were constructed and Tetrahymena 14S IAD fractions were used as a motor substratum for translocation assays using taxol-stabilized bovine brain MTs and darkfield microscopy. ATP, at 1 mM, was added, and tracking of individual MTs whose length was measured was followed at high resolution. Analysis followed the methods of Hamasaki et al. (1995).
RESULTS
PCR Cloning of the Centrin Gene Sequence from Tetrahymena Genomic DNA
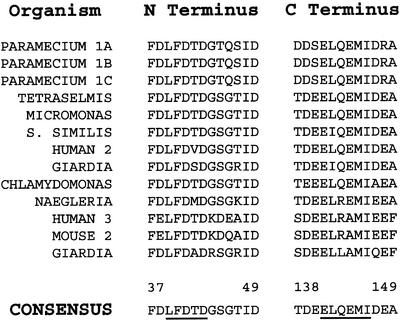
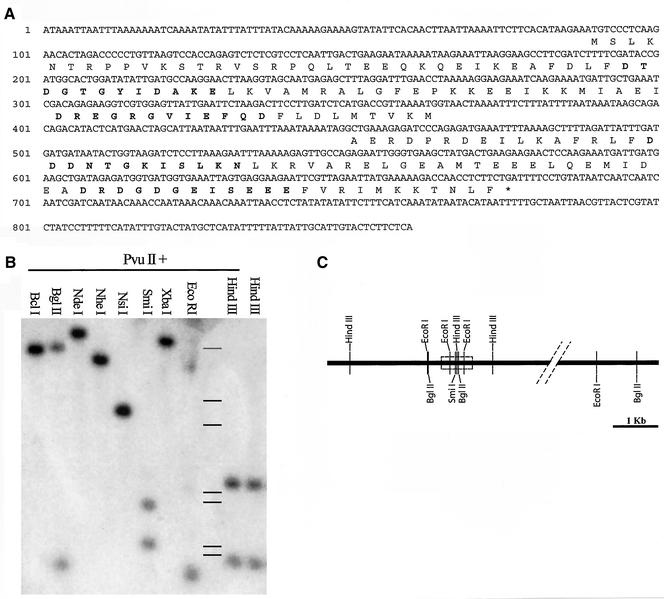
Alignment of 15 centrin amino acid sequences (Figure 1) published by the NCBI GenBank was used to construct four partially degenerate oligonucleotide primers (see MATERIALS AND METHODS) for direct PCR amplification of genomic Tetrahymena DNA. PCR amplification produced only a single detectable product that was sequenced directly, without subcloning. The PCR product was 428 nucleotides long and appeared to contain 343 nucleotides of the structural gene for centrin (65%). An intron of 85 nucleotides was found with standard splice sites (GT and AG, respectively), positioned eight amino acids following the second calcium-binding domain (EF hand) from the N-terminus. Our PCR clone included sequences that coded for three of four of the calcium-binding domains in centrin. The intron caused a shift of the reading frame in the final spliced PCR product. 5′ and 3′ RACE techniques were used to generate the complete coding region of the centrin gene. Centrin-specific primers designed using the 5′ and 3′ RACE sequences were used to PCR and sequence both the full-length cDNA (774 base pairs) and the full-length coding region of the genomic DNA (859 base pairs), shown in Figure 2A (EMBL accession no. AF141944). The Tetrahymena centrin gene has standard start (ATG) and stop (TGA) codons.
Figure 1.
Alignment of highly conserved centrin sequences from the N- and C-terminal regions of the molecule. The numbers above refer to amino acid positions in the derived consensus sequence. The underlined amino acids were used for degenerate primer design. Full sequences and accession numbers available on the EMBL website (http://www.ebi.ac.uk/embl/).
Figure 2.
(A) The Tetrahymena centrin genomic DNA sequence and the deduced amino sequence. Bold amino acids indicate EF hand regions. (B) Southern blot of Tetrahymena genomic DNA digested by the restriction enzymes indicated and hybridized with nick-translation reaction probe from the full-length genomic PCR product. In both single and double digests, the probe gives two bands after digestion with restriction enzymes with sites in the centrin gene as sequenced and otherwise only single bands, supporting the restriction map in 2C. Lines: Standards, top to bottom: 9, 6, 4, 2.2, 2, 1, 0.8 kb. (C) A restriction map of the centrin locus based on the Southern blot analysis. The boxed area represents the coding region for the molecule.
From Southern blot analysis, we have confirmed the presence or absence of a number of predicted restriction sites within the centrin gene (Figure 2B). A simplified restriction map is shown (Figure 2C). The data are consistent with the conclusion that there is a single centrin gene in Tetrahymena.
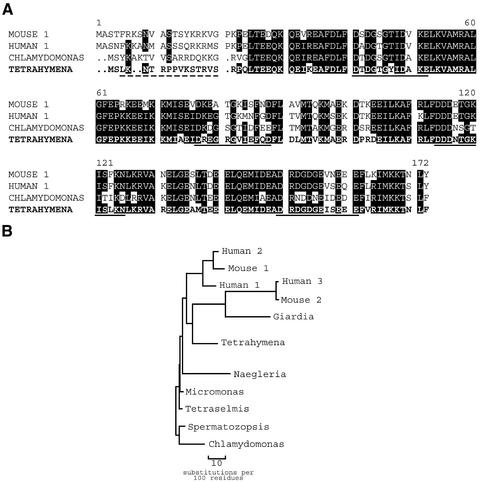
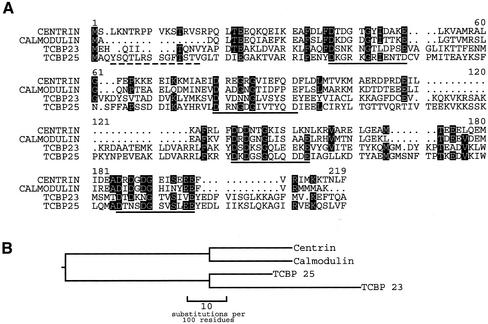
As cloned, Tetrahymena centrin has a molecular mass of 19.4 kDa with 75% identity to human centrin 1 and 71% identity to Chlamydomonas centrin (Figure 3A). The four EF hand regions are spread evenly throughout the molecule with intervals of 23 or 24 a.a.'s separating successive motifs. Only the N-terminal ca. 16 amino acids are gene specific, showing minimal identity to other centrins. A phylogenetic tree (Figure 3B) constructed from sequence data for various centrins shows that Tetrahymena centrin is on a branch related to mammalian centrins rather than algal centrins. The cloned Tetrahymena sequence was also compared with sequences of three other known Tetrahymena calcium-binding proteins: TCBP23, -25, and Tetrahymena CaM (Figure 4A). An unrooted tree comparing the molecules is shown in Figure 4B. Tetrahymena centrin and CaM are 52% identical. The N-terminus of centrin is absent in CaM; however, the four EF hand motifs are identically positioned in the two molecules. In contrast, TCBP23 and -25 show less identity to Tetrahymena centrin (19% and 23%, respectively). The N-terminus of centrin is different from the N-termini of the TCBPs. Although these molecules also possess four EF hand motifs, these motifs are skewed with respect to Tetrahymena centrin, so that with the best fit the third EF hand motif in TCBP23 and -25 is most homologous to the first EF hand motif of Tetrahymena centrin.
Figure 3.
(A) Comparison of Tetrahymena centrin with representative known centrin sequences. The four EF hand sequences are underlined. Dashed line indicates unique N-terminal sequence used for TcN antibody production. Shaded regions indicate identity of amino acid in at least three of the four sequences shown. (B) Unrooted phylogenetic tree showing the position of Tetrahymena centrin.
Figure 4.
(A) Comparison of Tetrahymena centrin with other cloned EF hand proteins from Tetrahymena. The four EF hand sequences are underlined. Dashed line indicates unique N-terminal sequence used for TcN antibody production. Shaded regions indicate identity of amino acid in at least three of the four sequences shown. (B) Unrooted phylogenetic tree showing the evolutionary relationship of the Tetrahymena EF hand proteins.
Recognition of Centrin by Specific Antibodies in Tetrahymena Whole Cell Lysates and Axonemes
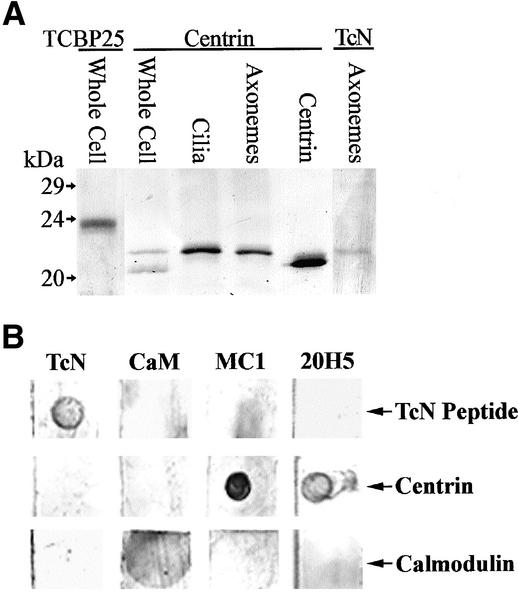
An antibody (TcN ab) directed against the unique N-terminal region of Tetrahymena centrin was produced. Tetrahymena crude whole cell lysates, isolated cilia and axonemes, and ciliary membrane-matrix fractions were blotted with MC1 antibody to mouse centrin, 20H5 antibody to Chlamydomonas centrin, TcN antibody or TCBP23 or -25 antibodies. Whole cells and isolated axonemes of Chlamydomonas and a centrin standard were run for comparison. Using 20H5, MC1 or TcN antibodies, centrin appears as one main component in Tetrahymena cilia and axonemes with a Mr of ∼21 kDa. In whole cells, a second centrin band at Mr ∼20 kDa, often seen with 20H5 in other systems (Lutz et al., 2001), is also present. Tetrahymena centrin is recognized by the centrin antibodies, which do not recognize CaM, TCBP23, or -25 (Figure 5A). CaM antibody recognizes CaM but does not recognize centrin. TcN antibody binds to TcN peptide and does not recognize Chlamydomonas centrin or bovine CaM; standard centrin antibodies, MC1 and 20H5, or CaM abs do not recognize the TcN peptide (Figure 5B).
Figure 5.
Immunoblot of centrin vs. TCBP25. (A) TCBP25 antibody recognizes a band Mr 24 kDa, whereas 20H5 centrin antibody recognizes bands at ca. Mr 20 and 21 kDa in whole cells. The 21-kDa band is also recognized in isolated cilia and axonemes. Similarly, TcN antibody recognizes only the 21-kDa band in axonemes. 20H5 antibody is blotted against Chlamydomonas centrin as a standard. (B) Dot blots for antibody specificity. Antibodies used in the localization studies were blotted against purified commercial bovine brain CaM, Chlamydomonas centrin, and the Tetrahymena centrin N-terminal peptide (TcN).
Localization of EF Hand Proteins in Tetrahymena
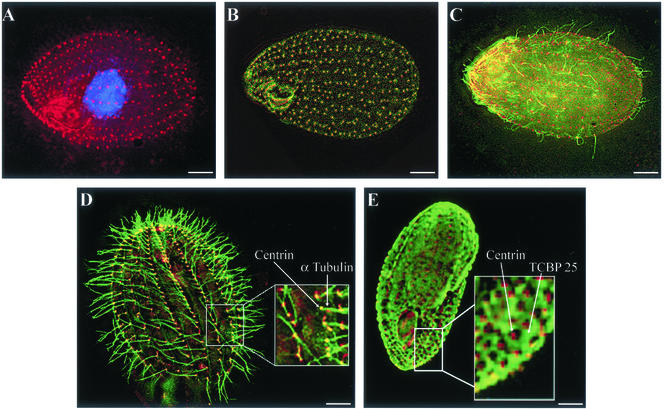
Indirect immunofluorescence with centrin antibodies confirmed earlier reports by Jerka-Dziadosz et al. (1995) that centrin localized preferentially to cortical basal bodies, to rootlet structures, to the oral apparatus, and to the apical filament ring. Figure 6 shows immunofluorescence reconstruction images of centrin localization in paraformaldehyde-fixed and Triton X-100–permeabilized Tetrahymena. Centrin labeling was observed consistently on both individual somatic kinetosomes and on the numerous kinetosomes found in the oral apparatus (Figure 6A). No difference in immunofluorescence labeling was observed in cells harvested in late exponential growth vs. starved cells or in strains SB255 vs. CU428. Centrin labeling almost exactly colocalizes with γ-tubulin labeling in the basal bodies (Figure 6B). In these preparations, CaM is found along ciliary axonemes, but centrin is not (Figure 6C). Figure 6D shows localization of centrin and α-tubulin. In this case α-tubulin labels the ciliary axoneme extending from the centrin-containing basal bodies.
Figure 6.
Immunolocalization of Tetrahymena EF hand proteins in permeabilized fixed whole cells. Bars, 10 μm. (A) Centrin localization to basal bodies and oral apparatus using TcN antibody. DAPI counterstain. (B) Colocalization of centrin (20H5 antibody Cy3-labeled red) with γ-tubulin in basal bodies and oral apparatus. (C) Colocalization of centrin (20H5 antibody) and CaM. CaM (FITC-labeled green) has a diffuse cortical and ciliary localization. (D) Colocalization of centrin (20H5 antibody) and α-tubulin. Centrin localizes to the basal body and ciliary rootlet with α-tubulin outlining each axoneme. Boxed region is 2× enlargement. (E) Colocalization of centrin (20H5 antibody) and TCBP25. TCBP25 localization defines a cortical lattice, but it is excluded from the region around the basal bodies. Boxed region is 3× enlargement.
TCBP23 and -25 are found in the cell cortex in a lattice surrounding the centrin containing basal bodies (Figure 6E). All centrin localizations have been controlled by parallel preparations without primary antibody; in these preparations no centrin localization was observed. For control experiments, Chlamydomonas centrin, bovine brain CaM or TcN peptide were used for competition with the centrin or CaM antibodies. Chlamydomonas centrin diminished CaM antibody labeling but abolished 20H5 and MC1 centrin labeling. TcN peptide abolished TcN antibody localization. Bovine brain CaM abolished CaM antibody labeling but had no effect on the 20H5 and MC1 centrin labeling.
Centrin Colocalization with α-Tubulin along the Length of the Axoneme
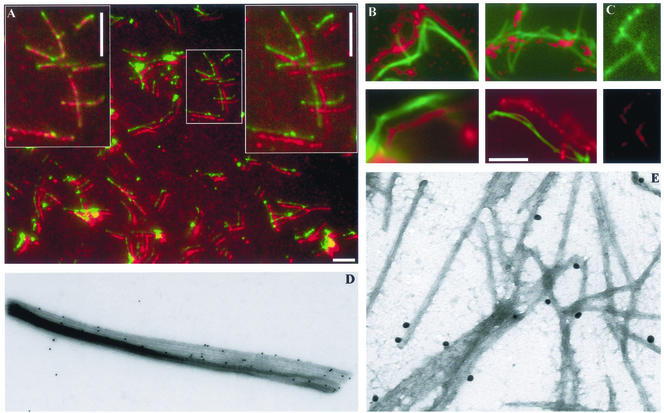
The lack of centrin localization along the axoneme in Figure 6 could be the result of difficulty of antibody penetration into the fixed cilium. To show whether centrin localization was present along the axoneme, more stringent preparative procedures were used. Cilia were isolated by fractionation, and their membranes were removed with detergent before centrin localization studies. After this treatment both α-tubulin and centrin are now localized along the isolated axonemes (Figure 7). Although merged images show discontinuities in centrin localization, with respect to α-tubulin, this is artifactual. To demonstrate colocalization along the entire axoneme, the centrin image was displaced by several pixels from the corresponding α-tubulin image; then, both images were continuous and superposable (Figure 7, A and B). TcN peptide abolished TcN antibody localization, without affecting the α-tubulin image (Figure 7C). Immunogold labeling at EM resolution confirms localization of label along the edges of splayed axonemal MTs (Figure 7, D and E). Therefore, centrin is present along the length of the axonemal microtubules, presumably as a component of an IAD.
Figure 7.
Immunolocalization of centrin in isolated axonemes. Bars, 5 μm. (A) Colocalization of centrin (20H5 antibody) with α-tubulin along the length of the axoneme. Box indicates axonemes enlarged in left and right panels. Left panel: direct overlap of red and green channels; right panel: displaced overlap. (B) Colocalization of centrin (TcN antibody) with α-tubulin; displaced overlap. (C) Localization after treatment with TcN antibody + peptide. Top panel: α-tubulin; bottom panel: centrin. (D and E) Immunogold localization of centrin (MC1 antibody) on splayed axonemes. Label is preferentially found along one edge of doublet MTs. Magnification, ×13,000 and ×50,000, respectively.
Centrin Copurifies with Inner Arm Dynein
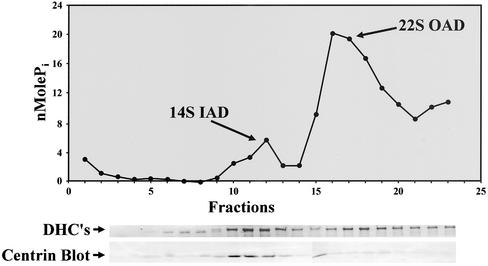
To probe whether centrin localized with a dynein subfraction from Tetrahymena, we extracted dyneins from isolated Tetrahymena axonemes, separated them over a sucrose gradient, and analyzed the resulting fractions (Figure 8). Fractions 10–12 and 16–19 contained high-molecular-weight dynein heavy-chain bands in SDS-PAGE, and they showed significant ATPase activity, corresponding to 14S IAD and 22S OAD, respectively. The fractions were immunoblotted against centrin or CaM antibodies. Centrin colocalized to fractions 10–12 with IAD; there was no CaM localization to these fractions.
Figure 8.
Centrin is associated with 14S IADs. Top panel: ATPase activity of sucrose density gradient fractions of crude dynein defines 14S and 22S dyneins. Middle panel: Dynein heavy chain (DHC) region of corresponding SDS-PAGE gel. Bottom panel: Immunoblot with 20H5 antibody.
Centrin Controls the Ca2+ Effect on the Velocity of IAD MT Translocation
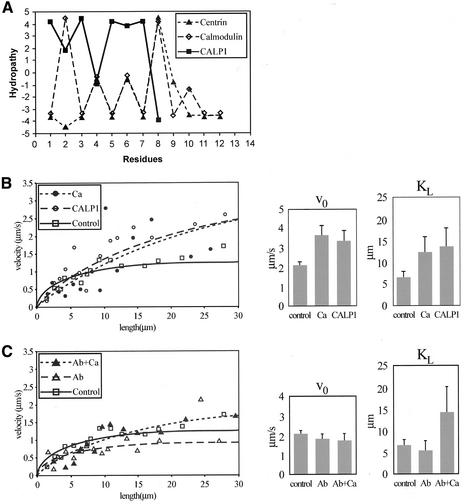
We hypothesized that centrin is the protein to which Ca2+ binds to exert its effect on ciliary motility. A likely means of control of motility is the regulation of MT sliding velocity (Satir, 1998). To explore the function of centrin in relation to the velocity of MT sliding produced by IAD, we utilized in vitro motility assays with a motility chamber constructed as in Figure 9A. IAD was bound to the surface of the chamber forming a substratum over which MTs of different lengths moved when activated with 1 mM Mg-ATP (Figure 9B). At appropriate times after motility was measured, Ca2+ was added to the chamber to adjust the concentration to pCa ∼5, or alternatively CALP1 was added to a 50 μM concentration. The hydropathy pattern of CALP1 is inverted with respect to the EF hand 4 region of centrin as well as CaM (Figure 10A), which suggests that CALP1 should mimic the effect of Ca2+. In some experiments antibodies to centrin were perfused into the chamber before the addition of Ca2+. Changes in MT translocation velocity were measured as a function of length (Figure 10, B and C). The data were then plotted in a Lineweaver-Burk type plot as described by Hamasaki et al. (1995) to determine v0, the maximum MT translocation velocity (analogous to Vmax) and KL, the MT length translocating at 0.5 v0 (analogous to Km).
Figure 9.
(A) Drawing of in vitro motility chamber. The chamber is constructed using a slide and coverslip. The chamber contains a 14S dynein substratum over which MTs are placed in motility buffer with appropriate Ca2+ reagents added. Motility was initiated by wicking in 1 mM Mg-ATP. Modified from Hamasaki et al. (1995). (B) Darkfield micrograph of MTs in motility chamber. Note varying lengths. Bar, 2 μm.
Figure 10.
(A) Hydropathy plot of CALP1 vs. CaM and Tetrahymena centrin EF hand 4. Residue 1 corresponds to centrin residue 146. Binding occurs when the hydropathy of the two regions is inverted (Villain et al., 2000). (B and C) Left: Plot of MT gliding velocity vs. MT length for various conditions of treatment of a single preparation. Trendlines arbitrarily set to a zero-zero intercept. A v0 and KL value is derived from each curve as in Hamasaki et al. (1995). Right: Average v0 and KL values (±SEM) for the conditions shown. t test: v0/KL Control vs. Ca2+ , p = 0.002/0.07; Ca2+ vs. CALP, p = 0.72/0.82; Control vs. antibody, p = 0.45/0.67; antibody vs. antibody+Ca2+ , p = 0.9/0.10; antibody+Ca2+ vs. antibody+CALP, p = 0.30/0.42; antibody+Ca2+ + antibody+CALP vs. Ca2+ + CALP, p = 0.05/0.85.
Figure 10, B and C, shows typical experimental measurements using one preparation (left panels) and average values of v0 and KL for 63 different experiments using 656 MTs (right panels). In the absence of Ca2+, the average v0 is ∼2 μm/s, but either Ca2+ (pCa 5) or CALP1 increases v0 ∼1.5 to 3-fold (Figure 10B). When TcN antibody is added, in the absence of Ca2+, there is little effect on v0. When Ca2+ is added, in the presence of TcN antibody, the rise in v0 is suppressed (Figure 10C). KL apparently increases in the presence of Ca2+ or CALP1 (Figure 10B), whereas there is no change of KL compared with controls when TcN antibody alone is added (Figure 10C). KL may also change when Ca2+ or CALP is added in the presence of TcN antibody. Changes in KL are seen in Figure 10, B and C (left panels); however, in our pooled experiments (Figure 10, B and C, right panels) KL changes are not statistically significant. Preliminary results with MC1 antibody are qualitatively similar, although some MTs nearly stop translocating in the presence of Ca2+ and MC1 antibody.
DISCUSSION
The cloning of Tetrahymena centrin, as reported here, is potentially important because this organism provides unique opportunities for assessing centrin functions. Tetrahymena has a single centrin gene. We have shown that centrin is localized to the Tetrahymena IADs, confirming earlier reports from LeDizet and Piperno (1995) from Chlamydomonas. This has made possible studies designed to elucidate centrin function in the ciliary axoneme. Although Tetrahymena centrin resembles other previously cloned Tetrahymena Ca2+-binding proteins, in particular by the presence of four EF hand regions, it is more closely related to centrins of other species, than to Tetrahymena CaM or TCBP23, -25. Although the other closely related Tetrahymena EF hand family proteins are present together with centrin in the cell, their specific localizations appear to be different; in particular, centrin and TCBP25 give complementary nonoverlapping localizations in the cell cortex, and similarly centrin and CaM have different localization within the ciliary axonemes. Although CaM is found at the ciliary membrane and is present along the axoneme (Yang et al., 2001), it is not directly part of the dynein arms. This suggests that each EF hand protein has a different function with respect to Ca2+ signal transduction, even although they lie within a few hundred nanometers or less of each other. The targeting of different EF hand proteins within such short distances is an intriguing problem.
Interestingly, the phylogenetic position of Tetrahymena centrin places it nearer mammalian than algal centrins. There are few surprises in the cloned sequence; only the N-terminal 16 amino acids differ significantly from other centrins. We have taken advantage of this to produce a peptide antibody specific for Tetrahymena centrin that does not recognize CaM. Likewise, the basal body and fibrous localizations are similar to localization of centrin in other organisms, although the details vary. For example, the conspicuous cortical centrin contractile lattice described by Garreau de Loubresse et al. (1991) and Klotz et al. (1997) for Paramecium seems absent in Tetrahymena. With this difference in mind, it would be interesting to compare Ca2+ induced cortical contraction in Paramecium and Tetrahymena.
Demonstrating centrin localization in the axoneme by light microscopic immunofluorescence in permeablized cells is difficult because as a component of the IAD, centrin is sequestered from easy antibody access. However, with isolated axonemes, localization can be demonstrated with a pixel displacement technique, which shows centrin colocalization with α-tubulin. Localization to the axoneme and IADs is confirmed by immunoblotting of axoneme and dynein fractions and by immunoelectron microscopy. The 21-kDa band recognized in the axoneme by all the centrin abs used in this study may reflect the presence of phosphorylated centrin (Lutz et al., 2001). Using isolated axonemes and IAD, we also have preliminary evidence that centrin is present in Paramecium. This suggests that the centrin subunit of an inner dynein arm, presumably IAD3 as described in Chlamydomonas (LeDizet and Piperno, 1995) is generally present in axonemes. There are a few reports of centrin-2 localization to human cilia (Laoukili et al., 2000), but most localization is confined to the basal body and the proximal transition zone (LeDizet et al., 1998). Because centrin is a Ca2+-binding protein, its localization to the ciliary axoneme may be significant for axonemal Ca2+ responses in Tetrahymena, including ciliary reversal or chemotaxis. In this regard, a knockout of Tetrahymena centrin should be able to clarify its role in ciliary responses, and might have more general implications.
The IADs are primarily responsible for ciliary beat form (Brokaw and Kamiya, 1987), which leads to the various behavioral responses. In the absence of changes in beat frequency, axonemal bending is proportional to the sliding velocity induced by IADs (Satir, 1998). Accordingly, one possible function of centrin in the IADs might be to modulate MT sliding velocity in the presence of Ca2+. This possibility has been explored using in vitro motility assays with isolated IADs. MT sliding velocity, measured as v0, was substantially increased in the presence of Ca2+. This increase was mimicked in the absence of Ca2+ when the EF hand binding peptide CALP1 was added. This indicates that Ca2+ was probably acting by binding directly to an EF hand containing protein, namely centrin, which has been shown to be a component of the IAD fractions used as substrata for these assays. To confirm this, we used the specific Tetrahymena anticentrin antibody, TcN. TcN had little effect on sliding velocity in the absence of Ca2+, but inhibited the activation of sliding velocity toward control values when Ca2+ was added. In human cilia, where Ca2+ increases ciliary beat frequency, Laoukili et al. (2000) found that antibodies to human centrin 2 significantly decrease beat frequency. Because only a fraction of the Tetrahymena IADs contain centrin, we would predict that KL would also increase in the presence of Ca2+ and CALP1. From our present data this is likely but not conclusive.
These results support the conclusion that Ca2+ binds to IAD centrin to increase MT sliding velocity directly, correspondingly increasing bend amplitude and changing axonemal beat form. This direct action is quite distinct from the indirect action of cAMP, which acts on an endogenous PKA to phosphorylate an OAD subunit in Tetrahymena (Christensen et al., 2001) In this respect, centrin must bind to and act on the IAD isoform in much the same way as CaM acts on various myosin isoforms. It may be that this is a more general function of centrin in cells.
ACKNOWLEDGMENTS
We thank Dr. Toshikazu Hamasaki for the helpful suggestions and Dr. Mitchell Bernstein and Dr. Nithila Isaac for initial encouragement with this project. Aashir Awan, Alok Prasad, and Claus A.F. Andersen provided valuable assistance. Many individuals, acknowledged in the text, provided special reagents. The work was supported in part by grants from the National Institute of Diabetes, Digestive and Kidney Diseases (DK41918 and DK41296). Charles Guerra was a student in the Sue Golding Graduate Division, supported by National Cancer Institute training grant CA 09475. Dr. Vagn Leick was supported by the Danish Research Council for Natural Sciences.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0298. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0298.
REFERENCES
- Asai DJ, Forney JD. Tetrahymena thermophila. In: Methods in Cell Biology. Vol. 62. San Diego, CA: Academic Press; 2000. p. 580. pps. [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskel. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Guerra C, Wada Y, Valentin T, Angeletti RH, Satir P, Hamasaki T. A regulatory light chain of ciliary outer arm dynein in Tetrahymena thermophila. J Biol Chem. 2001;276:20048–20054. doi: 10.1074/jbc.M008412200. [DOI] [PubMed] [Google Scholar]
- Cohen J, Beisson J. The Cytoskeleton. In: Görtz HD, editor. Paramecium. Vol. 21. Heidelberg: Springer Verlag; 1988. pp. 363–391. [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, McGrath KE, Callahan RC, Gorovsky MA. Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single alpha- and beta-tubulin. Cell Motil Cytoskel. 1993;25:243–53. doi: 10.1002/cm.970250305. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Klotz C, Vigues B, Rutin J, Beisson J. Ca2+-binding proteins and contractility of the infraciliary lattice in Paramecium. Biol Cell. 1991;71:217–225. [Google Scholar]
- Goodenough U. Motile detergent–extracted cells of Tetrahymena and Chlamydomonas. J Cell Biol. 1983;96:1610–1621. doi: 10.1083/jcb.96.6.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973;20:19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Hamasaki T, Holwill ME, Barkalow K, Satir P. Mechanochemical aspects of axonemal dynein activity studied by in vitro microtubule translocation. Biophys J. 1995;69:2569–2579. doi: 10.1016/S0006-3495(95)80128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Mengersen A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with CaM and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Jenkins LM, Nelson EM, Williams NE, Jaeckel-Williams R, Frankel J. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev Biol. 1995;169:644–661. doi: 10.1006/dbio.1995.1176. [DOI] [PubMed] [Google Scholar]
- Klotz C, Garreau de Loubresse N, Ruiz F, Beisson J. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil Cytoskel. 1997;38:172–186. doi: 10.1002/(SICI)1097-0169(1997)38:2<172::AID-CM6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, Bornens M, Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113:1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- Larsen J, Barkalow K, Hamasaki T, Satir P. Structural and functional characterization of Paramecium dynein: initial studies. J Protozool. 1991;38:55–61. doi: 10.1111/j.1550-7408.1991.tb04801.x. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Beck JC, Finkbeiner WE. Differential regulation of centrin genes during ciliogenesis in human tracheal epithelial cells. Am J Physiol. 1998;275:L1145–L1156. doi: 10.1152/ajplung.1998.275.6.L1145. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leick V, Koppelhus U, Rosenberg J. Cilia-mediated oriented chemokinesis in Tetrahymena thermophila. J Euk Microbiol. 1994;41:546–553. doi: 10.1111/j.1550-7408.1994.tb01515.x. [DOI] [PubMed] [Google Scholar]
- Lutz W, Lingle WL, McCormick D, Greenwood TM, Salisbury JL. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceeding centrosome duplication. J Biol Chem. 2001;276:20774–20780. doi: 10.1074/jbc.M101324200. [DOI] [PubMed] [Google Scholar]
- Maciejewski JJ, Vacchiano EJ, McCutcheon SM, Buhse HE., Jr Cloning and expression of a cDNA encoding a Vorticella convallaria spasmin: an EF hand calcium-binding protein. J Euk Microbiol. 1999;46:165–73. doi: 10.1111/j.1550-7408.1999.tb04601.x. [DOI] [PubMed] [Google Scholar]
- Madeddu L, Klotz C, Le Caer JP, Beisson J. Characterization of centrin genes in Paramecium. Eur J Biochem. 1996;238:121–128. doi: 10.1111/j.1432-1033.1996.0121q.x. [DOI] [PubMed] [Google Scholar]
- Maihle NJ, Satir BH. Calmodulin in the ciliates Paramecium tetraurelia and Tetrahymena thermophila. Ann NY Acad Sci. 1980;356:408–409. doi: 10.1111/j.1749-6632.1980.tb29650.x. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Onimaru H, Ohki K, Nozawa Y, Naitoh Y. Electrical properties of Tetrahymena, a suitable tool for studies on membrane excitation. Proc Jpn Acad. 1980;56:538–543. [Google Scholar]
- Routledge LM. Calcium-binding proteins in the vorticellid spasmoneme. J Cell Biol. 1978;77:358–370. doi: 10.1083/jcb.77.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Baron AT, Surek B, Melkonian M. Stritiated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL. Centrin, centrosomes, and meiotic spindle bodies. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1989;108:1751–1760. doi: 10.1083/jcb.108.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir B, Sale WS, Satir P. Membrane renewal after dibucaine deciliation of Tetrahymena. Freeze-fracture technique, cilia, membrane structure. Exp Cell Res. 1976;97:83–91. doi: 10.1016/0014-4827(76)90657-1. [DOI] [PubMed] [Google Scholar]
- Satir P. Mechanisms of ciliary motility: an update. Eur J Protistol. 1998;34:267–272. [Google Scholar]
- Schliwa M, van Blerkom J. Structural interaction of cytoskeletal components. J Cell Biol. 1981;90:222–235. doi: 10.1083/jcb.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemasa T, Ohnishi K, Kobayashi T, Takagi T, Konishi K, Watanabe Y. Cloning and sequencing of the gene for Tetrahymena calcium-binding 25-kDa protein (TCBP-25) J Biol Chem. 1989;264:19293–19301. [PubMed] [Google Scholar]
- Takemasa T, Takagi T, Kobayashi T, Konishi K, Watanabe Y. The third calmodulin family protein in Tetrahymena. Cloning of the cDNA for Tetrahymena calcium-binding protein of 23 kDa (TCBP-23) J Biol Chem. 1990;265:2514–2517. [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain M, et al. De novo design of peptides targeted to the EF hands of calmodulin. J Biol Chem. 2000;275:2676–2685. doi: 10.1074/jbc.275.4.2676. [DOI] [PubMed] [Google Scholar]
- Wada Y, Hamasaki T, Satir P. Evidence for a novel affinity mechanism of motor-assisted transport along microtubules. Mol Biol Cell. 2000;11:161–169. doi: 10.1091/mbc.11.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–1325. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]