Abstract
The yeast Saccharomyces cerevisiae has four genes, MCK1, MDS1 (RIM11), MRK1, and YOL128c, that encode homologues of mammalian glycogen synthase kinase 3 (GSK-3). A gsk-3 null mutant in which these four genes are disrupted showed growth defects on galactose medium. We isolated several multicopy suppressors of this growth defect. Two of them encoded Msn2p and phosphoglucomutase (PGM). Msn2p is a transcription factor that binds to the stress-response element (STRE). PGM is an enzyme that interconverts glucose-1 phosphate and glucose-6 phosphate and is regulated by Msn2p at the transcriptional level. Expression of the mRNAs of PGM2 and DDR2, whose promoter regions possess STRE sequences, on induction by heat shock or salt stress was reduced not only in an msn2 msn4 (msn2 homologue) double mutant but also in the gsk-3 null mutant. STRE-dependent transcription was greatly inhibited in the gsk-3 null mutant or mck1 mds1 double mutant, and this phenotype was suppressed by the expression of Mck1p but not of a kinase-inactive form of Mck1p. Although Msn2p accumulated in the nucleus of the gsk-3 null mutant as well as in the wild-type strain under various stress conditions, its STRE-binding activity was reduced in extracts prepared from the gsk-3 null mutant or mck1 mds1 double mutant. These results suggest that yeast GSK-3 promotes formation of a complex between Msn2p and DNA, which is required for the proper response to different forms of stress. Because neither Msn2p–GSK-3 complex formation nor GSK-3–dependent phosphorylation of Msn2p could be detected, the regulation of Msn2p by GSK-3 may be indirect.
INTRODUCTION
The serine/threonine kinase glycogen synthase kinase 3 (GSK-3) was first described in a metabolic pathway for glycogen synthase regulation that is sensitive to insulin-mediated inhibition (Plyte et al., 1992). GSK-3 has subsequently been shown to regulate several physiological responses, including protein synthesis, gene expression, subcellular localization of proteins, and protein degradation in mammalian cells by phosphorylating many substrates, including neuronal cell adhesion molecule, neurofilament, synapsin I, tau, transcription factors, adenomatous polyposis coli gene product, β-catenin, and cyclin D1 (Plyte et al., 1992; Cohen and Frame, 2001). GSK-3 is highly conserved through evolution and plays a fundamental role in cellular responses. Xenopus GSK-3 functions as a member of the Wnt signaling pathway, determines cell fate, and regulates axis formation during early development (He et al., 1995; Yost et al., 1996). The Drosophila zeste-white3/shaggy gene product is homologous to GSK-3β (Ruel et al., 1993) and is required at several developmental stages during fly embryogenesis (Simpson et al., 1988; Perrimon and Smouse, 1989). A Dictyostelium homologue of GSK-3 has been found to be important for cellular differentiation (Harwood et al., 1995).
In Saccharomyces cerevisiae, there are four genes, MCK1, MDS1/RIM11, MRK1, and YOL128c, that encode homologues of mammalian GSK-3. Mck1p plays a role in mitotic chromosomal segregation specific to CDEIII function (Shero and Hieter, 1991), acts in the transcription of IME1 at the beginning of meiosis (Neigeborn and Mitchell, 1991), and is important for inducing the cell cycle delay in response to Ca2+ (Mizunuma et al., 2001). Mds1p/Rim11p plays a role in expression of meiotic genes by phosphorylating Ime1p and Ume6p (Bowdish et al., 1994; Xiao and Mitchell, 2000). Thus, S. cerevisiae GSK-3 seems to play important roles in both meiosis and mitosis. It is possible that it has additional functions, because mammalian GSK-3 has multiple substrates and functions (see above). To look for new functions of yeast GSK-3, we have generated the mck1 mds1 double-null and mck1 mds1 mrk1 yol128c quadruple-null (gsk-3 null) mutants (Andoh et al., 2000). Both of these GSK-3 mutants show temperature sensitivity. We have screened for rog mutations (revertant of gsk-3), which suppress the temperature sensitivity of the mck1 mds1 double-null mutant, and designated one of them rog1. Rog1 degradation depends on both GSK-3 and Rsp5, which is a HECT-type ubiquitin ligase (E3). Although it has been demonstrated that mammalian GSK-3 triggers phosphorylation and ubiquitination and subsequent degradation of proteins such as β-catenin and cyclin D1 (Aberle et al., 1997; Diehl et al., 1998; Ikeda et al., 1998), our observations provide the first evidence that yeast GSK-3 also regulates protein stability.
S. cerevisiae has become an important model organism for studies of how eukaryotic cells respond to stresses (Estruch, 2000). A cis-regulatory element mediating transcriptional induction by various forms of stress was identified in the promoter of CTT1, a catalase-encoding gene, and DDR2, a DNA damage–responsive gene, and this element was designated the stress-response element (STRE) (Wieser et al., 1991; Kobayashi and McEntee, 1993; Marchler et al., 1993). Subsequently, STRE sequences have been identified in many stress-induced genes (Treger et al., 1998). Two trans-acting factors, Msn2p and Msn4p, are involved in STRE-mediated gene expression (Martinez-Pastor et al., 1996). MSN2, which encodes a Cys2His2 zinc-finger protein, was originally isolated as a multicopy suppressor of the raffinose utilization defect shown by mutants with a thermosensitive allele of SNF1 (Estruch and Carlson, 1993). MSN4 was isolated on the basis of its sequence homology with MSN2 (Estruch and Carlson, 1993). Msn2p and Msn4p bind to STRE both in vitro and in vivo and are required for the induction of an STRE-LEU2-lacZ reporter gene in response to different forms of stress (Martinez-Pastor et al., 1996; Schmitt and McEntee, 1996). Msn2p and Msn4p are distributed throughout the cytoplasm in unstressed cells. In response to stresses, they are translocated to the nucleus (Görner et al., 1998). The cAMP-dependent protein kinase (PKA) and target of rapamycin (TOR) pathways regulate the subcellular localization of Msn2p and Msn4p (Görner et al., 1998; Beck and Hall, 1999). An inverse correlation is found between PKA activity and the nuclear localization of Msn2p and Msn4p. Rapamycin, acting through the TOR pathway, induces the nuclear accumulation of Msn2p and Msn4p. The transcription mediated by Msn2p and Msn4p is dependent on their nuclear localization, but it is not known whether nuclear translocation of Msn2p and Msn4p is the only regulatory mechanism involved in STRE-mediated activation. There may be additional mechanisms of regulating the activities of Msn2p and Msn4p.
Here, we show that GSK-3 is not necessary for nuclear translocation of Msn2p but that it is required for Msn2p- and STRE-dependent transcription and formation of a complex between Msn2p and STRE. These results indicate that GSK-3 is important for Msn2p-dependent transcription of stress-response genes in yeast.
MATERIALS AND METHODS
Materials and Chemicals
The yeast strain CEPA (Estruch and Carlson, 1993) and a yeast genomic library in the multicopy, URA3- marker vector YEp24 were kind gifts from Drs. F. Estruch (University of Valencia, Burjassot, Spain) and Y. Ohya (University of Tokyo, Tokyo, Japan), respectively. pTS009 (a single-copy, TRP1-marker vector possessing two HA epitopes in YCplac22), pTS904CU, pTS904EU (Sasaki et al., 2000), pRS316-GFP (green fluorescent protein) (a single-copy, URA3-marker vector), and PGM18/17 (Marchler et al., 1993) were gifts from Dr. Y. Kikuchi (University of Tokyo, Tokyo, Japan), and pASZ11 (a single-copy, ADE2-marker vector) was from Dr. R. Akada (University of Yamaguchi, Ube, Japan). pBTM116HA (a multicopy, TRP1-marker vector) was a gift from K. Tanaka (University of Hokkaido, Sapporo, Japan). pBSKS(+) was obtained from Toyobo Co. (Tokyo, Japan). The anti-Myc antibody was purified from 9E10 cells (ATCC, Manassas, VA). [α-32P]dCTP and [α-32P]dATP were purchased from Amersham Bioscience Corp. (Piscataway, NJ). Other materials and chemicals were obtained from commercial sources.
Mutants
Yeast strains used in this study are listed in Table 1. From spores of the MCK1/mck1::TRP1 MDS1/mds1::HIS3 YOL128c/yol128c::LEU2 MRK1/mrk1::URA3 diploid cells in the W303 strain background (Andoh et al., 2000), a Trp+ His+ haploid cell and a Trp+ His+ Leu+ haploid cell were obtained and named YTA002W and YTA004W, respectively. To generate strains W303a stre, YTA002W stre, YTA003W stre, and CEPA stre, the NcoI-cut PGM18/17 fragment was integrated into the chromosomal URA3 locus in the corresponding strains. The mck1 mds1 msn2 msn4 quadruple mutant was created by crossing strain YTA004W to CEPA stre, sporulating the resulting diploid, and isolating a Trp+ Ura+ His+ Leu− haploid cell from a tetrad that segregated 2 Trp+ His+ : 2 Trp− His−. The MCK1/mck1::LEU2 MDS1/mds1::HIS3 diploid cells derived from L40 (Vojtek et al., 1993) were sporulated, and wild-type (LTA001) and mck1::LEU2 mds1::HIS3 (LTA002) haploid segregants were selected.
Table 1.
Yeast strains used in this study
| Strain | Genotype | |
|---|---|---|
| W303a | MATa his3 leu2 ura3 trp1 ade2 | This study |
| YTA002W | MATa his3 leu2 ura3 trp1 ade2 mck1∷TRP1 mds1∷HIS3 | This study |
| YTA003W | MATα his3 leu2 ura3 trp1 ade2 mck1∷TRP1 mds1∷HIS3 mrk1 yol128c∷LEU2 | Andoh et al. (2000) |
| YTA004W | MATα his3 leu2 ura3 trp1 ade2 mck1∷TRP1 mds1∷HIS3 yol128c∷LEU2 | This study |
| CEPA | MATa SUC2 ade2 his3 leu2 trp1 ura3 msn2Δ-3∷HIS3 msn4-1∷TRP1 | Estruch et al. (1993) |
| W303a stre | MATa his3 leu2 ura3 trp1 ade2 URA3∷STRE-LEU2-lacZ | This study |
| YTA002W stre | MATa his3 leu2 ura3 trp1 ade2 mck1∷TRP1 mds1∷HIS3 URA3∷STRE-LEU2-lacZ | This study |
| YTA003W stre | MATα his3 leu2 ura3 trp1 ade2 mck1∷TRP1 mds1∷HIS3 mrk1 yol128c∷LEU2 URA3∷STRE-LEU2-lacZ | This study |
| CEPA stre | MATa SUC2 ade2 his3 leu2 trp1 ura3 msn2Δ-3∷HIS3 msn4-1∷TRP1 URA3∷STRE-LEU2-lacZ | This study |
| YYH001 stre | MATα SUC2 ade2 his3 leu2 trp1 ura3 mck1∷TRP1 mds1∷HIS3 msn2Δ-3∷HIS3 msn4-1∷TRP1 URA3∷STRE-LEU2-lacZ | This study |
| L40 | MATa trp1 leu2 his3 ade2 LYS2∷lexA-HIS3 URA3∷lexA-lacZ | Vojtek et al. (1993) |
| LTA001 | MATα trp1 leu2 his3 ade2 LYS2∷lexA-HIS3 URA3∷lexA-lacZ | This study |
| LTA002 | MATa trp1 leu2 his3 ade2 mck1∷LEU2 mds1∷HIS3 LYS2∷lexA-HIS3 URA3∷lexA-lacZ | This study |
Plasmid Constructions
All fragments amplified by PCR were produced by using genomic DNA of strain W303a as a template, and the correctness of sequences was confirmed by sequence analysis. pRS315-MCK1 was constructed by insertion of the 1.4-kilobase (kb) SalI-NcoI fragment of YCp50-MCK1 (Andoh et al., 2000) and the 1.4-kb NcoI-XbaI fragment of pGAL-MCK1, which contained the 1.6-kb BamHI-SalI fragment of pTA002 (Andoh et al., 2000) in pYES2 (Invitrogen, Groningen, Netherlands), into the SalI and XbaI sites of pRS315 (Sikorski and Hieter, 1989). Mck1KN, an inactive form of Mck1p, was generated by changing Lys68 to Met. pRS315-MCK1KN was constructed from pRS315-MCK1 by mutating 5′-AAA-3′ (nucleotides +202 to +204) to 5′-ATG-3′. pASZ11-MCK1 and pASZ11-MCK1KN were constructed by insertion of the 1.4-kb SalI-NcoI fragments of pRS315-MCK1 and pRS315-MCK1KN, respectively, and the 1.7-kb NcoI-EcoRI fragment of YCp50-MCK1, into the SalI and EcoRI sites of pASZ11. To generate plasmids expressing HA-tagged Mck1p, the PCR product containing full-length MCK1 was inserted into pTS009, in which two HA tags were inserted between the SmaI and KpnI sites of YCplac22, to create pTS009-MCK1-HA. The MCK1-HA fragment was digested with HindIII and KpnI from pTS009-MCK1-HA and inserted into the HindIII and KpnI sites of pYES2BS, which was made from pYES2 by digestion with BamHI and SphI and end-filled, followed by self-ligation, to generate pYES2BS-MCK1-HA. The 1.5-kb SalI-NcoI fragment of YCp50-MCK1 and the 0.8-kb NcoI-XbaI fragment of pYES2BS-MCK1-HA were inserted into the SalI and XbaI sites of pRS315 and YEplac181 (Gietz and Sugino, 1988) to generate pRS315-MCK1HA and YEplac181-MCK1HA, respectively. To construct plasmids expressing Msn2p-Myc, PCR was performed with the primers 5′-AGAAAGCTTGGATTCATGACGGTCGACCATGA-3′ and 5′-GGGCTGCAGTGCCCGGGCAATGTCTCCATGTTTTTTATG-3′. The PCR product was digested with KpnI and PstI, and the 1.7-kb KpnI-PstI fragment and the 2.1-kb SphI-KpnI fragment of MSN2 from YEp24–85, which was isolated from the library in our screening as described below, were inserted into the PstI and SphI sites of vector pTS904CU to generate pTS904CU-MSN2. pTS904EU-MSN2 was then constructed by inserting the 3.8-kb SphI-PstI fragment of pTS904CU-MSN2 into the SphI and PstI sites of vector pTS904EU. pRS316-MSN2-GFP was constructed by inserting the 3.8-kb HindIII-SmaI fragment of pTS904EU-MSN2 into vector pRS316-GFP. pBSKS(+)-PGM2 was constructed by inserting the 1.1-kb SalI-EcoRI fragment produced by PCR with the primers 5′-TTAGTCGACTAGTTATTGGCCAGCAT-3′ and 5′-AGTACGGCCGTACTTTG-3′ into the SalI and EcoRI sites of pBSKS(+). pBSKS(+)-DDR2 was constructed by inserting the 0.5-kb HindIII fragment produced by PCR with the primers 5′-AGTTTAGCCGCTCAAGC-3′ and 5′-ACTAAGCTTCCTATAGATGGAATATC-3′ into the HindIII site of pBSKS(+). To construct pBTM116HA-MSN2ΔZn, the 1.5-kb BglII-ClaI fragment from pTS904EU-MSN2 and the 0.6-kb ClaI-PstI PCR fragment of MSN2ΔZn produced by PCR with the primers 5′- AATCCTTCAACAGCGATC-3′ and 5′-AATCTGCAGTTAGTGGAACGGTTTCTCCTC-3′ were inserted into the BamHI and PstI sites of pBTM116HA.
Screening of Yeast Genomic DNA Library
Strain YTA003W (gsk-3 null mutant) was transformed with the yeast genomic DNA library by the usual lithium-acetate method. Transformants were selected on 5% galactose (SG) plates (0.17% yeast nitrogen base without ammonium sulfate, 0.5% ammonium sulfate, 5% galactose, 0.3% sucrose, and supplements) for 3 d at 26°C. Plasmid DNAs from transformants were recovered by passage through bacteria. The plasmids were retested by transforming YTA003W.
Stress Conditions
Strains without plasmids were grown in YPD medium (2% glucose, 2% bactopeptone, and 1% yeast extract); strains with plasmids were grown in synthetic complete (SC) medium (0.17% yeast nitrogen base without ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, and supplements). In either case, cells were grown to an OD600 of 0.1–0.3 at 26°C and then subjected to stress treatments as follows (except for the viability measurements shown in Figure 2; see below). For heat-shock stress, cells were incubated at 37°C for 10 min to 1 h. For salt stress, cells were incubated in medium containing 400 mM NaCl for 10 min to 1 h. For glucose depletion, cells were washed once with synthetic medium without glucose and incubated in the absence of glucose for 10 min at 26°C.
Figure 2.
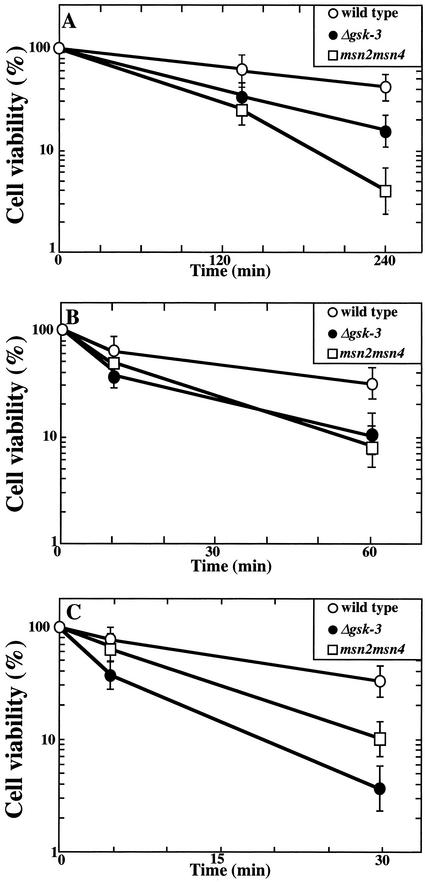
Similar pleiotropic stress sensitivity of the gsk-3 null and msn2 msn4 double mutants. After cells of strains W303a (wild-type, open circles), YTA003W (Δgsk-3, closed circles), and CEPA (msn2 msn4 mutant, squares) were incubated in SC at 45°C (A), in YPD containing 2 M NaCl at 26°C (B), or in YPD including 5 mM hydrogen peroxide at 26°C (C) for the indicated time, the cells were plated to determine viability (see MATERIALS AND METHODS). The results shown are means ± SEM of three independent experiments.
Viability Measurements
For heat-shock stress, cells were grown to an OD600 of 0.3 in SC medium at 26°C, transferred to a water bath at 45°C, and incubated for 4 h. For salt stress, cells were grown to an OD600 of 1.0 in YPD medium at 26°C, diluted to an OD600 of 0.2 in YPD containing 2 M NaCl, and incubated at 26°C for 1 h. For oxidative stress, cells were grown to an OD600 of 0.3 at 26°C, hydrogen peroxide was added to the culture medium at a final concentration of 5 mM, and the cells were incubated for 30 min with vigorous agitation at 26°C. Viability was measured by plating the appropriate dilution of cells on YPD plates at 30°C, in duplicate, and incubating for 2–3 d. It was expressed as a percentage of the initial colony-forming units before stresses.
Preparation of RNA and Northern Blotting Analysis
After being exposed to stress treatments, cells were harvested and frozen in liquid nitrogen. RNA was extracted by a hot phenol method as described (Köhrer and Domdey, 1991). Denatured RNA (20 μg) was electrophoresed on 1% agarose gels containing formaldehyde, blotted onto nitrocellulose membranes, and hybridized to multiprimed [α-32P]dCTP–labeled probes. The following gene probes were used: the 0.5-kb SalI and NsiI-cut PGM2 fragment obtained from pBSKS(+)-PGM2 and the 0.5-kb HindIII-cut DDR2 fragment obtained from pBSKS(+)-DDR2.
Stress-dependent Transcription Assay
After being exposed to various stress conditions for 1 h, transformants were harvested and frozen in liquid nitrogen. To measure stress-mediated transcription, β-galactosidase activity induced by STRE was assayed. The cells were suspended in 150 μl of cold Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 100 mM MgSO4) containing 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml antipain, 1 mg/ml aprotinin, 1 mg/ml leupeptin, and 1 mg/ml pepstatin A, and broken by agitation with glass beads by use of a vortex mixer for 5 min at 4°C. Then, 150 μl of Z buffer was added and the homogenate was again mixed vigorously. The homogenate was centrifuged at 14,000 × g for 5 min, and the supernatant was recovered and centrifuged again at 14,000 × g for 5 min. β-Galactosidase activity of the resultant supernatant fraction was assayed using 4 mg/ml 0-nitrophenyl-β-d-galactopyranoside as substrate (Miller, 1972). The activity was normalized by the protein concentration, which was determined with BSA as standard (Lowry et al., 1951).
GFP Fluorescence Microscopy
After being subjected to stress treatments, transformants were fixed with 4% paraformaldehyde for 1 h. The cells were washed with PBS three times, and then 50 vol PBS was added to the cells. Aliquots were then viewed with a fluorescence microscope (Olympus IX 70). Images were scanned with a CCD camera (Leica DC 250) and analyzed with Leica Qfluoro (version 1.0) software. Nuclei were stained by the addition of 2 mg/ml 4,6-diamidino-2-phenylindol (DAPI) DNA dye to the cultures.
DNA Binding Assays
DNA binding assays were performed as described (Schmitt and McEntee, 1996). After cells were grown in YPD or with SC-Ura with 0.5% casamino acids at 26°C to an OD600 of 2–4, the cells were suspended in gel shift buffer [50 mM HEPES-NaOH, pH 8.0, 0.4 M (NH4)2SO4, 1 mM EDTA, and 5% glycerol] containing 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml antipain, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, 100 mM NaF, 1 mM sodium orthovanadate, and 25 mM β-glycerophosphate and disrupted mechanically with glass beads. The cell extracts were immediately dialyzed against dialysis buffer (25 mM HEPES-NaOH, pH 8.0, 75 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, and 12% glycerol). The extract (60 μg of protein) was incubated with 0.2 ng of DNA probe (20,000–40,000 cpm) in 40 μl of binding buffer (25 mM HEPES-NaOH, pH 8.0, 75 mM KCl, 1 mM dithiothreitol, 0.05% Nonidet P-40, and 5% glycerol) and separated by electrophoresis on a 4% polyacrylamide gel. The gel was dried and subjected to autoradiography. To make the DNA probe, two complementary oligonucleotides from the DDR2 promoter sequence 5′-AATTCTG-TCTTTTCTCACCCCTTATGGGGAC-3′ and 5′-TCGAGTCCCCA-TAAGGGGTGAGAAAAGACA-3′ were annealed at 85°C and radiolabeled by end-filling with the Klenow fragment with [α-32P]dATP. Unlabeled competitor DNA was added at 400-fold molar excess. As mutant oligonucleotides, two complementary oligonucleotides, 5′-AATTCTGTCTTTTCTCACCACTTATGGGGAC-3′ and 5′-TCGAGTCCCCATAAGTGGTGAGAAAAGACA-3′, were used.
Assay for Transcriptional Activation by LexA-MSN2ΔZn
Yeast strains LTA001 and LTA002 (containing LexA-lacZ) were transformed with plasmid pBTM116HA-MSN2ΔZn or pBTM116HA. After the cells were grown in SC-Trp to an OD600 of 0.1–0.2 at 26°C, β-galactosidase activity was measured.
Other Methods
For immunoprecipitation assays, transformants were harvested and frozen in liquid nitrogen after being exposed to various stress conditions for 10 min. Cells were suspended in 125 μl of cold lysis buffer (50 mM Tris-HCl, pH 7.5, 0.3 M mannitol, 0.1 M KCl, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml antipain, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, 100 mM NaF, 1 mM sodium orthovanadate, and 25 mM β-glycerophosphate) and broken by vortex mixer with glass beads for 5 min at 4°C. Then, 125 μl of lysis buffer was added, and the homogenate was again mixed vigorously. The homogenate was centrifuged at 14,000 × g for 5 min, and the supernatant was immunoprecipitated with the anti-Myc antibody. After SDS-PAGE, blots of the immunoprecipitates were probed with the anti-Myc and anti-HA antibodies.
To examine the phosphorylation of Msn2p in intact cells in response to stress, cells were lysed by incubation with 1.8 M NaOH and 1% 2-mercaptoethanol (final concentrations) for 10 min on ice after exposure to stress treatments. Proteins were then precipitated by adding trichloroacetic acid to a final concentration of 25%. The precipitates were centrifuged at 14,000 × g for 5 min and resuspended in 30 μl of sample buffer and 20 μl of 1 M Tris base (Volland et al., 1992). After SDS-PAGE, blots of the lysates were probed with the anti-Myc antibody.
RESULTS
Isolation of Multicopy Suppressors of the gsk-3 Null Mutant
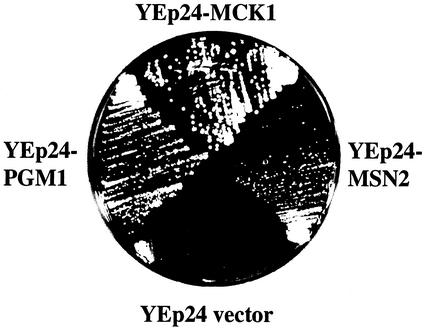
In a previous study (Andoh et al., 2000), we generated the mck1 mds1 mrk1 yol128c quadruple-null mutant (gsk-3 null mutant) and the mck1 mds1 double-null mutant to examine functions of GSK-3 in yeast. We found that the gsk-3 null mutant and the mck1 mds1 double-null mutant grow extremely poorly on galactose plates at 26°C. To select multicopy suppressors of the gsk-3 null mutant, this strain was transformed with a library of yeast genomic DNA clones. From ∼80,000 Ura+ transformants, 87 colonies were isolated. Most plasmid DNAs from these original 87 colonies contained the MCK1 or MDS1 gene. The remaining seven plasmid DNAs did not contain a GSK-3 gene and were found to suppress a galactose-sensitive phenotype on reintroduction into the gsk-3 null mutant. Two of them contained MSN2 and PGM1. MSN2 encodes a zinc-finger transcription activator for genes involved in various stress responses (Estruch and Carlson, 1993; Martinez-Pastor et al., 1996). PGM1 encodes a phosphoglucomutase that interconverts glucose-1 phosphate and glucose-6 phosphate. Interestingly, it has been reported that expression of PGM2, a PGM1 homologue, is regulated by Msn2p (Treger et al., 1998). As shown in Figure 1, the presence of MSN2 or PGM1 in high copy number permitted growth of the gsk-3 null mutant on a galactose plate, as did MCK1, indicating that MCK1, MSN2, and PGM1 interact genetically.
Figure 1.
Suppression by MSN2 and PGM1 of growth defect of the gsk-3 null mutant on a galactose plate. Cells of strain YTA003W (gsk-3 null mutant, Δgsk-3) transformed with YEp24-MSN2 or YEp24-PGM1 were streaked on SG-Ura plates and incubated at 26°C for 3 d. Cells of strain YTA003W transformed with YEp24-MCK1 or YEp24 were streaked as positive and negative controls, respectively.
Similar Pleiotropic Stress Sensitivity of the gsk-3 and msn2 msn4 Mutants
Msn4p is a homologue of Msn2p. Msn2p and Msn4p seem to be functionally redundant, because double but not single mutants exhibit pleiotropic stress sensitivity to conditions including carbon-source starvation, heat shock, high osmolarity, and oxidation (Martinez-Pastor et al., 1996). Furthermore, both are required for metabolic adaptation to glucose limitation and growth on alternative carbon sources (Martinez-Pastor et al., 1996). Indeed, growth inhibition by carbon-source limitation (substitution of glucose with galactose) was observed in the gsk-3 null (Figure 1) and the msn2 msn4 double mutant (Estruch and Carlson, 1993). However, although the gsk-3 null mutant showed a growth defect under moderate temperature (37°C) and NaCl (0.4 M) stress, the msn2 msn4 double mutant did not (Martinez-Pastor et al., 1996). Therefore, we examined the effects of severe stresses on these mutants. The tolerance for stresses was analyzed by measuring the viability of exponentially growing cells after treatment with heat shock at 45°C, 2 M NaCl, and 5 mM hydrogen peroxide (Figure 2). The gsk-3 null and msn2 msn4 mutants exhibited a lower survival rate than the wild-type. These phenotypes with respect to survival against stresses of the gsk-3 null mutant and the msn2 msn4 mutant suggest that both GSK-3 and Msn2p/Msn4p are involved in stress tolerance.
Requirement of GSK-3 for Stress-dependent Transcription
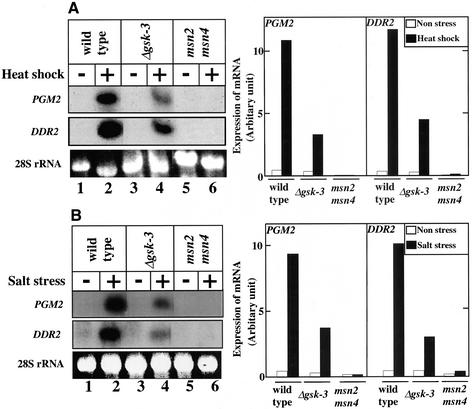
Msn2p and Msn4p play a role in the transcriptional activation of genes such as HSP12, CTT1, and DDR2 in the protective response to different types of stress (Martinez-Pastor et al., 1996). Stress-dependent PGM2 mRNA induction is abolished in the msn2 msn4 double mutant (Treger et al., 1998). Therefore, we compared the transcriptional levels of PGM2 and DDR2 after heat-shock and salt stresses by Northern blot analysis of the wild-type strain, the gsk-3 null mutant, and the msn2 msn4 double mutant. Consistent with previous observations (Martinez-Pastor et al., 1996; Treger et al., 1998), both genes showed defective induction in the msn2 msn4 mutant (Figure 3, A and B, lanes 1, 2, 5, and 6). Furthermore, expression of PGM2 and DDR2 mRNA induced by high temperature and NaCl was also decreased in the gsk-3 null mutant (Figure 3, A and B, lanes 1–4). These results suggest that GSK-3 is at least partly involved in stress-dependent and Msn2p- and Msn4p-mediated transcriptional activation.
Figure 3.
Requirement for GSK-3 in the stress-dependent transcription of PGM2 and DDR2. Exponentially growing cells of strains W303a (wild-type, lanes 1 and 2), YTA003W (Δgsk-3, lanes 3 and 4), and CEPA (msn2 msn4, lanes 5 and 6) were treated with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) heat for 10 min (A) or salt for 10 min (B) as described in MATERIALS AND METHODS. Total RNA (20 μg) prepared from the cells was electrophoresed on a 1% agarose gel containing formaldehyde and probed with PGM2 and DDR2 fragments. The application and transfer of equal amounts of RNA were verified by ethidium bromide staining of 28S rRNA. After exposure, each film was divided into three to be arranged (left panels). The intensities of bands were analyzed by NIH Image software (right panels).
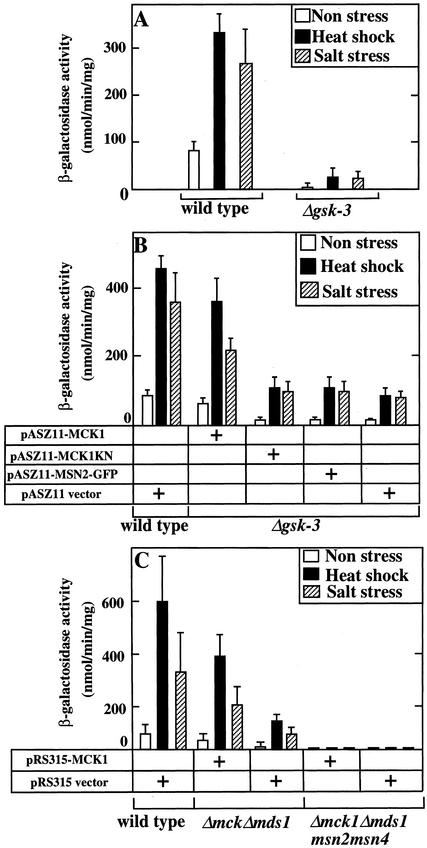
Because Msn2p and Msn4p are involved in the stress-induced gene expression driven by STRE (Martinez-Pastor et al., 1996), we asked whether the stress-induced gene expression conferred by an STRE fused to a heterologous LEU2-lacZ gene is also affected in the gsk-3 null mutant. To this end, the wild-type strain and the gsk-3 null mutant carrying a single chromosomally integrated STRE-LEU2-lacZ reporter gene were generated. After the cells were subjected to high temperature or NaCl, β-galactosidase activity was determined (Figure 4A). In the wild-type cells, the transcription of β-galactosidase was increased in response to these stresses. In the gsk-3 null mutant, the basal level of transcription was dramatically reduced, and transcription under stress conditions was also lower than that of the wild-type strain (Figure 4A). These results suggest that GSK-3 is necessary for the gene expression mediated by Msn2p and Msn4p through STRE. To examine whether GSK-3 actually regulates Msn2p- and Msn4p-driven transcription, Mck1p, one of the yeast GSK-3 family members, was expressed by an original promoter (single copy) in the gsk-3 null mutant (Figure 4B). The activity of β-galactosidase in the gsk-3 null mutant was restored to 50–75% of the wild-type level by expression of Mck1p. The reason that the activity was not completely restored in the gsk-3 null mutant may be that other GSK-3 family members are also involved in STRE-mediated gene expression. Expression of a kinase-negative form of Mck1p (Mck1KN) did not rescue the transcriptional activity in the gsk-3 null mutant (Figure 4B), suggesting that the kinase activity of Mck1p is necessary for STRE-mediated gene expression.
Figure 4.
Requirement for GSK-3 in STRE-mediated transcription. (A) Stress-induced transcription in the gsk-3 null mutant. After cells of strains W303a stre (wild-type) and YTA003W stre (Δgsk-3) were exposed to heat shock or salt stress for 1 h, β-galactosidase activities of the cells were assayed and expressed in Miller units (Miller, 1972). (B) Effects of Mck1p on stress-induced transcription in the gsk-3 null mutant. Cells of strains W303a stre and YTA003W stre transformed with the indicated plasmids were exposed to heat or salt stress for 1 h. (C) Effects of Mck1p on stress-induced transcription in the Δmck1 Δmds1 msn2 msn4 quadruple mutant. Cells of strains W303a stre (wild-type), YTA002W stre (Δmck1 Δmds1 double mutant), and YYH001 stre (Δmck1 Δmds1 msn2 msn4) transformed with the indicated plasmids were exposed to heat or salt stress for 1 h. The results shown are means ± SEM of three independent experiments. Note that the experiments shown in A used YPD medium, whereas those shown in B and C used synthetic media. The differences in the absolute values of transcription observed may reflect this difference in medium.
To examine whether Msn2p and Msn4p function downstream of GSK-3, we generated the mck1 mds1 msn2 msn4 quadruple mutant. The mck1 mds1 double-null mutant was used instead of the gsk-3 null mutant because the msn2 and msn4 mutations could be introduced more easily and because the double mutant showed similar phenotypes to the gsk-3 null mutant with respect to the growth defect caused by various forms of stress. In both unstressed and stressed conditions, the mck1 mds1 double-null mutant showed low transcriptional activity of the β-galactosidase gene compared with the wild-type, and the mck1 mds1 msn2 msn4 quadruple mutant did not exhibit β-galactosidase activity under any conditions (Figure 4C). Expression of Mck1p restored activity of β-galactosidase in the mck1 mds1 double-null mutant but not in the mck1 mds1 msn2 msn4 quadruple mutant. These results indicate that Mck1p is required for STRE-LEU2-lacZ transcription through Msn2p and Msn4p and suggest that Msn2p and Msn4p function downstream of GSK-3.
Effect of GSK-3 on the Stress-induced Nuclear Translocation of Msn2p
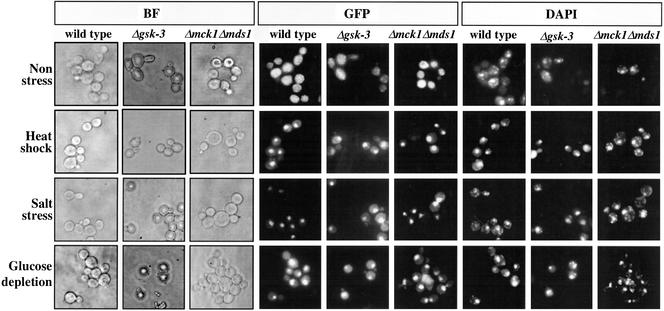
Msn2p is translocated to the nucleus in response to various types of stress (Görner et al., 1998). Thus, we next examined whether GSK-3 regulates stress-induced nuclear localization of Msn2p. To this end, the wild-type strain, the gsk-3 null mutant, and the mck1 mds1 double-null mutant were transformed with pRS316-MSN2-GFP, which is under control by the MSN2 promoter, to express a single copy of MSN2-GFP. Under the unstressed conditions, Msn2p-GFP was distributed diffusely throughout the cytoplasm and was partly excluded from the nucleus in all of the strains (Figure 5). Consistent with previous observations (Görner et al., 1998), Msn2p-GFP was observed in the nuclei of most cells of the wild-type strain after treatment with high temperature, NaCl, or glucose depletion (Figure 5). Similar nuclear localization of Msn2p-GFP was also observed in the gsk-3 null mutant and the mck1 mds1 double-null mutant after the same stress treatments (Figure 5). These results suggest that GSK-3 is not necessary for the stress-induced nuclear localization of Msn2p.
Figure 5.
Effect of GSK-3 on the stress-induced nuclear translocation of Msn2p. Cells of strains W303a (wild-type), YTA003W (Δgsk-3), and YTA002W (Δmck1 Δmds1) transformed with pRS316-MSN2-GFP were examined after non stress or after 10 min of heat, salt, or glucose-depletion stress. GFP was visualized directly after fixation. Chromosomal DNA appears as a diffuse spot in DAPI-stained cells. BF, bright-field microscopy. The results shown are representative of three independent experiments.
To exclude the possibility that expression of Msn2p-GFP might suppress the requirement for GSK-3 in nuclear accumulation of Msn2p, we examined the effects of Msn2p-GFP on other phenotypes of the gsk-3 null mutant. As expected, expression of Msn2p-GFP did not rescue the growth defect of the gsk-3 null mutant on galactose plates or STRE-dependent gene expression of the gsk-3 null mutant (Figure 4B).
Involvement of GSK-3 in the Formation of a Complex between Msn2p and STRE
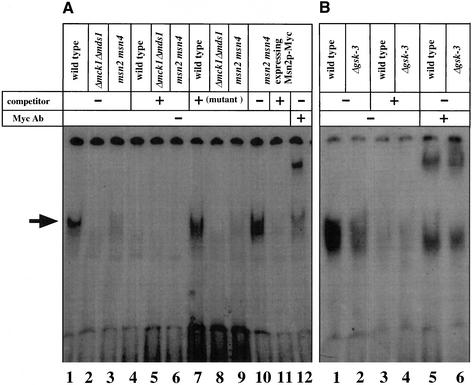
Msn2p and Msn4p each contain two Cys2 His2 zinc fingers at the C terminus, and this domain binds to STRE directly (Martinez-Pastor et al., 1996; Schmitt and McEntee, 1996). Because we found that GSK-3 is not involved in the nuclear translocation of Msn2p, we next examined whether GSK-3 regulates the binding of Msn2p to STRE (Figure 6). Extracts of the wild-type strain, the mck1 mds1 double-null mutant, and the msn2 msn4 double mutant were analyzed by a gel shift assay with a labeled oligonucleotide that includes the sequence of a functional STRE from the DDR2 promoter (base pairs −187 to −165) (Kobayashi and McEntee, 1993). The STRE-binding activity was observed in the wild-type strain (Figure 6A, lane 1). This binding was efficiently competed by an excess of unlabeled oligonucleotide but not by that of mutant oligonucleotide (Figure 6A, lanes 4 and 7). The extracts prepared from the mck1 mds1 double-null and msn2 msn4 double mutants lacked the STRE-binding activity (Figure 6A, lanes 2, 3, 5, 6, 8, and 9). When Msn2p-Myc was expressed in the msn2 msn4 double mutant, a band with mobility similar to that of the band detected in the wild-type strain was observed, and this band also disappeared when excess unlabeled oligonucleotide was added (Figure 6A, lanes 10 and 11). Furthermore, the band was reduced and its migration was retarded by addition of the anti-Myc antibody (Figure 6A, lane 12). Therefore, the STRE-binding activity in the wild-type strain was a result of Msn2p (and/or Msn4p). Similar experiments were carried out with the gsk-3 null mutant. The STRE-binding activity of exogenous Msn2p-Myc was observed in the wild-type strain (Figure 6B, lane 1). This band was competed by unlabeled oligonucleotide, and its migration was retarded by the anti-Myc antibody (Figure 6B, lanes 3 and 5). The STRE-binding activity of Msn2p-Myc was reduced in the gsk-3 null mutant (Figure 6B, lanes 2, 4, and 6). These results suggest that yeast GSK-3 is important for formation of a complex between Msn2p and STRE.
Figure 6.
Involvement of GSK-3 in formation of the complex including Msn2p and STRE. (A) Yeast extracts (60 μg of proteins) from cells of strains W303a (wild-type) (lanes 1, 4, and 7), YTA002W (Δmck1 Δmds1) (lanes 2, 5, and 8), or CEPA (msn2 msn4) (lanes 3, 6, and 9) were incubated with a 32P-labeled oligonucleotide including the STRE sequence without unlabeled oligonucleotide (lanes 1–3), with a 400-fold molar excess of unlabeled oligonucleotide (lanes 4–6), or with a 400-fold molar excess of unlabeled mutant oligonucleotide (lanes 7–9). Yeast extracts (60 μg of proteins) from cells of strain CEPA (msn2 msn4) transformed with pTS904CU-MSN2 (lanes 10–12) were incubated with 32P-labeled oligonucleotide in the presence (lane 11) or absence (lane 10) of a 400-fold molar excess of unlabeled oligonucleotide or in the presence of the anti-Myc antibody (0.5 μg/μl) (lane 12). The position of the Msn2p specific complex is indicated by an arrow. Some faint bands observed below the Msn2p-specific complex in lanes 2, 3, 5, 6, 8, and 9 may be a result of DNA-binding protein(s) other than Msn2p and Msn4p. Ab, antibody. (B) Yeast extracts (60 μg of proteins) from cells of strains W303a (wild-type) (lanes 1, 3, and 5) or YTA003W (Δgsk-3) (lanes 2, 4, and 6) transformed with pTS904CU-MSN2 were incubated with a 32P-labeled oligonucleotide without unlabeled oligonucleotide (lanes 1 and 2), with a 400-fold molar excess of unlabeled oligonucleotide (lanes 3 and 4), and with the anti-Myc antibody (0.5 μg/μl) (lanes 5 and 6). The results shown are representative of three independent experiments.
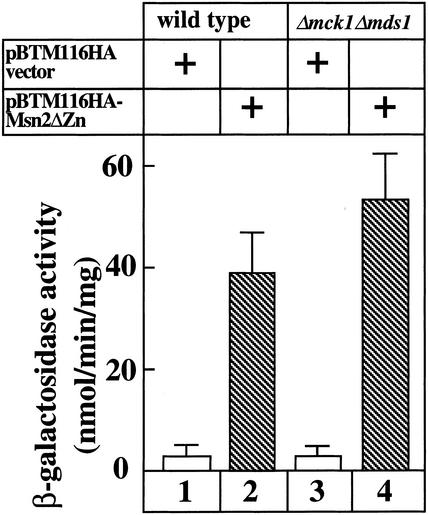
To examine whether GSK-3 stimulates the activity of the transcriptional activator domain, the DNA binding domain of Msn2p was artificially changed. We deleted the C-terminal zinc-finger domain from Msn2p and fused the residual Msn2p to LexA (pBTM116HA-MSNΔZn). The wild-type strain and the mck1 mds1 double-null mutant containing a lacZ reporter gene with the LexA site were transformed with pBTM116HA-MSNΔZn. Immunoblot analysis confirmed that the LexA-Msn2ΔZn protein was expressed to the same level in the wild-type strain and the mck1 mds1 double-null mutant. LexA-Msn2ΔZn protein activated transcription from the LexA site to similar levels in the wild-type and mck1 mds1 mutant strains (Figure 7). These results suggest that GSK-3 is not required for activation of the transcriptional activator domain of Msn2p but is required in the binding of the zinc-finger domain to STRE.
Figure 7.
Transcriptional activation by LexA-Msn2ΔZn protein in the mck1 mds1 double-null mutant. Cells of strains LTA001 (lanes 1 and 2) and LTA002 (lanes 3 and 4) were transformed with pBTM116HA-MSN2ΔZn or pBTM116HA vector. After the cells were grown, β-galactosidase activity was measured. The results shown are the mean ± SEM of three independent experiments.
Possible Mechanism of Regulation of Msn2p by GSK-3
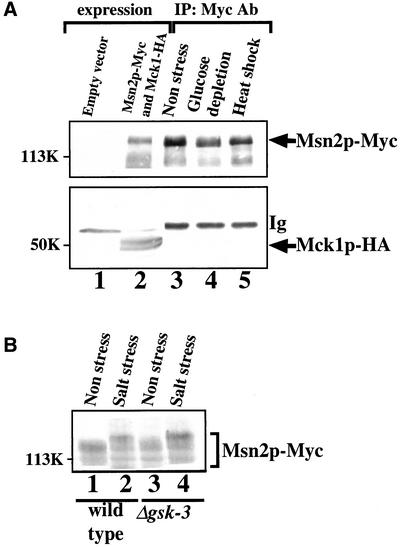
There are several consensus sequences (SXXXS, where S is Ser and X is any amino acid) for phosphorylation by GSK-3 in Msn2p, and the DNA binding site of Msn2p contains two possible phosphorylation sites (S655FKRS and S688DNLS). We examined whether GSK-3 might interact with and phosphorylate Msn2p. When Msn2p-Myc and Mck1p-HA were expressed in the mck1 mds1 double-null mutant and cell extracts were immunoprecipitated with the anti-Myc antibody, Mck1p-HA was not detected in the Msn2p-Myc immune complex either from unstressed or stressed cells (Figure 8A). Mck1p-HA immunoprecipitated from yeast phosphorylated GST-Msn2p purified from Escherichia coli directly in vitro, but the stoichiometry was ∼5%, suggesting that Msn2p is not a good substrate for Mck1p under these conditions. This does not necessarily imply that Mck1p does not phosphorylate Msn2p in intact cells, because the phosphorylation of some substrates by mammalian GSK-3 requires prior phosphorylation by a distinct kinase (Plyte et al., 1992; Cohen and Frame, 2001). Therefore, we examined whether yeast GSK-3 phosphorylates Msn2p in intact cells. It has been shown that heat shock and glucose starvation induce a mobility shift of Msn2p on an SDS gel, which reflects the phosphorylation of Msn2p (Garreau et al., 2000). After the wild-type strain and the gsk-3 null mutant had been subjected to salt stress, the yeast extracts were electrophoresed and probed with the anti-Myc antibody to detect Msn2p. Salt stress induced a similar mobility shift of Msn2p-Myc in both the wild-type and the gsk-3 null mutant strains (Figure 8B). Thus, no role for GSK-3 in the phosphorylation of Msn2p was detectable in these experiments.
Figure 8.
Attempts to detect GSK-3-Msn2p complexes or GSK-3–dependent Msn2p phosphorylation. (A) Attempt to detect Mck1p-Msn2p complexes. Cells of strain YTA002W (Δmck1Δmds1) transformed with pTS904EU-MSN2 and YEplac181-MCK1-HA (lanes 2–5) were treated with glucose depletion (lane 4) or heat shock (lane 5) for 10 min. Some cells remained unstressed (lane 3). The cell extracts were probed directly with the anti-Myc and anti-HA antibodies (Ab) or immunoprecipitated with the anti-Myc antibody. The immunoprecipitates were probed with the anti-Myc and anti-HA antibodies. (B) Attempts to detect GSK-3–dependent phosphorylation of Msn2p in intact cells. Cells of strains W303a (wild-type) (lanes 1 and 2) and YTA003W (Δgsk-3) (lanes 3 and 4) transformed with pTS904CU-MSN2 were untreated (lanes 1 and 3) or treated with salt stress (lanes 2 and 4). The cell extracts were probed with the anti-Myc antibody. The mobility shift of Msn2p-Myc under stress conditions might not have been seen clearly in A because of the difference in polyacrylamide gel concentrations (10% in A and 6% in B). The results shown are representative of three independent experiments.
DISCUSSION
Galactose metabolism in yeast is mediated by the Leloir pathway. This pathway converts galactose to glucose-1 phosphate, which is further converted to glucose-6 phosphate to enter glycolysis. A key enzyme involved in this process is phosphoglucomutase, which catalyzes the interconversion of glucose-1 phosphate and glucose-6 phosphate. S. cerevisiae contains two genes encoding isoforms of phosphoglucomutase, PGM1 and PGM2. Expression of PGM2, encoding the major isoform of PGM, is greatly increased in galactose medium (Oh and Hopper, 1990). In contrast, PGM1 expresses the minor isoform of PGM at a relatively low and constitutive level (Oh and Hopper, 1990). Yeast strains that lack both enzymes are unable to grow when galactose is the sole carbon source (Boles et al., 1994). The upstream region of PGM2 has five putative STRE sequences (Treger et al., 1998), and the msn2 msn4 mutant is defective in anaerobic growth on galactose (Estruch and Carlson, 1993). In this study, we have demonstrated a growth defect of the gsk-3 null mutant in galactose medium and isolated MSN2 and PGM1 as multicopy suppressors of this phenotype. We also isolated SEC53, encoding phosphomannomutase, as a multicopy suppressor of the gsk-3 null mutant (our unpublished results). Overexpression of SEC53 restored growth of the pgm1 pgm2 double mutant on galactose medium (Boles et al., 1994). Phosphomannomutase catalyzes the reversible conversion not only of mannose-1 phosphate and mannose-6 phosphate but also of glucose-1 phosphate and glucose-6 phosphate (Boles et al., 1994). These results suggest that the growth defect of the gsk-3 mutant on galactose medium is caused by its low PGM level. As described below, PGM expression is indeed low in the gsk-3 mutant. When GSK-3 is lacking, overexpression of Msn2p, PGM, or Sec53p could restore growth in galactose medium by increasing an activity to catalyze the reversible transfer of phosphate between the first and terminal carbons of glucose phosphate.
Three lines of evidence suggest that GSK-3 is necessary for Msn2p-dependent transcription. First, the gsk-3 null mutant showed reduced cell viability in response to different types of stress, including heat, salt, and oxidative stresses, as also observed for the msn2 msn4 mutant (Martinez-Pastor et al., 1996). Second, induction of the mRNAs of PGM2 and DDR2, which contain STREs in their promoter regions, by treatment with heat or NaCl was greatly reduced in the gsk-3 null mutant. This finding is similar to previous observations that heat induction of transcripts of PGM2 is completely abolished in the msn2 msn4 double mutant (Treger et al., 1998). Finally, in the gsk-3 null mutant, expression of an STRE-LEU2-lacZ reporter gene was decreased, and the GSK-3 isoform Mck1p but not kinase-negative Mck1p rescued this phenotype. These results indicate that GSK-3 mediates Msn2p- and STRE-dependent gene expression in yeast.
How does GSK-3 regulate Msn2p? In response to stress, Msn2p is translocated to the nucleus (Görner et al., 1998), but whether this process is sufficient to stimulate gene expression has not yet been addressed. Under nonstress conditions, PKA phosphorylates the nuclear localization signal (NLS) of Msn2p, resulting in the retention of Msn2p in the cytoplasm (Görner et al., 1998, 2002). Srb10p, a cyclin-dependent protein kinase, directly phosphorylates Msn2p, and Msn2p is localized to the nucleus of unstressed srb10 mutants, suggesting that Srb10p-dependent phosphorylation suppresses nuclear localization of Msn2p (Chi et al., 2001). Glucose depletion induces dephosphorylation of the NLS of Msn2p and its nuclear translocation (Görner et al., 1998). Other types of stress, including heat and NaCl, do not affect the PKA-dependent phosphorylation of the Msn2p NLS, but instead modulate the nuclear export signal, resulting in the nuclear accumulation of Msn2p (Görner et al., 2002). Thus, phosphorylation of Msn2p is important for the regulation of its subcellular localization. However, it is unlikely that GSK-3 is involved in these processes of stress-induced nuclear translocation of Msn2p, because heat shock, salt stress, and glucose depletion induced seemingly normal nuclear accumulations of Msn2p in the gsk-3 null and mck1 mds1 double mutants. These results also clearly show that nuclear translocation of Msn2p is not sufficient for stress-induced gene expression. It has been shown that Mck1p binds to and directly inhibits, but does not phosphorylate, the catalytic subunits of PKA (Rayner et al., 2002). PKA is important for retention of Msn2p in the cytoplasm. Because we showed that Msn2p is still present in the cytoplasm of the gsk-3 null mutant in the absence of stress, it remains to be clarified whether this new action of Mck1p on PKA is related to stress-induced nuclear translocation of Msn2p.
Our results have demonstrated that GSK-3 is necessary for the formation of a complex between Msn2p and STRE. Moreover, the transcriptional activation activity of a LexA–Msn2ΔZn fusion protein is not affected by GSK-3. Therefore, it is likely that GSK-3 primarily regulates the complex formation between Msn2p and STRE but does not stimulate the activity of the transcriptional activation domain. Nitrogen limitation stimulates the interaction of Ume6p and Ime1p, two subunits of a heteromeric transcriptional activator, and activates meiotic gene expression in yeast. It has been shown that Rim11p (Mds1p) phosphorylates Ume6p and Ime1p and that this phosphorylation promotes formation of a complex between Ume6p and Ime1p and hence meiotic gene activation (Xiao and Mitchell, 2000). Thus, direct phosphorylation of transcription factors by yeast GSK-3 may be important for the regulation of their transcriptional activity. Although our results did not show that Mck1p phosphorylates Msn2p significantly, we cannot exclude the possibility that GSK-3 may affect the phosphorylation states of Msn2p under the appropriate conditions. Alternatively, it is possible that Mck1p phosphorylates another protein that regulates the transcriptional activity of Msn2p by enhancing the formation of a complex between Msn2p and STRE. Further experiments will be necessary for a full understanding of the regulation of gene expression by GSK-3.
ACKNOWLEDGMENTS
We are grateful to Drs. F. Estruch, Y. Ohya, Y. Kikuchi, and R. Akada for their strains and plasmids. We thank to Drs. Y. Kikuchi, T. Miyakawa, K. Matsumoto, K. Tanaka, and D. Kaida for helpful discussion. This work was supported by a grant-in-aid for scientific research on priority areas (C) from the Ministry of Education, Science, and Culture, Japan (2000, 2001).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0247. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0247.
REFERENCES
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh T, Hirata Y, Kikuchi A. Yeast glycogen synthase kinase 3 is involved in protein degradation in cooperation with Bul1, Bul2, and Rsp5. Mol Cell Biol. 2000;20:6712–6720. doi: 10.1128/mcb.20.18.6712-6720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Boles E, Liebetrau W, Hofmann M, Zimmermann FK. A family of hexosephosphate mutases in Saccharomyces cerevisiae. Eur J Biochem. 1994;220:83–96. doi: 10.1111/j.1432-1033.1994.tb18601.x. [DOI] [PubMed] [Google Scholar]
- Bowdish KS, Yuan HE, Mitchell AP. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau H, Hasan RN, Renault G, Estruch F, Boy-Marcotte E, Jacquet M. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology. 2000;146:2113–2120. doi: 10.1099/00221287-146-9-2113. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ, Plyte SE, Woodgett J, Strutt H, Kay RR. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet J-P, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mizunuma M, Hirata D, Miyaoka R, Miyakawa T. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 2001;20:1074–1085. doi: 10.1093/emboj/20.5.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Oh D, Hopper JE. Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol. 1990;10:1415–1422. doi: 10.1128/mcb.10.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Smouse D. Multiple functions of a Drosophila homeotic gene, zeste-white 3, during segmentation and neurogenesis. Dev Biol. 1989;135:287–305. doi: 10.1016/0012-1606(89)90180-2. [DOI] [PubMed] [Google Scholar]
- Plyte SE, Hughes K, Nikolakaki E, Pulverer BJ, Woodgett JR. Glycogen synthase kinase-3: functions in oncogenesis and development. Biochim Biophys Acta. 1992;1114:147–162. doi: 10.1016/0304-419x(92)90012-n. [DOI] [PubMed] [Google Scholar]
- Rayner TF, Gray JV, Thorner JW. Direct and novel regulation of cAMP-dependent protein kinase by Mck1p, a yeast glycogen synthase kinase-3. J Biol Chem. 2002;277:16814–16822. doi: 10.1074/jbc.M112349200. [DOI] [PubMed] [Google Scholar]
- Ruel L, Bourouis M, Heitzler P, Pantesco V, Simpson P. Drosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch. Nature. 1993;362:557–560. doi: 10.1038/362557a0. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Toh-E A, Kikuchi Y. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol Cell Biol. 2000;20:7971–7979. doi: 10.1128/mcb.20.21.7971-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero JH, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P, El Messal M, Moscoso del Prado J, Ripoll P. Stripes of positional homologies across the wing blade of Drosophila melanogaster. Development. 1988;103:391–401. [Google Scholar]
- Treger JM, Schmitt AP, Simon JR, McEntee K. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:26875–26879. doi: 10.1074/jbc.273.41.26875. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Volland C, Garnier C, Haguenauer-Tsapis R. In vivo phosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- Wieser R, Adam G, Wagner A, Schüller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12406–12411. [PubMed] [Google Scholar]
- Xiao Y, Mitchell AP. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol Cell Biol. 2000;20:5447–5453. doi: 10.1128/mcb.20.15.5447-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]