Abstract
RNA binding proteins often contain multiple arginine glycine repeats, a sequence that is frequently methylated by protein arginine methyltransferases. The role of this posttranslational modification in the life cycle of RNA binding proteins is not well understood. Herein, we report that Sam68, a heteronuclear ribonucleoprotein K homology domain containing RNA binding protein, associates with and is methylated in vivo by the protein arginine N-methyltransferase 1 (PRMT1). Sam68 contains asymmetrical dimethylarginines near its proline motif P3 as assessed by using a novel asymmetrical dimethylarginine-specific antibody and mass spectrometry. Deletion of the methylation sites and the use of methylase inhibitors resulted in Sam68 accumulation in the cytoplasm. Sam68 was also detected in the cytoplasm of PRMT1-deficient embryonic stem cells. Although the cellular function of Sam68 is unknown, it has been shown to export unspliced human immunodeficiency virus RNAs. Cells treated with methylase inhibitors prevented the ability of Sam68 to export unspliced human immunodeficiency virus RNAs. Other K homology domain RNA binding proteins, including SLM-1, SLM-2, QKI-5, GRP33, and heteronuclear ribonucleoprotein K were also methylated in vivo. These findings demonstrate that RNA binding proteins are in vivo substrates for PRMT1, and their methylation is essential for their proper localization and function.
INTRODUCTION
Protein arginine methylation is a common posttranslational modification in higher eukaryotes, but its precise role is not well understood. Arginine methylation has been shown to affect several cellular processes, including intracellular localization, protein–protein interactions as well as transcription (Gary and Clarke, 1998; McBride and Silver, 2001; Stallcup, 2001). In yeast, Npl3p, Hrp1p, and Nab2p are shuttling proteins involved in the transport of mRNAs across the nuclear membrane (Anderson et al., 1993; Lee et al., 1996). Arginine methylation of these two heteronuclear ribonucleoproteins (hnRNPs) is required for their nuclear export (Shen et al., 1998; McBride et al., 2000; Green et al., 2002). We have recently shown that arginine methylation negatively affects the binding of proline-rich ligands to Src homology 3 (SH3), but not WW, domain protein modules (Bedford et al., 2000). The methylation of STAT1 is required for transcriptional activation by IFNα/β by displacing PIAS1 (Mowen et al., 2001). Arginine methylation can also have a positive effect on protein–protein interactions. The spliceosomal snRNP proteins SmD1, D3 and B/B′ contain symmetrically dimethylated arginines (sDMAs), and this modification is necessary for enhanced interaction with SMN (Brahms et al., 2000, 2001; Friesen et al., 2001a,b), the product of the spinal muscular atrophy gene (Lefebvre et al., 1995). Transcription can also be regulated by arginine methylation through the modification of histones and cofactors (Chen et al., 1999a; Wang et al., 2001; Xu et al., 2001).
The enzymes responsible for protein arginine methylation have been classified in three groups (Gary and Clarke, 1998). Type I enzymes promote the formation of both NG-monomethylated and asymmetric ω-NG,NG-dimethylated arginines (aDMAs). Type II enzymes catalyze the formation of monomethylated and sDMAs. The type III enzyme found in yeast catalyzes the monomethylation of the δ-guanidino nitrogen atom of the arginine residue. The known mammalian type I enzymes are protein arginine N-methyltransferase (PRMT)1, PRMT3, coactivator-associated arginine methyltransferase 1 (CARM1), and PRMT6 (Lin et al., 1996; Tang et al., 1998; Schurter et al., 2001; Frankel et al., 2002). These PRMTs are ubiquitously expressed and may serve multiple functions. PRMT1 is thought to be the major PRMT accounting for >90% of the generation of aDMAs (Tang et al., 2000) and has been shown to be required for early mouse postimplantation development (Pawlak et al., 2000). PRMT1 was first isolated through its interaction with BTG1 and TIS21, two genes associated with cell quiescence (Lin et al., 1996). Most of the proteins identified as being targets of PRMT1 are methylated within glycine-arginine rich (GAR) domains (Najbauer et al., 1993). Many proteins involved in RNA metabolism contain such regions with clustered arginine residues in an Arg-Gly-Gly motif (RGG box) or RG repeats (Burd and Dreyfuss, 1994). Heteronuclear ribonucleoproteins (hnRNPs) represent a major protein family that contains methylated arginines (Liu and Dreyfuss, 1995).
The K homology (KH) motif is a type of RNA binding domain found in a large family of proteins associated with RNA metabolism (Gibson et al., 1993; Siomi et al., 1993). A subfamily of KH domain proteins contains an extended KH domain in a larger protein module called the GRP33, Sam68, and GLD-1 (GSD) domain (Jones and Schedl, 1995). This family of proteins includes the Src substrate associated in mitosis of 68 kDa (Sam68; Wong et al., 1992; Fumagalli et al., 1994; Taylor and Shalloway, 1994), Sam68 like mammalian protein 1 and 2 (SLM-1 and SLM-2; Di Fruscio et al., 1999), Artemia salina glycine rich protein (GRP33), Caenorhabditis elegans germ-line defective 1 (GLD-1; Jones and Schedl, 1995), and QUAKING (QKI; Ebersole et al., 1996). Although the cellular function of Sam68 is unknown, it can functionally substitute for Rev in the export of unspliced human immunodeficiency virus (HIV) RNAs (Reddy et al., 1999). The majority of KH domain RNA binding proteins contain RGG boxes and RG-rich regions, making them potential arginine methyltransferase targets.
Herein, we report that Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K are methylated in vivo. Sam68 is asymmetrically methylated by PRMT1 in vivo, as assessed by using a novel antiasymmetrically dimethylated arginine antibody and ES cells harboring a prmt1 null mutation. At steady state, Sam68 is predominantly nuclear, with no cytoplasmic staining (Sam68/SLM nuclear bodies [SNBs]; Chen et al., 1999b). In PRMT1-deficient cells or cells treated with methylase inhibitors a significant fraction of Sam68 resided in the cytoplasm. The methylation of Sam68 is required to enhance the export of unspliced HIV RNAs. Our findings demonstrate the necessity of arginine methylation in the localization and function of KH domain RNA binding proteins.
MATERIALS AND METHODS
Antibodies
Peptides were synthesized at W.M. Keck Biotech Resource Center (New Haven, CT). Polyclonal antibodies were generated by using New Zealand White rabbits injected with peptides coupled to keyhole limpet hemocyanin (Sigma-Aldrich, St. Louis, MO). The peptides used to generate antibodies against asymmetrically dimethylated arginines (UBI, Upstate USA, Waltham, MA) and PRMT1 were as follows: peptide 24 (KGRGRGRGRGPPPPPRGRGRGRG), where all arginine residues are asymmetrically dimethylated, and PRMT1 (KTGEEIFGTIGMRPNAKNNRD). The 9E10 anti-myc monoclonal antibody (mAb) was from the American Type Culture Collection (Manassas, VA). The polyclonal antibodies were affinity purified over the antigenic peptide coupled to Affigel beads, eluted with 100 mM glycine pH 2.5, and concentrated by using Centricon columns (Millipore, Bedford, MA). The anti-Sam68 polyclonal antibody AD-1 has been described previously (Chen et al., 1999b). The anti-SMN mAb was purchased from Transduction Laboratories (Lexington, KY).
Enzyme-linked Immunosorbent Assay (ELISA)
ELISA plates (Costar, Cambridge, MA) were coated with the indicated quantity of peptide in 50 μl of 50 mM carbonate buffer, incubated at 37°C for 30 min, and blocked with blocking buffer (1% bovine serum albumin, 5% sucrose in phosphate-buffered saline [PBS]). Primary antibodies were diluted at 1:1000 in dilution buffer (1% bovine serum albumin, 0.5% ovalbumin, 10 mM Tris pH 7.4, 150 mM NaCl) and added to the corresponding well followed by incubation at 37°C for 30 min. The plate was washed extensively with PBS containing 0.1% Tween. Goat anti-rabbit antibodies covalently coupled to horseradish peroxidase (Cappel Laboratories, Durham, NC) was incubated at 1:1000 in dilution buffer at 37°C for 15 min. The plate was washed and developed using BM Blue POD substrate (Roche Diagnostics, Indianapolis, IN) and quantitated by using spectrophotometry at 405 nm.
DNA Constructs
PRMT1 was amplified from a mouse cDNA library by using EcoRI oligonucleotides (5′-GGA GAA TTC CTG TGG CCA GGC GGA AAG-3′) and (5′-CCG GAA TTC AGC GCA TCC GGT AGT CGG-3′), subcloned into myc-pcDNA (Chen et al., 1997) and verified by DNA sequencing. The T7 promoter expression vectors encoding Myc-epitope–tagged Sam68, SΔN, SΔN:Δ280–339, SΔN:Δ294–339, SΔN:ΔC, QKI-5, GRP33, SLM-1, SLM-2, and hnRNP K have been described previously (Chen et al., 1997; Di Fruscio et al., 1999). Mammalian expression vectors for SΔN:Δ280–339, SΔN:Δ294–339, and SΔN:ΔC were generated by subcloning the EcoRI fragment in myc-pcDNA. The Sam68- and PRMT1-GST fusion protein were expressed in bacteria by subcloning the full-length EcoRI fragment from myc-Sam68 into pGEX-KG.
Protein Expression
For Figures 1, B and C, 2B, and 3E, proteins were expressed in HeLa cells by using the vaccinia virus T7 expression system and lysed in a 1% Triton-based lysis buffer as described previously (Chen et al., 1997). In other cases, HeLa cells were transfected with LipofectAMINE PLUS (Invitrogen, Carlsbad, CA) according to manufacturer protocol. For immunoprecipitations, cell lysates were incubated on ice with the primary antibody for 1 h. Then 20 μl of a 50% protein A-Sepharose slurry was added and incubated at 4°C for 30 min with constant end-over-end mixing. The beads were washed twice with lysis buffer and once with PBS. Protein samples were analyzed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Immunoblotting was performed using the anti-myc (9E10) or anti-PRMT1 antibodies. The designated primary antibody was followed by goat anti-mouse or goat anti-rabbit antibodies conjugated to horseradish peroxidase (ICN Pharmaceuticals, Costa Mesa, CA) and chemiluminescence was used for protein detection (DuPont, Wilmington, DE).
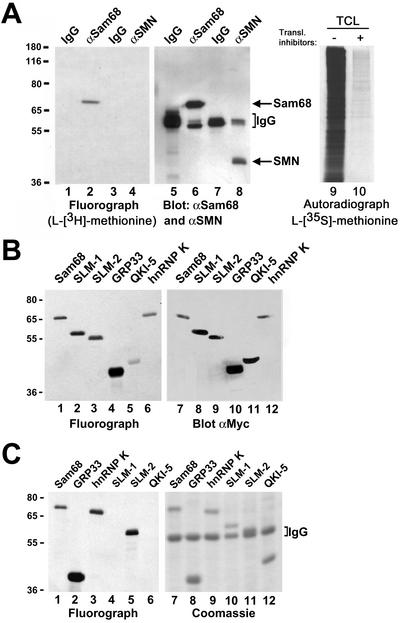
Figure 1.
In vivo methylation of Sam68 and other KH domain RNA binding proteins. (A) In vivo methylation labeling was performed by metabolically labeling cells with [l-[methyl-3H]methionine for 3 h in the presence of cycloheximide and chloramphenicol. The cell lysates were immunoprecipitated with anti-Sam68 (mAb 7-1) and anti-SMN antibodies. The 3H-labeled proteins were separated by SDS-PAGE and visualized by fluorography (lane 1–4; exp. 24 h), and the proteins were visualized by immunoblotting with anti-Sam68 and anti-SMN antibodies (lanes 5–8). The migration of Sam68 and SMN is shown. The heavy chain of IgG is indicated with a bracket. TCLs of HeLa cells metabolically labeled with l-[35S]methionine in the absence (lane 9; −) or presence (lane 10; +) of protein synthesis inhibitors were separated by SDS-PAGE and visualized by autoradiography (exp. 24 h). (B) Metabolic labeling for the in vivo methylation was performed 24 h after transfection of myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K expression vectors in HeLa cells. The cell lysates were immunoprecipitated with anti-myc antibodies and the bound 3H-labeled proteins were separated by SDS-PAGE and visualized either by fluorography (lanes 1–6; exp. 12 h) or by immunoblotting with the anti-myc antibody (lanes 7–12). (C) Endogenous protein methyltransferase activity coimmunoprecipitates with KH domain-containing proteins. HeLa cells were transfected with myc-epitope–tagged Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K. The cells were lyzed and myc immunoprecipitations were incubated with [3H-methyl]S-adenosyl-l-methionine in vitro. The immunoprecipitated proteins were visualized by Coomassie staining (lanes 7–12) or by fluorography (lanes 1–6; exp. 6 h). The migration of the heavy chain of IgG is indicated.
Mass Spectrometry
Endogenous Sam68 was immunopurified from 5 × 108 HeLa cells by using 1 mg of polyclonal anti-Sam68 antibody (AD1) coupled to 1 g of protein A-Sepharose (Sigma-Aldrich). After extensive washings with lysis buffer, 1× PBS, the bound proteins were eluted with 500 μl of 1× PBS containing 250 μM of the immunogenic Sam68 peptide. Eluted proteins were resolved by SDS-PAGE and revealed by Coomassie Blue R-250 staining. The apparent band at 68 kDa was excised, in-gel digested with trypsin, and sent to the University of Calgary Mass Spectrometry Proteomics Facility for MALDI-TOF analysis on a Voyager DE-STR mass spectrometer.
Methylation Assays
Myc-tagged proteins (PRMT1, Sam68, SLM-1, SLM-2, GRP33, hnRNP K, or QKI-5) were expressed in HeLa cells and immunoprecipitated with anti-myc antibodies. The immune complex was incubated with 0.55 μCi of [methyl-3H]S-adenosyl-l-methionine (3H-SAM; PerkinElmer Life Sciences, Boston, MA) in the presence of 25 mM Tris-HCl at pH 7.5 for 1 h at 37°C in a final volume of 30 μl. Reactions were stopped by adding 20 μl of 2× SDS-PAGE sample buffer, followed by heating at 100°C for 10 min. Samples were loaded on 10% SDS-polyacrylamide gels and stained with Coomassie Blue. The destained gels were soaked in EN3HANCE (PerkinElmer Life Sciences) according to manufacturer instructions and visualized by fluorography. In vivo methylation labeling was performed by metabolically labeling cells with l-[methyl-3H]methionine directly in methionine-free medium for 3 h in the presence of cycloheximide and chloramphenicol as described previously (Liu and Dreyfuss, 1995). l-[35S]methionine was also used as a control under the same conditions. The cell lysates were immunoprecipitated and the 3H-labeled proteins were visualized by fluorography. ES cells were obtained from Earl H. Ruley (Vanderbilt University, Nashville, TN) and were cultured in the absence of a feeding layer in DMEM (no. 11960-044; Invitrogen) supplemented with 15% fetal bovine serum, 2% l-glutamine, 1.2% minimal essential medium nonessential amino acids (no. 11140-050; Invitrogen), 1.2% Pen/Strep, 1× 2-β-mercaptoethanol, and 0.5 ng/ml leukemia inhibitory factor.
Immunofluorescence Confocal Microscopy
HeLa cells were cultured directly on coverslips in a six-well dish. Cells plated on coverslips were incubate for 24 h with the vehicle (DMSO) or with the methyltransferase inhibitor adenosine-2′,3′-dialdehyde (AdOx) (Sigma-Adrich) at a final concentration of 1 mM. Transfection of HeLa cells for immunofluorescence was achieved using LipofectAMINE PLUS (Invitrogen) according to the manufacturer protocol by using 2 μg of DNA. Cells were fixed with 1% paraformaldehyde in 1× PBS at pH 7.4 for 5 min at room temperature (RT) and permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT. The cells were incubated with the primary antibodies at RT for 1 h in PBS. The cells were washed with 0.1% Triton X-100 in PBS and incubated with the appropriate secondary antibodies in PBS. Goat anti-rabbit coupled to Alexa 488 (Molecular Probes, Eugene, OR) and goat anti-mouse coupled to Alexa 546 (Molecular Probes) were used as secondary antibodies. The cells were washed again, mounted onto glass slides, and visualized with a confocal microscope (Carl Zeiss, Thornwood, NY).
Rev Assays
COS cells were transfected with a total of 1 μg of DNA containing 0.2 μg of Rev response element (RRE) chloramphenicol acetyltransferase (CAT) reporter plasmid pDM128, 0.7 μg of Rev expression vector B1-SVH6Rev, 0.7 μg of Rev mutant B1-SVH6RevM10, or 0.7 μg of pcDNA-Sam68, along with increasing concentrations of AdOx (0, 100, and 250 μM). The β-galactosidase expression vector pCH110 (0.1 μg; Amersham Biosciences, Piscataway, NJ) was included in all transfections for measuring the efficiency of transfection. Forty-eight hours after transfection, the cells were collected and resuspended in 150 μl of 0.25 M Tris-HCl, pH 7.8. The cell extracts were prepared by three freeze-thaw cycles, followed by a brief centrifugation to remove cell debris. CAT activity was normalized to the β-galactosidase activity and did not exceed twofold.
RESULTS
Arginine Methylation of Sam68 and Other KH Domain RNA Binding Proteins
Metabolic labeling of HeLa cells by using l-[methyl-3H]methionine was performed in the presence of translation inhibitors to examine whether Sam68 was methylated in vivo. The presence of translation inhibitors ensured that the labeling was due to posttranslational methylation, not a low level of translational incorporation of l-[methyl-3H]methionine in newly synthesized proteins (Liu and Dreyfuss, 1995). Sam68 and a control known not to be methylated, SMN, were immunoprecipitated from cells that were metabolically labeled. The immunoprecipitated proteins were resolved by SDS-PAGE and visualized by fluorography. The fluorograph revealed that Sam68 was labeled in HeLa cells and that SMN was not (Figure 1A, lanes 1–4). Immunoblotting with anti-Sam68 and anti-SMN antibodies confirm that Sam68 and SMN were immunoprecipitated, respectively (Figure 1A, lane 5–8). Performing the metabolic labeling with l-[35S]methionine in the presence of protein synthesis inhibitors revealed that virtually no label is incorporated under these conditions (Figure 1A, lane 10). This control verifies that the protein synthesis inhibitors are functioning properly. These findings demonstrate that Sam68 is methylated in vivo.
Many other KH domain RNA binding proteins contain RG-rich sequences that are potential sites of methylation by PRMTs. The ability of other KH domain RNA binding proteins to become methylated in HeLa cells was examined by a transient transfection assay followed by metabolic labeling with l-[methyl-3H]methionine. Expression vectors encoding myc-epitope–tagged Sam68 (positive control), SLM-1, SLM-2, GRP33, the nuclear QKI-5 isoform, and hnRNP K were transfected in HeLa cells. Anti-myc immunoprecipitates were examined for incorporation of 3H-methyl groups (Figure 1B, lanes 1–6) and protein expression was assessed by immunoblotting total cell lysates (Figure 1B, lanes 7–12). Sam68, SLM-1, SLM-2, GRP33, QKI-5, and hnRNP K were methylated (Figure 1B, lanes 1–6). These findings suggest that many KH domain proteins are methylated in vivo.
PRMTs are known to frequently associate with their substrates (Gary and Clarke, 1998). The ability of KH domain proteins to coimmunoprecipitate PRMT activity was examined in myc immunoprecipitates of cells transiently transfected with myc-epitope–tagged KH domain RNA binding proteins. Anti-myc immunoprecipitates were incubated in the presence of [methyl-3H]S-adenosyl-l-methionine and the ability of the KH domain RNA binding proteins to serve as an exogenous substrates was examined. The proteins were resolved by SDS-PAGE, stained with Coomassie Blue for protein expression (Figure 1C, lanes 7–12), and the methylated proteins were visualized by fluorography (Figure 1C, lanes 1–6). Sam68, GRP33, hnRNP K, and SLM-2 coimmunoprecipitated protein methyltransferase activity, whereas SLM-1 and QKI-5 did not (Figure 1C, lanes 1–6). The reason for the absence of protein methyltransferase activity in SLM-1 and QKI-5 is unknown. Our findings demonstrate that Sam68, GRP33, hnRNP K, and SLM-2 are methylated in vitro by associated protein methyltransferases.
Association of Sam68 and Other KH Domain RNA Binding Proteins with PRMT1
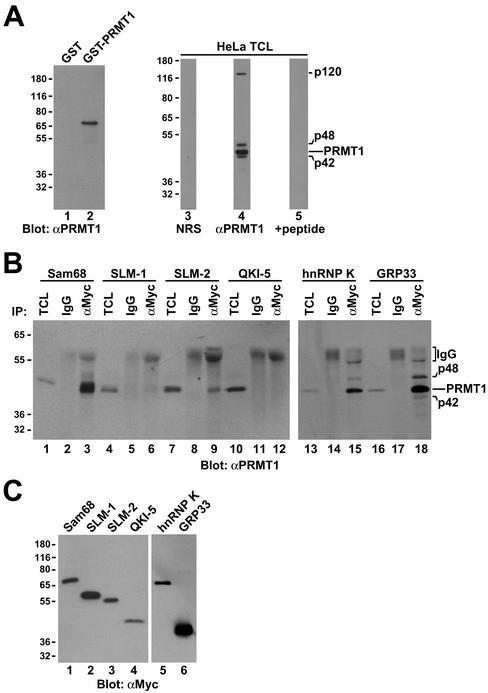
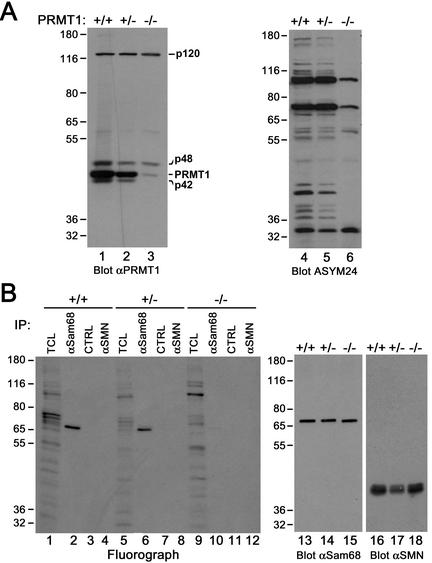
We have previously reported that the RG repeats flanking the P3 and P4 domains in Sam68 are asymmetrically dimethylated by glutathione S-transferase (GST)-PRMT1 in vitro (Bedford et al., 2000). To determine whether the protein methyltransferase activity associated with KH-containing RNA binding proteins could be attributed to endogenous PRMT1, we carried out coimmunoprecipitation experiments. A polyclonal PRMT1 antibody was generated using a peptide derived from a unique sequence of PRMT1. The antibody recognized recombinant GST-PRMT1, but not GST by immunoblotting (Figure 2A). The antibody recognized a major protein species of 45 kDa in HeLa cells, corresponding to the predicted molecular mass of human PRMT1 (Figure 2A, lane 4). The antibody also recognized proteins with molecular masses of 120, 48, and 42 kDa that were competed away using the immunogenic peptide (Figure 2A, lane 5). The p48 and p42 may represent PRMT1 isoforms, however, the identity of p120 is unknown.
Figure 2.
Association of Sam68 and other KH domain RNA binding proteins with PRMT1. (A) Purified recombinant GST or GST-PRMT1 were separated by SDS-PAGE and immunoblotted with anti-PRMT1 antibodies (lanes 1 and 2). The band at ∼70 kDa represents GST-PRMT1. HeLa cell extracts were separated by SDS-PAGE and immunoblotted with NRS, anti-PRMT1 antibodies, and anti-PRMT1 immunoabsorbed with the antigenic peptide. The migration of the four immunogenic proteins (p120, p48, p45, and p42) recognized by anti-PRMT1 antibodies is indicated on the right (lanes 3–5). The predominant band of ∼45 kDa most likely represents PRMT1 as predicted. The migration of the molecular mass markers in kilodaltons is shown on the left. (B) Myc-tagged Sam68, SLM-1, SLM-2, GRP33, hnRNP K, and QKI-5 were expressed in HeLa and immunoprecipitated with control IgG or anti-myc antibodies. Aliquots of TCLs (10% input) and the immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted with anti-PRMT1 antibodies. The migration of the PRMT1 immunogenic proteins and the heavy chain of IgG are indicated. Longer exposure of lanes 1–12 also reveals p42 and p48 (our unpublished data). (C) Equivalent expression of myc-epitope–tagged KH domain RNA proteins was confirmed by immunoblotting an aliquot of TCLs.
Myc-tagged Sam68, SLM-1, SLM-2, QKI-5, hnRNP K, and GRP33 were expressed in HeLa cells and cell lysates were immunoprecipitated with control mouse IgG or anti-myc antibodies. The presence of coimmunoprecipitating PRMT1 was detected by immunoblotting with anti-PRMT1 antibodies (Figure 2B). Endogenous PRMT1 coimmunoprecipitated with Sam68, SLM-2, hnRNP K, and GRP33 (Figure 2B, lanes 3, 9, 15, and 18). P120, p48, and p42 were visible in total cell lysates (TCL) and anti-myc immunoprecipitations upon longer exposures (our unpublished data). Little or no PRMT1 was detected in SLM-1 and QKI-5 immunoprecipitates (Figure 2B, lanes 6 and 12), consistent with Figure 1C. Equivalent expression of the myc-tagged proteins was assessed by immunoblotting with anti-myc antibodies (Figure 2C). These findings show that the KH domain RNA binding proteins Sam68, SLM-2, hnRNP K, and GRP33 interact with endogenous PRMT1.
Endogenous and Transfected Sam68 Contain Asymmetrical Dimethylated Arginines
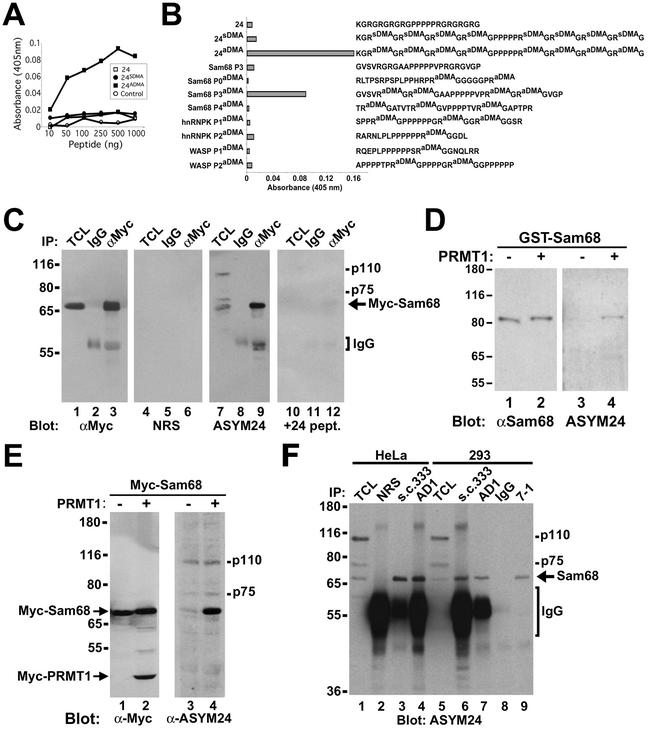
The association of Sam68 with PRMT1, a protein methyltransferase that mediates the generation of aDMAs (Lin et al., 1996), suggests that Sam68 may contain aDMA in vivo. To examine whether Sam68 contained aDMAs in vivo, we generated an antibody that recognizes aDMA-containing polypeptides (see MATERIALS AND METHODS). Rabbits were immunized with peptide #24aDMA covalently coupled to keyhole limpet hemocyanin. The affinity-purified polyclonal antibody, named ASYM24, recognized strongly the aDMA-containing peptide #24aDMA, but showed no reactivity to the unmethylated peptide #24 or to a symmetrical dimethylarginine-containing peptide #24sDMA (Figure 3, A and B; peptides 24aDMA, 24 and 24sDMA, respectively). We examined whether ASYM24 could also recognize any peptide containing aDMA. ASYM24 was generated using a peptide with neighboring proline motifs. Three proteins known to have this type of arrangement are Sam68, hnRNP K, and Wiscott-Aldrich Syndrome protein (WASP). Peptides from these proteins were generated containing aDMA and tested by ELISA with ASYM24. ASYM24 only recognized a peptide containing aDMAs corresponding to the proline motif P3 of Sam68 (Figure 3B). The unmethylated Sam68 P3 peptide was not recognized (Figure 3B). These results demonstrate that ASYM24 is an aDMA-specific antibody that has selectivity toward some polypeptides containing aDMA and recognizes a methylated sequence neighboring the Sam68 proline motif P3.
Figure 3.
Sam68 is asymmetrically dimethylated in vivo as detected by using novel anti-aDMA-specific antibodies. (A) Generation of an anti-aDMA antibody. Peptide (24aDMA; see B for sequence) was used as an antigenic peptide to inject rabbits. The specificity of the purified polyclonal antibody (ASYM24) was examined by ELISA with a control unrelated peptide (AD1 peptide), unmethylated “backbone” peptide 24, or an identical peptide containing symmetrical dimethylated arginines (24sDMA). The quantity of peptide bound to the plate was plotted against the immunoreactivity of the ASYM24 antibody. Each point represents the average of three separate experiments. (B) Specificity of the antibody was examined by using a variety of unrelated aDMA containing peptides with neighboring proline-rich sequences. One microgram of the indicated peptides was coated per ELISA well. WASP P1 and P2 denote the Wiscott-Aldrich Syndrome protein proline motifs 1 and 2 containing aDMA. hnRNPK P1 and P2 denote hnRNPK proline motifs 1 and 2 containing aDMA. Sam68 P0, P3, and P4 denote the Src substrate activated in mitosis of 68-kDa proline motifs 0, 3, and 4 containing aDMA. The unmethylated P3 peptide was also used as control (Sam68 P3). Each bar denotes greater than n > 8 from three separate experiments. (C) Myc-tagged Sam68 was transfected in HeLa cells and immunoprecipitated with control (IgG) or anti-Myc antibodies. Aliquots of TCLs (10% input) and the bound proteins were separated by SDS-PAGE and visualized by immunoblotting with anti-myc, NRS, ASYM24, and ASYM24 antibodies absorbed with the antigenic peptide (+24 pept.). The migration of Sam68, the heavy chain of IgG, and endogenous proteins (p110 and p75) is indicated. (D) GST-Sam68 fusion protein was either left untreated (−) or incubated with GST-PRMT1 under in vitro methylation conditions in the presence of “cold” S-adenosyl-l-methionine. An aliquot of each reaction was resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-Sam68 AD1 antibodies (lanes 1 and 2) and ASYM24 (lanes 3 and 4). The molecular mass markers are shown on the left in kilodaltons. (E) HeLa cells were cotransfected with expression vectors encoding myc-Sam68 and myc-PRMT1. The cells were lysed in sample buffer and the proteins separated by SDS-PAGE and immunoblotted with either anti-myc or ASYM24 antibodies. The migration of myc-PRMT1, myc-Sam68, p110, and p75 is indicated. (F) HeLa cells or HEK293 (293) cells were lysed and immunoprecipitated with control antibodies (NRS and mouse IgG) and anti-Sam68 antibodies (pAb s.c.333, pAb AD1, mAb 7-1). The proteins were separated by SDS-PAGE and immunoblotted with ASYM24. The migration of Sam68, the heavy chain of IgG, p110, and p75 is shown on the right. The molecular mass markers are shown on the left.
To determine whether full-length Sam68 is recognized by ASYM24, myc-tagged Sam68 was expressed in HeLa cells. Proteins from cell lysates were immunoprecipitated with control IgG or anti-myc antibodies. Immunoprecipitated Sam68 was indeed recognized by ASYM24 but not by normal rabbit serum (NRS) or ASYM24 preadsorbed with the immunogenic peptide (+24 pept.; Figure 3C, lanes 4–12). ASYM24 also recognized unknown endogenous proteins in HeLa cells with molecular masses of 110 and 75 kDa (lane 7). Immunoblotting with anti-myc antibodies was carried out to control for protein expression and efficiency of the immunoprecipitation procedure (Figure 3C, lanes 1–3). To examine whether PRMT1 could increase the ASYM24 immunoreactivity of Sam68, recombinant GST-Sam68 was purified from bacteria, an organism that does not contain arginine methyltransferase activity (Gary and Clarke, 1998). The GST-Sam68 fusion protein was left untreated (mock) or methylated in vitro with purified GST-PRMT1 in the presence of unlabeled methyl donor S-adenosyl-l-methionine. The level of methylation was examined by immunoblotting with the aDMA-specific antibody ASYM24 (Figure 3D). ASYM24 recognized the methylated GST-Sam68, but not the unmethylated GST-Sam68 (Figure 3D, lanes 3 and 4). Anti-Sam68 AD1 antibody confirmed equal levels of GST-Sam68 (Figure 3D, lanes 1 and 2). To verify that this was also true in vivo, PRMT1 was coexpressed with Sam68 in HeLa cells. Again, the expression of PRMT1 dramatically increased the ability of Sam68 to be recognized by ASYM24 (Figure 3E, compare the 68-kDa protein in lanes 3 and 4), further suggesting that PRMT1 is able to methylate Sam68 in vivo. Finally, we wanted to address whether endogenous Sam68 contained aDMAs. Lysates were prepared from HeLa and HEK293 cells and immunoprecipitations were performed with three distinct anti-Sam68 antibodies (polyclonal sc333, AD1, and monoclonal 7-1). Immunoblotting revealed that endogenous Sam68 from HeLa and human embryonic kidney (HEK)293 is also recognized by ASYM24 (Figure 3F, lanes 1–9), suggesting that the arginines neighboring the proline motif P3 of Sam68 are asymmetrically dimethylated in vivo. The endogenous bands of 110 and 75 kDa recognized by ASYM24 did not coimmunoprecipitate with Sam68 and are likely to represent other asymmetrically dimethylated proteins in the cell (Figure 3F).
To determine whether other regions of Sam68 were methylated, matrix-assisted laser desorption ionization/time of flight (MALDI-TOF) mass spectrometry was carried out with endogenous Sam68 purified from HeLa cells. Three domains of Sam68 harbor arginine-glycine motifs that could potentially be methylated (Table 1; P0, P3, and P4). By knowing that methylation of arginines prevents cleavage by trypsin (Baldwin and Carnegie, 1971), predicted peptide sizes that would result from monomethylated or dimethylated arginines were calculated and compared with the peptide masses identified by MALDI-TOF mass spectrometry. Arginines at position 45 and 52 were found to be dimethylated (Table 1). However, some peptides contained dimethylated R45 but unmodified R52, perhaps indicating that R45 is methylated first, followed by R52. No peptides containing only monomethylated arginines were identified for this region. These two arginines are the only arginines followed by two glycines, which is characteristic of the RGG motif found in hnRNPs. Arginines 310, 315, 320, and 325, which are located in between the proline motifs P3 and P4 were found to be monomethylated (Table 1). No peptides containing dimethylated arginines were identified for this region, indicating that those arginines are mostly found monomethylated endogenously. Interestingly, these arginines are each distanced by exactly four amino acids and are part of a distinct RG(A/T)XV motif. Finally, arginine 304, the second arginine in the RGRG motif downstream of the proline motif P3, was found to be dimethylated (but never monomethylated), whereas arginine 302 was unmodified. The absence of peptides containing methylated arginines from the region upstream of proline motif P3 is likely due to technical limitations (i.e., inability to detect very small or very large peptides). Overall, this analysis has revealed that Sam68 is endogenously methylated at least on seven of the 14 arginines found in an arginine-glycine context.
Table 1.
| P0 Region |
| 1MQRRDDPAARMSRSSGRSGSMDPSGAHPSVRQTPSRQPPLPHRSRGGGGGSRGG54 |
| P1 Region P2 Region |
| 55ARASPATQPPPLLPPSATGPDATVGGPAPTPLLPPSATASVKMEPENKYLPELMA109 |
| 110EKDSLDPSFTHAMQLLTAEIEKIQKGDSKKDDEENYLDLFSHKNMKLKERVLIPV164 |
| 165KQYPKFNFVGKILGPQGNTIKRLQEETGAKISVLGKGSMRDKAKEEELRKGGDP218 |
| 219KYAHLNMDLHVFIEVFGPPCEAYALMAHAMEEVKKFLVPDMMDDICQEQFLEL271 |
| P3 Region |
| 272SYLNGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGTPVRGAITRG326 |
| P4 Region P5 Region |
| 327ATVTRGVPPPPTVRGAPAPRARTAGIQRIPLPPPPAPETYEEYGYDDTYAEQSYEG382 |
| 383YEGYYSQSQGDSEYYDYGHGEVQDSYEAYGQDDWNGTRPSLKAPPARPVKGA437 |
| 438YREHPYGRY443 |
| Observed mass | Predicted peptide mass | Amino acid sequence | Residue numbers in Sam68 protein | Region | Modification |
|---|---|---|---|---|---|
| Da | Da | ||||
| 974.3173 | 974.4930 | SRGGGGGSR | 44–52 | P0 | Arg 45 is dimethylated |
| 1187.6421 | 1187.5730 | SRGGGGGSRGGAR | 44–56 | P0 | Arg 45 and 52 are dimethylated |
| 823.4907 | 823.4590 | GRGVGPPR | 303–310 | P3 | Arg 304 is dimethylated |
| 1092.6650 | 1092.6490 | GVGPPRGALVR | 305–315 | P4 | Arg 310 is monomethylated |
| 1039.4719 | 1039.6220 | GALVRGTPVR | 311–320 | P4 | Arg 315 is monomethylated |
| 1041.6316 | 1041.6010 | GTPVRGAITR | 316–325 | P4 | Arg 320 is monomethylated |
| 1116.6402 | 1116.6330 | GAITRGATVTR | 321–331 | P4 | Arg 325 is monomethylated |
RG Sequences Neighboring Sam68 Proline Motif P3 Are Methylated
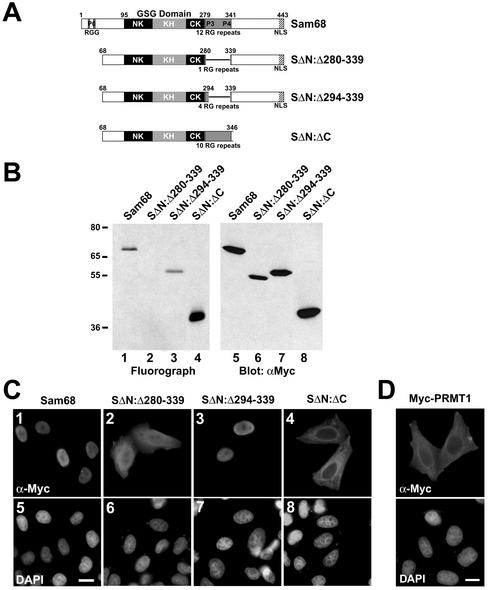
To address whether the RG repeats upstream of the proline motif P3 are methylated in vivo, we performed the methylation labeling assay by using Sam68 deletion mutants (Figure 4A). A Sam68 protein was engineered that removed all the RG repeats surrounding P3 except one (SΔN:Δ280–339). An expression vector encoding SΔN:Δ280–339 was transfected in HeLa cells, the cells were metabolically labeled and the level of in vivo methylation assessed by immunoprecipitating with anti-myc antibodies followed by SDS-PAGE and fluorography. SΔN:Δ280–339 was unable to incorporate 3H-methyl groups (Figure 4B, lane 2). Note that all arginines found to be methylated in the analysis by mass spectrometry are missing in the deletion mutant SΔN:Δ280–339, which is not methylated at all upon transfection. Two additional constructs were generated: one that added four RG repeats N-terminal to the proline motif P3 (SΔN:Δ294–339) and the other that restored the entire RG-rich region between proline motifs P3 and P4 (SΔN:ΔC). Both SΔN:Δ294–339 and SΔN:ΔC proteins were methylated in vivo, indicating that at least one of the RGs upstream of P3 is methylated (Figure 4B, lanes 3 and 4). Equivalent expression of myc-tagged proteins was confirmed by anti-myc immunoblotting of HeLa total cell lysates (Figure 4B, lanes 5–8). The deletion analysis has demonstrated that the RGs upstream of P3 (aa 280–294) are sufficient to observe in vivo methylation.
Figure 4.
Sam68 RG-rich sequences are methylated in vivo and regulate protein localization. (A) Schematic diagram representing the various Sam68 proteins is shown. The position of the KH domain and the neighboring N-terminal of KH (NK) and C-terminal of KH (CK) are indicated. The GSG domain encompasses the NK, KH, and CK domains. The gray boxes represent regions where RG repeats are present in Sam68. The C-terminal nuclear localization signal (NLS) of Sam68 is represented by a stippled box. (B) In vivo methylation labeling was performed 24 h after DNA transfection of the expression vectors encoding the proteins depicted in A. The cell lysates were immunoprecipitated with anti-myc antibodies and the 3H-labeled proteins were separated by SDS-PAGE and visualized by fluorography (lanes 1–4; exp. 12 h) and by immunoblotting by using anti-myc antibodies (lanes 5–8). (C) HeLa cells plated on glass coverslips were transfected with myc-epitope–tagged Sam68 (1 and 5), the Sam68 deletion protein that is no longer methylated (SΔN:Δ280–339; 2 and 6), a slightly longer deleted mutant that is methylated (SΔN:Δ294–339; 3 and 7), and a Sam68 missing the C terminus, including the NLS (SΔN:ΔC; 4 and 8). Twenty-four hours after transfection, the cells were fixed and immunostained by using anti-myc antibodies followed by a secondary antibody conjugated to Alexa 546 (red; 1–4). Nuclei were visualized using the DNA-specific stain 4,6-diamidino-2-phenylindole (blue; 5–8).(D) HeLa cells plated on glass coverslips were transfected with myc-epitope–tagged PRMT1 followed by indirect immunofluorescence by using anti-myc epitope-tagged antibodies followed by a secondary antibody conjugated to Alexa 546 (red). Nuclei were visualized using the DNA specific stain 4,6-diamidino-2-phenylindole.
Methylation of Sam68 Is Required for Its Nuclear Localization
To determine whether methylation regulates Sam68 localization, HeLa cells were transfected with the Sam68 expression vectors encoding the various Sam68 deletion proteins described above. The cells were fixed, permeabilized, and the localization of myc-epitope–tagged Sam68 was detected by indirect immunofluorescence. The unmethylated Sam68 protein (SΔN:Δ280–339) had a whole cell distribution and behaved somewhat like a Sam68 devoid of its C-terminal nuclear localization signal (SΔN:ΔC; Figure 4C). Interestingly, the addition of four RG repeats that results in the methylation of Sam68 (SΔN:Δ294–339) was sufficient to concentrate Sam68 in the nucleus, as observed with the wild-type protein (Figure 4C). These results uncover a direct correlation between the methylation of Sam68 (amino acids 280–294) and the cellular localization of the protein. Moreover, the fact that the cytoplasmic Sam68 protein (SΔN:ΔC) was methylated suggests that methylation of Sam68 can occur in the cytoplasm.
PRMT1 is thought to reside predominantly in the nucleus (Tang et al., 1998); however, it has been identified through purification from a cytoplasmic macromolecular complex (Lin et al., 1996). To further examine the localization of PRMT1, we transfected HeLa cells and performed indirect immunofluorescence by using anti-myc antibodies (Figure 4D). PRMT1 was localized mostly in the cytoplasm, although some of the cells showed both cytoplasmic and nuclear staining. The anti-PRMT1 antibodies were also used for immunofluorescence studies and were unable to stain endogenous PRMT1 (our unpublished data), perhaps indicating that our peptide antibody does not recognize the native folded protein or that PRMT1 is present in large complexes inaccessible to our antibody. A GFP-PRMT1 was recently reported to reside in both cellular compartments (Frankel et al., 2002).
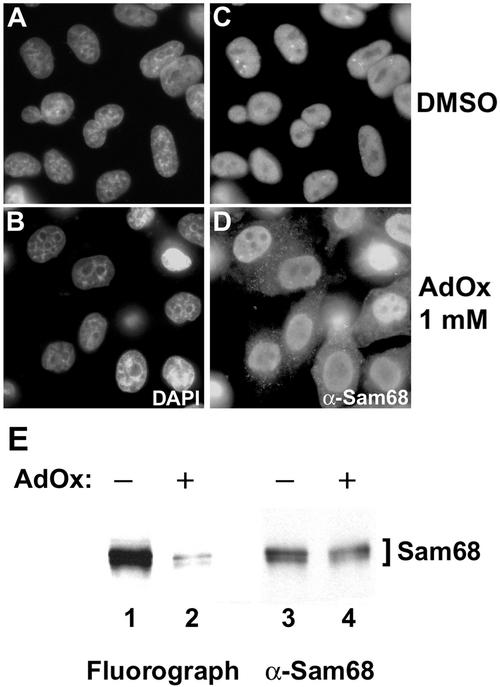
To confirm that methylation of endogenous Sam68 is required for its nuclear localization, we examined whether methylase inhibitors could affect Sam68 cellular distribution. HeLa cells were treated with the drug vehicle dimethyl sulfoxide (DMSO, Figure 5, A and C) or with 1 mM AdOx (Figure 5, B and D) for 24 h. The cells were fixed, permeabilized, and immunofluorescence was performed using the anti-Sam68 AD1 antibody (Figure 5, C and D). The treatment of Sam68 with DMSO did not alter the localization of Sam68 in the nucleoplasm and in Sam68/SLM nuclear bodies (SNBs; Chen et al., 1999b). The methylase inhibitor AdOx increased the cytoplasmic staining of Sam68 and SNBs were not visible (Figure 5D). Strikingly, the distribution of Sam68 in both the cytoplasm and the nucleus was comparable with the nonmethylated mutant SΔN:Δ280–339. To show that Sam68 methylation was effectively reduced by the AdOx treatment, we performed metabolic labeling to assess the levels of protein methylation (Figure 5E). A drastic reduction of Sam68 methylation was observed in the presence of AdOx and no change in protein level was observed (Figure 5E). Comparable results were obtained using another general methylation inhibitor 5′-deoxy-5′-methylthioadenosine (our unpublished data). These results suggest that arginine methylation is essential for the normal localization of Sam68.
Figure 5.
Sam68 accumulates in the cytoplasm in the presence of methylase inhibitors. HeLa cells were treated with either DMSO (A and C) or with the methyltransferase inhibitor AdOx at 1 mM (B and D) for 24 h. The cells were fixed and immunostained with the anti-Sam68 AD1 antibody followed by a secondary antibody conjugated to Alexa 488 (green; C and D). Nuclei were visualized by the DNA-specific stain 4,6-diamidino-2-phenylindole (blue; A and B). (E) HeLa cells were incubated with AdOx for 24 h preceding an in vivo methylation labeling. Immunoprecipitations were performed with anti-Sam68 AD1 antibodies. Samples were separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, sprayed with Enhance, and exposed to film overnight. The 3H-labeled proteins were visualized by fluorography (left), and the same membrane was immunoblotted with anti-Sam68 AD1 antibodies (right).
Abrogation of Sam68 Arginine Methylation in PRMT1 −/− Embryonic Cells
To provide definitive proof for the involvement of PRMT1 in the methylation of Sam68 in vivo, we made use of embryonic stem (ES) cells in which insertion of a gene trap retrovirus into the second intron of the prmt1 gene has created essentially a null mutation (Pawlak et al., 2000). Ruley and coworkers reported that low levels of prmt1 transcripts (∼1% of wild-type levels) can be detected in −/− ES cells (Pawlak et al., 2000). Consistent with this observation, our PRMT1 antibody permitted the detection of very low residual levels of PRMT1 in −/− ES cells (Figure 6A, compare lanes 1 and 3). Among the additional polypeptides that our antibody recognizes, p42 and p48 are also reduced, but p120 levels remain identical (Figure 6A, lanes 1–3), suggesting it represents a cross-reacting band that is not related to PRMT1. Numerous proteins can be detected in wild-type ES cells by immunoblotting with our ASYM24 aDMA-specific antibody (Figure 6A, lane 4). As expected if PRMT1 activity is greatly reduced, the majority of these polypeptides are no longer recognized by ASYM24 in PRMT1 −/− ES cells (Figure 6A, lane 6).
Figure 6.
In vivo methylation of Sam68 is abrogated in prmt1 −/− ES cells. (A) Wild-type ES cells (+/+) as well as ES cells heterozygotes (+/−) or homozygotes (−/−) for a null mutation in the prmt1 gene were harvested in lysis buffer and subjected to immunoblotting by using our anti-PRMT1 and ASYM24 antibodies. Molecular weight markers (in kilodaltons) are shown on the left of each panel. (B) In vivo methylation labeling was performed by metabolically labeling cells with l-[methyl-3H]methionine for 3 h in the presence of cycloheximide and chloramphenicol. The cell lysates were immunoprecipitated with anti-Sam68 (AD1), normal rabbit serum (CTRL), or anti-SMN antibodies. The 3H-labeled proteins in TCLs (10% input) and in immunoprecipitates were separated by SDS-PAGE and visualized by fluorography (lane 1–12; exp. 24 h). Immunoblotting was performed on total cell lysates by using anti-Sam68 (lanes 13–15) and anti-SMN (lanes 16–18) antibodies to confirm equal levels of proteins.
Metabolic labeling using l-[methyl-3H]methionine was performed in the presence of translation inhibitors to examine the methylation state of Sam68 in these cells where PRMT1 activity was greatly reduced. Immunoprecipitations were carried out with normal rabbit serum (CTRL), anti-Sam68, or anti-SMN antibodies. The immunoprecipitated proteins were resolved by SDS-PAGE and visualized by fluorography. The fluorograph revealed that Sam68 methylation was severely abrogated in PRMT1 −/− ES cells (Figure 6B, compare lanes 2 and 10). As expected and shown in Figure 1A, SMN did not incorporate any [3H]methionine (Figure 6B, lanes 4, 8, and 12). Immunoblotting with anti-Sam68 and anti-SMN antibodies confirmed that Sam68 and SMN were present in equal amounts in each cell extracts (Figure 6B, lanes 13–15 and 16–18, respectively). These findings clearly establish PRMT1 as an enzyme responsible for the methylation of Sam68 in vivo.
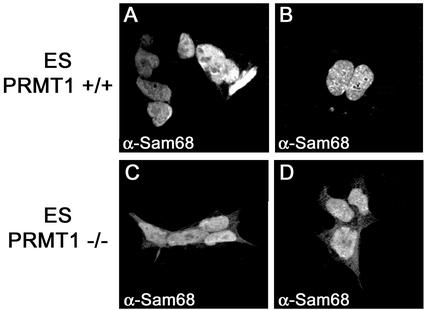
We next examined the cellular localization of Sam68 in PRMT1-deficient ES cells because Sam68 is no longer methylated in these cells (Figure 6). Indirect immunofluorescence was performed as described above by using the anti-Sam68 AD1 antibody and the cells were visualized using a confocal microscope (Carl Zeiss). ES cells carrying both wild-type alleles of PRMT1 (+/+) showed a normal nuclear localization of Sam68 (Figure 7, A and B). In contrast, some cytoplasmic staining of Sam68 was detectable in PRMT1 −/− cells (Figure 7, C and D). These results confirm that Sam68 arginine methylation by PRMT1 influences its cellular localization.
Figure 7.
Sam68 accumulates in the cytoplasm of PRMT1 −/− ES cells. Wild-type ES cells (PRMT1 +/+; A and B) or ES cells homozygotes (PRMT1 −/−; C and D) for a null mutation in the prmt1 gene were fixed and immunostained using the anti-Sam68 AD1 antibody followed by a secondary antibody conjugated to Alexa 488. Cells were then visualized using a confocal microscope (Carl Zeiss).
Methylation Is Necessary for Ability of Sam68 to Function in Nuclear Export of HIV RNAs
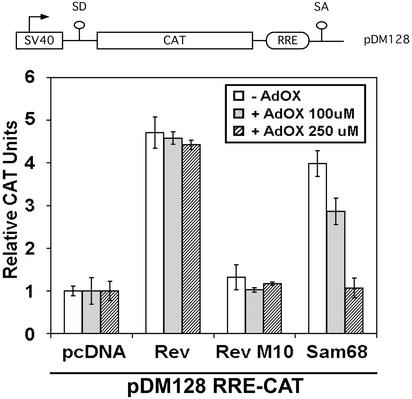
Sam68 has been shown to function as a cellular homologue of Rev in accelerating the transport of HIV RNAs from the nucleus to the cytoplasm (Reddy et al., 1999). We examined whether arginine methylation could regulate this function of Sam68 by determining whether methylase inhibitors could modulate RRE-directed reporter gene expression (Figure 8). Transport to the cytoplasm and expression of the intronic CAT gene is Rev and RRE dependent, as described previously (Hope et al., 1990). COS7 cells were transfected with the RRE-CAT reporter plasmid in the presence of either an empty plasmid (pcDNA), expression vectors for Rev, an inactive mutant of Rev (Rev M10), and Sam68. After transfection, cells were treated with increasing concentrations of AdOx or DMSO and harvested for CAT activity (Figure 8). The transfection of Sam68 or Rev with the RRE-CAT reporter resulted in an approximately fivefold increase in CAT activity relative to an empty plasmid or RevM10 (Figure 8). The presence of the methylase inhibitor AdOx decreased CAT activity in a dose-dependent manner in Sam68, but not Rev-transfected cells (Figure 8). Sam68 deletion mutants (SΔN:Δ280–339 and SΔN:Δ294–339) were unable to substitute for Rev in this assay, although they did not act as dominant negative as was reported for the SΔN:ΔC mutant (our unpublished data; Reddy et al., 1999). These findings suggest that arginine methylation of Sam68 regulates its ability to export RRE-containing HIV RNAs.
Figure 8.
Methylation is necessary for the ability of Sam68 to function in the nuclear export of HIV RNAs. A schematic diagram of the Rev responsive element reporter CAT construct is shown. The splice acceptor and donor sites are indicated as SA and SD. CAT indicates the chloramphenicol acetyltransferase cDNA and RRE is the HIV Rev response element. COS7 cells were transfected with the RRE-CAT reporter plasmid pDM128 in the presence of either an empty plasmid (pcDNA), Rev, an inactive mutant of Rev (Rev M10), or Sam68. After transfection, cells were treated with increasing concentration of AdOx or with the vehicle alone (DMSO; −AdOx). CAT activity was normalized to β-galactosidase activity. Each bar represents CAT activity from at least three separate experiments.
DISCUSSION
Herein, we report that the KH domain containing Sam68 harbors asymmetric dimethylarginines near its proline motif P3 in vivo by using a novel anti-aDMA–specific antibody. PRMT1 interacts with Sam68 and is a major PRMT responsible for Sam68 methylation in vivo. Arginine methylation was necessary to localize Sam68 to the nucleoplasm, where it normally resides. The methylation of Sam68 was required for its ability to facilitate the export of unspliced HIV RNAs into the cytoplasm. We demonstrate that other KH domain RNA binding proteins, including SLM-1, SLM-2, QKI-5, hnRNP K, and GRP33 are also methylated in vivo. Our findings suggest that arginine methylation may be an essential maturation step for the proper localization and function of RG-rich–containing RNA binding proteins.
Sam68 and Other KH Domain-containing Proteins Interact with PRMT1
The association of KH domain RNA binding proteins with PRMT1 demonstrates a link between RNA and PRMT1. The disruption of ribonucleoprotein complexes by RNase treatment increases the accessibility of proteins to in vitro methylation by PRMT1 (Frankel and Clarke, 1999). Moreover, the yeast Hmt1p/Rmt1p was identified in a screen for interactors of poly-(A)–associated proteins (Henry and Silver, 1996). These observations suggest that PRMTs are present in ribonucleoprotein complexes with their substrates.
SLM-1 and QKI-5 are methylated in vivo, but did not coimmunoprecipitate a protein methyltransferase activity and did not interact with PRMT1. Thus, SLM-1 and QKI-5 may be the target for a different PRMT that may not require a tight interaction with its substrates for activity. The fact that SLM-1 did not associate with PRMT1 is surprising because it shares a high degree of homology with Sam68 and SLM-2 and thus has a comparable number of RG repeats. Examining the RG sequences in SLM-1, compared with those of Sam68 and SLM-2 might provide new insights about the preferred motif of PRMT1 vs. other PRMTs. We have found that QKI-5 is weakly methylated in vivo. But what seems to be weak in vivo methylation could in fact be the lower amount of incorporated labeled methyl groups due to the few RG repeats found in QKI-5.
By using biochemical methods, it has been previously shown in insect cells that Sam68 contained methylated arginines in its sequence (Wong et al., 1992). We now provide evidence by using a novel aDMA-specific antibody that mammalian Sam68 harbors aDMA in vivo. The sites of arginine methylation are two hnRNP-like RGG motifs at position 45 and 52 in the N-terminal region as well as multiple RG repeats surrounding proline motif P3 as determined by mass spectrometry. Furthermore, the deletion analysis revealed that at least one arginine is methylated between amino acids 280–294 upstream of proline motif P3. All these methylation sites fit the PRMT1 GAR and RxR consensus sequence (Gary and Clarke, 1998; Smith et al., 1999). Interestingly, four RG repeats (310, 315, 320, and 325) located between proline motifs P3 and P4 were found to be monomethylated. It remains to be determined whether these represent precursors or intermediates leading to dimethylation. Arginine 3 of histone H4 has been shown to contain monomethylated arginines at steady state, and PRMT1 is thought to be the major, if not exclusive, methyltransferase responsible for this modification (Strahl et al., 2001).
Arginine Methylation Affects Sam68 Cellular Distribution
The present study shows that arginine methylation of Sam68 is a prerequisite for its exclusive nuclear localization. HnRNP A2, another RNA binding protein methylated by PRMT1, accumulates in the cytoplasm in the presence of general methylation inhibitors and its nuclear localization requires an intact RGG domain (Nichols et al., 2000). Similarly, nuclear accumulation of high-molecular-weight fibroblast growth factor-2 could also be linked to methylation (Pintucci et al., 1996). These observations suggest that arginine methylation of RNA binding proteins regulates their nucleocytoplasmic transport. Indeed, arginine methylation facilitates the nuclear export of Npl3p, Nab2p, and Hrp1p in yeast and the methyltransferase activity of Hmt1p is required for this effect (Shen et al., 1998; McBride et al., 2000; Green et al., 2002). Moreover, arginine methylation of Npl3p RGG motif interferes with its Sky1p-mediated phosphorylation, which in turn prevents the interaction of Npl3p with the nuclear import receptor Mtr10p (Yun and Fu, 2000). This provides an indirect mechanism by which arginine methylation can affect nuclear import. The reason for Sam68 cytoplasmic accumulation is unknown, but it can be reasoned that 1) arginine methylation is cotranslational and newly synthesized Sam68 accumulates in the cytoplasm, or 2) unmethylated Sam68 is imported at a slower rate than it is exported. The use of protein synthesis inhibitors with methylase inhibitors rule out the first possibility. Thus, the likely possibility is that the nucleocytoplasmic transport of Sam68 is altered by methylation. This is consistent with the fact that Sam68 cannot functionally export HIV RNAs in the presence of methylase inhibitors.
Arginine Methylation: A Maturation Step for RNA Binding Proteins?
The presence of conserved RGG boxes and RG-rich repeats in RNA binding proteins is thought to be necessary to mediate interactions with RNA. Indeed, this has been demonstrated for several RNA binding proteins, including Sam68, FMRP, hnRNP A1, and U (Kiledjian and Dreyfuss, 1992; Chen et al., 2001; Darnell et al., 2001). A major family of proteins containing aDMAs is the hnRNP RNA binding proteins (Liu and Dreyfuss, 1995) and the presence of aDMAs in other RNA binding proteins is rapidly being discovered. What may be the role of arginine methylation in the function and/or turnover of RNA binding proteins? It is conceivable that arginine methylation could modulate the contribution of RNA binding by the arginine-rich regions. Methylation does not change the positive charge of the arginines, but increases hydrophobicity and might prevent hydrogen bonding and/or introduce steric constraints that should reduce charge-induced interactions with RNA (Calnan et al., 1991). In agreement with this prediction, a reduction in hnRNP A1 binding to nonspecific single-stranded nucleic acid was observed subsequent to its arginine methylation (Rajpurohit et al., 1994). However, other studies have reported no significant effect of arginine methylation on RNA binding (Valentini et al., 1999; Raman et al., 2001). We have observed a modest effect of arginine methylation on Sam68 RNA binding activity in vitro, by using gel mobility shift assays and poly (U) homopolymer binding assays (our unpublished data). However, no difference in Sam68 binding to poly (U)-Sepharose could be detected between PRMT1 +/+ and PRMT1 −/− ES cell extracts (our unpublished data). Nonetheless, arginine methylation might affect interactions with specific RNA targets or structures that would have evolved to better accommodate the bulkier methyl groups. It is becoming increasingly clear that arginine methylation will not be a quick and rapid mode of regulation such as phosphorylation. Our data with Sam68 support the hypothesis that arginine methylation may be a maturation step for certain proteins such as RNA binding proteins. We define maturation as a step necessary for the proper localization of a protein and a step that is required to enhance its function. This maturation step may be either cotranslational or coupled to the nuclear import machinery. Because arginine methylation is most likely irreversible (Gary and Clarke, 1998), it may be a constitutive pathway for the maturation of proteins. Interestingly, four of the 12 neighboring Sam68 RG repeats were sufficient to properly localize Sam68 in the nucleus, implying that partial methylation may be sufficient to target the protein to its proper localization. The methylation of the RG repeats of Sam68 may be regulated and occur initially in the cytoplasm and then in the nucleus. Moreover, the level of RG methylation of certain proteins may reflect the “age” of the protein. The presence of multiple neighboring RG repeats also implies that methylation may be a processive event, especially because PRMTs associate with their substrates. The maturation hypothesis our data supports may apply to histones and Sm proteins.
In summary, this study demonstrates that Sam68 is asymmetrically dimethylated on several arginine residues by PRMT1 in vivo. This modification is required for proper Sam68 localization in the nucleus and for one of its functions, the ability to substitute for Rev in exporting unspliced HIV RNAs. These results suggest a general role for arginine methylation in the maturation and function of RNA binding proteins.
ACKNOWLEDGMENTS
We thank Dave Schriemer (University of Calgary, Alberta, Calgary, Canada) for the mass spectrometry analysis and Yi Zhang and Earl H. Ruley for kindly providing PRMT1 −/− ES cells. This work was supported by grant 011291 from the National Cancer Institute of Canada with funds from the Canadian Cancer Society of Canada and grant MT13377 from the Canadian Institutes of Health Research. F.-M.B. and J.C. are recipients of a studentship and a postdoctoral fellowship from the National Cancer Institute of Canada, respectively. M.T.B. is supported by a Welch Foundation grant (G-1495). S.R. is a recipient of a Canadian Institutes of Health Research Investigator award.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0484. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0484.
REFERENCES
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin GS, Carnegie PR. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171:579–581. doi: 10.1126/science.171.3971.579. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999a;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen T, Boisvert F-M, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999b;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Côté J, Carvajal HV, Richard S. Identification of Sam68 arginine glycine-rich sequences capable of conferring non-specific RNA binding to the GSG domain. J Biol Chem. 2001;276:30803–30811. doi: 10.1074/jbc.M102247200. [DOI] [PubMed] [Google Scholar]
- Chen T, Damaj B, Herrerra C, Lasko P, Richard S. Self-association of the single-KH domain family members Sam68, GRP33, GLD-1 and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M, Chen T, Richard S. Two novel Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci USA. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K. The quaking gene unites signal transduction and RNA binding in the developing nervous system. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Frankel A, Clarke S. RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem Biophys Res Commun. 1999;259:391–400. doi: 10.1006/bbrc.1999.0779. [DOI] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001a;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001b;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+ RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope TJ, Huang X, McDonald D, Parslow TG. Steroid-receptor fusion of the HIV type I Rev trans-activator: mapping of the cryptic functions of the arginie rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Schedl T. Mutations in GLD-1, a female germ cell-specific tumor suppressor gene in C. elegans, affect a conserved domain also found in Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A, Silver P. State of the Arg: protein methylation at arginines comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- McBride AE, Weiss VH, Kim HK, Hogle JM, Silver PA. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. J Biol Chem. 2000;275:3128–3136. doi: 10.1074/jbc.275.5.3128. [DOI] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFN-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Najbauer J, Johnson BA, Young AL, Aswad DW. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J Biol Chem. 1993;268:10501–10509. [PubMed] [Google Scholar]
- Nichols RC, Wang XW, Tang J, Hamilton BJ, High FA, Herschman HR, Rigby WFC. The RGG domain in hnRNP A2 affects subcellular localization. Exp Cell Res. 2000;256:522–532. doi: 10.1006/excr.2000.4827. [DOI] [PubMed] [Google Scholar]
- Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintucci G, Quarto N, Rifkin DB. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol Biol Cell. 1996;7:1249–1258. doi: 10.1091/mbc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit R, Paik WK, Kim S. Effect of enzymic methylation of heterogeneous ribonucleoprotein particle A1 on its nucleic-acid binding and controlled proteolysis. Biochem J. 1994;304:903–909. doi: 10.1042/bj3040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman B, et al. N(omega)-arginine dimethylation modulates the interaction between a Gly/Arg-rich peptide from human nucleolin and nucleic acids. Nucleic Acids Res. 2001;29:3377–3384. doi: 10.1093/nar/29.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TR, Xu W, Mau JKL, Goodwin CD, Suhasini M, Tang H, Frimpong K, Rose DW, Wong-Staal F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- Strahl BD, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–19876. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D. An RNA-binding protein associated with src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Weiss VH, Silver PA. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA. 1999;5:272–280. doi: 10.1017/s1355838299981633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Methylation of histone H4 at arginine 3 facilitates transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wong G, Muller O, Clark R, Conroy L, Moran MF, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B.M., Montminy, M., and Evans, R.M. (2001). A transcriptional switch mediated by cofactor methylation. Science 8, Sciencexpress 10.1126. [DOI] [PubMed]
- Yun CY, Fu X-D. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–717. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]