Abstract
The core of the cytochrome c oxidase complex is composed of its three largest subunits, Cox1p, Cox2p, and Cox3p, which are encoded in mitochondrial DNA of Saccharomyces cerevisiae and inserted into the inner membrane from the inside. Mitochondrial translation of the COX1, COX2, and COX3 mRNAs is activated mRNA specifically by the nuclearly coded proteins Pet309p, Pet111p, and the concerted action of Pet54p, Pet122p, and Pet494p, respectively. Because the translational activators recognize sites in the 5′-untranslated leaders of these mRNAs and because untranslated mRNA sequences contain information for targeting their protein products, the activators are likely to play a role in localizing translation. Herein, we report physical associations among the mRNA-specific translational activator proteins, located on the matrix side of the inner membrane. These interactions, detected by coimmune precipitation and by two-hybrid experiments, suggest that the translational activator proteins could be organized on the surface of the inner membrane such that synthesis of Cox1p, Cox2p, and Cox3p would be colocalized in a way that facilitates assembly of the core of the cytochrome c oxidase complex. In addition, we found interactions between Nam1p/Mtf2p and the translational activators, suggesting an organized delivery of mitochondrial mRNAs to the translation system.
INTRODUCTION
Translational control and mRNA localization, achieved via a variety of mechanisms, are important for the delivery of certain cytoplasmically synthesized proteins to their functional destinations within cells of animals (Johnstone and Lasko, 2001; Palacios and Johnston, 2001), plants (Choi et al., 2000), and yeast (Zoladek et al., 1995; Lithgow et al., 1997; Corral-Debrinski et al., 2000). In general, these strategies seem to reinforce targeting information present within the protein products themselves. In S. cerevisiae, for example, nuclearly coded mRNAs for several mitochondrial proteins bearing mitochondrial import signals seem to be translated preferentially by cytoplasmic ribosomes tightly associated with the organelles, facilitating their localization (Marc et al., 2002). This targeting is dependent upon signals in the mRNA 3′-untranslated regions.
Translation within the mitochondrial matrix of most, if not all, mRNAs encoded in mitochondrial DNA depends upon mRNA-specific translational activators that recognize targets in the mRNA 5′-untranslated leaders (UTLs) and seem to mediate mRNA interactions with mitochondrial ribosomes (Fox, 1996a). All but one of the major proteins encoded by yeast mitochondrial genes are integral membrane proteins that are assembled with nuclear gene products to form respiratory chain complexes in the inner membrane (Pon and Schatz, 1991). The core of the cytochrome c oxidase complex of mammals and yeast comprises three mitochondrially coded subunits, Cox1p, Cox2p, and Cox3p, and is surrounded by imported subunits coded by nuclear genes (Tzagoloff and Dieckmann, 1990; Tsukihara et al., 1996). Translation of each mitochondrially coded mRNA is specifically activated by distinct nuclear gene products: Pet309p for COX1 (Manthey and McEwen, 1995), Pet111p for COX2 (Mulero and Fox, 1993a,b), and Pet54p, Pet122p, and Pet494p for COX3 (Costanzo and Fox, 1988; Brown et al., 1994). Pet309p, detected as an overproduced epitope-tagged protein, is an integral inner membrane protein partially exposed on the intermembrane space side (outside) (Manthey et al., 1998). Pet111p, detected as an epitope-tagged protein at wild-type levels, is an integral inner membrane protein facing the matrix (Green-Willms et al., 2001). Pet54p is a peripheral inner membrane protein, whereas Pet122p and Pet494p, detected as overproduced proteins, behaved like integral inner membrane proteins (McMullin and Fox, 1993). The topology of the COX3 mRNA-specific proteins has not been previously investigated.
The fact that several mRNA-specific translational activator proteins were found to be bound to the inner membrane suggested that they could localize translation to sites where the mitochondrial gene products could be efficiently assembled into respiratory complexes (Costanzo and Fox, 1990; Michaelis et al., 1991; McMullin and Fox, 1993; Fox, 1996a). Consistent with this hypothesis, topological information required for efficient assembly of two mitochondrially coded subunits of the cytochrome c oxidase complex, Cox2p and Cox3p, has been shown to reside in untranslated portions of their mRNAs (Sanchirico et al., 1998). In addition to their apparent role in localizing translation on the matrix side of the inner membrane, the COX2 and COX3 mRNA-specific translational activators are also present at levels that limit expression of their target mitochondrial genes (Steele et al., 1996; Green-Willms et al., 2001).
Although it is easy to imagine a biological rationale for using a single mechanism to both regulate the expression level of an organellar gene and to target its product to the inner membrane, possible rationales for mRNA-specific functions are less obvious. Why should the mRNAs specifying the three core subunits of the cytochrome c oxidase complex, Cox1p, Cox2p, and Cox3p, require three distinct nuclearly encoded translational activators? One attractive hypothesis is that these mRNA-specific translational activators could be organized on the surface of the inner membrane such that they would promote adjacent translation of the COX1, COX2, and COX3 mRNAs, and thereby facilitate assembly of the cytochrome oxidase core.
A clear prediction of this hypothesis is that the translational activators for the three cytochrome oxidase subunits should physically interact with each other. In this study, we have tested this prediction by two-hybrid analysis and coimmune precipitation experiments. We found evidence for a network of interactions among these translational activator proteins, suggesting that they could be organized in the mitochondrial inner membrane to colocalize synthesis of the three core cytochrome c oxidase subunits. Furthermore, we found evidence that Nam1p/Mtf2p, a protein involved in mRNA transactions that interacts with the mitochondrial RNA polymerase (Lisowsky and Michaelis, 1989; Wallis et al., 1994; Rodeheffer et al., 2001; Bryan et al., 2002), also interacts with the translational activators. This suggests an organized flow of information from mitochondrial DNA to the inner mitochondrial membrane.
MATERIALS AND METHODS
Yeast Strains, Media, and Standard Methods
Saccharomyces cerevisiae strains used in this study are listed in Table 1. All strains are isogenic to the wild-type strain D273-10B (ATCC 25657) except PJ69-4a (James et al., 1996). Cells were grown in rich medium YPD or YPR (1% yeast extract, 2% bacto-peptone, 2% glucose or 2% raffinose) and in nonfermentable medium YPEG (1% yeast extract, 2% bacto-peptone, 20 mg adenine/l, 3% ethanol, 3% glycerol). Synthetic complete (SC) media have been described previously (Sherman, 1991) except that nutritional supplements lacking specific factor(s) were purchased from Bio 101 (Vista, CA). Transformation of plasmids and polymerase chain reaction (PCR) products into yeast cells were accomplished by using EZ-Transformation kit (Zymo Research, Orange, CA). To construct the nam1 deletion strain SN24, a disruption cassette containing the URA3 gene flanked by 45 base pairs of sequence homologous to the NAM1 coding region was amplified by PCR, purified, and transformed into SB09. The transformants were selected on SC-uracil media and then printed on YPEG plates to identify cells incapable of respiratory growth, because NAM1 is essential for respiration. Deletion of NAM1 was confirmed by PCR.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MCC100 | MATa ura3-52 pet494-41 [ρ+] | M. C. Costanzo |
| CAB21-2 | MATa ura3-52 PET494∷3xHA [ρ+] | This study |
| CAB13 | MATa ade2 ura3-52 trp1-Δ1 pet122-6 [ρ+] | This study |
| CAB14 | MATa ade2 ura3-52 PET122∷3xHA∷TRP1 [ρ+] | This study |
| CAB30 | MATa ura3-52 PET494∷3xHA PET122∷3xHA [ρ+] | This study |
| SN25 | MATa PET494∷3xHA PET122∷3xHA PET54∷3xMYC [ρ+] | This study |
| SN28 | MATα ura3-52 trp1-Δ1 [rho+] PET54∷3xMYC [ρ+] | This study |
| SN32 | MATa ura3-52 PET494∷3xHA PET122∷3xHA PET309∷3xMYC [ρ+] | This study |
| SN33 | MATa ura3-52 PET494∷3xHA PET122∷3xHA PET111∷3xMYC [ρ+] | This study |
| PJ69-4A | MATa ura3-52 leu2-3, 112 trp1-901 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2∷GAL1-HIS3 met2∷GAL7-lacZ [ρ+] | James et al., (1996) |
| PTY11 | MATα ura3-52 trp1-Δ1 [ρ+] | Green-Willms et al., (1998) |
| SB09 | MATα ade2 PET309∷3xHA ura3-52 [ρ+] | S. A. Broadley |
| SN24 | MATα ade2 PET309∷3xHA nam1Δ∷URA3 [ρ+] | This study |
Epitope Tagging
The coding region of PET122 from +170 to +961 base pairs (relative to the ATG start codon) was amplified by PCR by using upstream primer 5′-GGG AAT TCC CAT GCC GAC ACT ATA GC-3′ and downstream primer 5′-CCC CAT GGT GTT GAT TTC AAA TCC TCT-3′. The amplified DNA fragment was subcloned at NcoI-EcoRI sites of the p3XHA plasmid (Tyers et al., 1992). The resulting plasmid, pPET122HA, contains an insertion of the hemagglutinin (HA)-cassette (encoding three tandem copies of HA-epitope) at the C terminus of the PET122. pPET122HA was linearized with NruI and then transformed into strain CAB13. Trp+ transformants were printed on YPEG plates to check restoration of respiratory growth. The integration of PET122-HA was confirmed by PCR for strain CAB14.
Pet494p is inactivated by some modifications to its C terminus (our unpublished data). We therefore inserted an HA-cassette at +358 base pairs relative to PET494 ATG start codon, a region of the coding sequence known to tolerate sequence changes (Costanzo et al., 1986). A 4.7-kb HindIII-EcoRI fragment containing full-length PET494 was cloned in pBluescript KS (Stratagene, La Jolla, CA) that had the polylinker XhoI site was removed by digestion with ApaI and SalI, followed by Klenow treatment and religation. The HA-cassette from p3XHA was amplified by PCR by using upstream primer 5′-GGA ATT CCG GCT CGA GGC ACT GAG CAG CGT AAT CTG G-3′ and downstream primer 5′-GGA ATT CCG GCT CGA GTA CCC ATA CGA TGT TCC TG-3′ and then cloned into pCB4 at the XhoI site. The resulting plasmid pPET494HA contains an in-frame fusion of the HA-cassette with the coding region of PET494 at +358 base pairs (relative to ATG initiation codon). Recombinant plasmid pPET494HA was digested with HindIII and EcoRI and a 2.9-kb insert containing HA-tagged PET494 was purified and cotransformed with YEp24 (Botstein et al., 1979) into MCC100. Transformants were selected on SC-uracil plates and subsequently printed on YPEG plates to score restoration of functional PET494. Integration of the PET494-HA fragment was verified by PCR in CAB21-2.
To incorporate three copies of HA or MYC tags at the C terminus of other proteins, an HA-URA3-HA or MYC-URA3-MYC cassette flanked by 45 base pairs of sequence homologous to the gene of interest was amplified by PCR by using the appropriate primer pair (Schneider et al., 1995). The resulting PCR products were transformed into desired strains and integration of the MYC or HA-cassette at the desired genomic locus was identified by PCR. Transformants were streaked on medium containing 5′-fluoroorotic acid to select for the loss of selected marker URA3. In-frame fusion of the HA or MYC-cassette was confirmed by sequencing in each case.
Two-Hybrid Analysis
The multicopy expression vectors pGBDU-C1, pGAD-C1 (James et al., 1996), pGAD424, pGBT9 (Bartel et al., 1993), and pGAD2F (Chien et al., 1991) used in this study have been described elsewhere. The PET309 coding region was cloned in the pGADC1 and pGBDUC1 two-hybrid vectors at SalI site to create pSN20 and pSN21 containing in-frame fusion of PET309 coding region with the activation domain (AD) and DNA-binding domain (BD) of GAL4, respectively. The partial PET111 coding region (from +112 to +2434 relative to the PET111 initiation codon) was cloned into pGBT9 at the BamHI site to create pNSG5, containing an in-frame fusion of the PET111 coding region with the BD of GAL4 (Green-Willms, unpublished data). The XmaI-PstI fragment from pNSG5 was cloned into pGAD424. The resulting plasmid, pSCS2, contains an in-frame fusion of the PET111 coding region to the AD of GAL4.
Plasmids carrying coding regions of the PET54 (pNGB8), PET122 (pNGB11), and PET494 (pNGB39) were described previously (Brown et al., 1994). DNA fragments similar to those present in the pNGB8, PNGB11, and pNGB39 were cloned into the binding domain plasmid pGBT9 at the BamHI site. The resulting plasmids pNGB67 and pNGB68 contain fusion of the BD of GAL4 with the full-length coding region of PET54 or PET122, respectively. pNGB70 expresses a hybrid protein consisting of the Gal4p BD fused to an amino terminal truncated form of the Pet494p (lacking 146 residues). The correct orientation and in-frame translational fusion in each case were confirmed by sequencing. Plasmids pACT and pACT-NAM1 were gifts from Dr. G.S. Shadel (Emory University, Atlanta, GA).
All AD plasmids used in this study carried the LEU2 marker gene. The binding domain plasmids based on pGBT9 carried tryptophan (TRP1) selection marker, and pGBDU-C1 based plasmids carried URA3 marker gene. To test interactions a plasmid pair was transformed into the yeast two-hybrid strain PJ69-4a and double transformants were selected on SC media lacking appropriate nutrients depending upon the selection markers on the two plasmids. The growth of the double transformants was tested on SC-histidine containing 2.5 mM 3-aminotriazole and SC-adenine.
Mitochondrial Isolation, Purification, Subfractionation, and Western Analysis
Mitochondria were prepared from late exponential phase cells grown at 30°C in YPR. Mitochondrial isolation, purification, mitoplasting, proteinase K treatment, SDS-PAGE, and Western analyses were carried out as described previously (Glick, 1995; Glick and Pon, 1995; He and Fox, 1997, 1999). Anti-HA-horseradish peroxidase (HRP) (3F10) and anti-MYC-HRP (9E10) were purchased from Roche Diagnostics (Indianapolis, IN). Polyclonal anti-Nam1p was a gift from N. Bonnefoy (CNRS, Gif-sur-Yvette, France), and anti α-ketoglutarate dehydrogenase was a gift from B. Glick (University of Chicago, Chicago, IL). The enhanced chemiluminescence plus detection system (Amersham Biosciences, Piscataway, NJ) was used for detection of proteins on Western blots.
Cross-Linking and Coimmunoprecipitations
Mitochondria (500 μg-1 mg of protein) were resuspended at 1 mg/ml in cross-linking buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 0.6 M sorbitol, 1 mM EDTA, and protease inhibitors cocktail (Roche Diagnostics). Samples were incubated with 1 mM dithiobis(succinimidylpropionate) (DSP) or 2 mM dithio-bis-maleimidoethane (DTME)-membrane permeable, thiol-reversible chemical cross-linkers, at 25°C for 30 min or 1 h, respectively, followed by addition of 100 mM Tris-HCl, pH 8.0 to quench the excess cross-linker. The mitochondria were washed once in 0.6 M sorbitol 50 mM HEPES pH 7.4 and then gently resuspended in solubilization buffer. In experiments where digitonin was used mitochondria were solubilized (1 mg/ml) in immunoprecipitation (IP) buffer 1 (150 mM KOAc, 50 mM HEPES, pH 7.4, 2 mM MgOAc, 2 mM ATP, protease inhibitors cocktail, 1% digitonin) at 4°C for 30 min. Alternatively, mitochondria were solubilized (2.5 mg/ml) with Triton X-100 by using IP buffer 2 (50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, protease inhibitors cocktail) at 4°C for 30 min. The soluble fraction was clarified by centrifugation at 80,000 × g for 20 min and supernatant was incubated with agarose beads (Sepharose CL-4B; Sigma-Aldrich, St. Louis, MO) at 4°C for 1 h under gentle shaking conditions. The supernatant was then incubated with either anti-HA (3F10) affinity matrix (Roche Diagnostics) or anti-MYC (9E10)-agarose conjugate (Santa Cruz Biotechnology, Santa Cruz, CA) for 4 h at 4°C under gentle shaking conditions. Precipitates from digitonin-solubilized mitochondria were washed three times with the IP buffer 1 and once with wash buffer 1 (150 mM KOAc, 50 mM HEPES, pH 7.4, 2 mM MgOAc, 2 mM ATP, 0.1% NP-40, 0.1% Triton X-100). Precipitates from Triton X-100–solubilized mitochondria were washed four times with wash buffer 2 (50 mM HEPES, pH 7.4, 50 mM NaCl, 10% glycerol, protease inhibitors, 0.05% NP-40, 0.1% Triton X-100). Proteins were eluted from beads with SDS sample buffer containing 150 mM dithiothreitol by boiling for 3 min and were analyzed by Western blotting.
RESULTS
Interacting Components of the COX3 mRNA-specific Activator Are on Inner Surface of Mitochondrial Inner Membrane
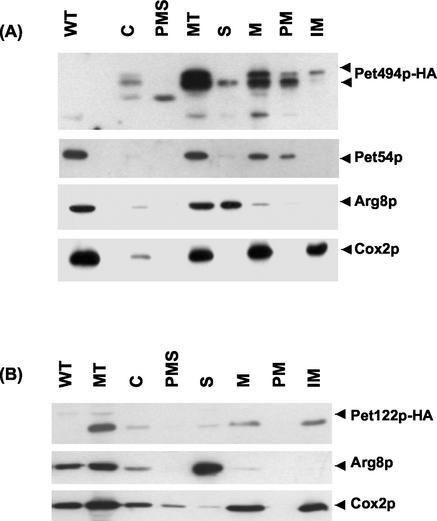
We have previously found that both Pet494p and Pet122p were associated with the mitochondrial inner membrane when overproduced (McMullin and Fox, 1993). However, firm conclusions regarding localization of a protein and its in vivo associations cannot be drawn from studying cells overproducing it. We therefore tagged Pet122p and Pet494p with the HA-epitope by modification of their respective chromosomal genes (see MATERIALS AND METHODS), and examined the submitochondrial location of these proteins along with wild-type Pet54p. Purified mitochondria were fractionated after disruption by osmotic shock and sonication to separate membrane proteins from soluble proteins. Western analysis revealed that Pet122p-HA and most of the Pet494p-HA were recovered in the pellet (Figure 1, A and B), indicating membrane association of these proteins. Pet494p-HA is present as a series of species apparently generated by proteolysis. Full-length Pet494p-HA (∼57 kDa) is present only in the membrane fraction; however, a small proportion of a shorter form (∼53 kDa) is also present in the soluble fraction. Cox2p and Pet54p were also found in the membrane fraction, whereas Arg8p fractionated with the soluble proteins as expected (Figure 1, A and B). In previous similar experiments, we recovered roughly half of Pet54p in the soluble fraction (McMullin and Fox, 1993). However, we are unable to reproduce this result for unknown reasons.
Figure 1.
COX3 mRNA-specific activators Pet54p, Pet122p, and Pet494p are bound to the mitochondrial inner membrane. Mitochondria were purified from yeast strains bearing PET494-HA (CAB21–2) (A) and PET122-HA (CAB14) (B). Aliquots (50 μg) of total cell extract (C), postmitochondrial supernatant (PMS), and total mitochondrial proteins (MT), as well as total mitochondrial protein from an untagged wild-type strain PTY11 (WT) were analyzed on Western blots along with submitochondrial fractions. Mitochondrial proteins were separated into soluble (S) and membrane fractions (M). Membranes were extracted with alkaline carbonate to separate alkaline-soluble peripheral (PM) and alkaline-insoluble integral (IM) membrane proteins. The Western blots were probed with anti-HA-HRP to detect HA-tagged Pet122p or Pet494p. The filters were stripped and reprobed with antibody against Pet54p; Cox2p, an integral membrane; and Arg8p, a soluble matrix protein.
The membrane fraction was then extracted with alkaline sodium carbonate to separate peripheral membrane proteins from integral membrane proteins. Pet54p and most of the Pet494p-HA could be solubilized with alkaline carbonate (Figure 1A). However, a significant fraction of full-length Pet494p-HA (Figure 1A), and all of the Pet122p-HA (Figure 1B) could not be solubilized by alkaline carbonate extraction, indicating that they are integral membrane proteins. The known integral membrane protein Cox2p remained completely associated with the membranes in this experiment, whereas the soluble matrix protein Arg8p did not (Figure 1, A and B).
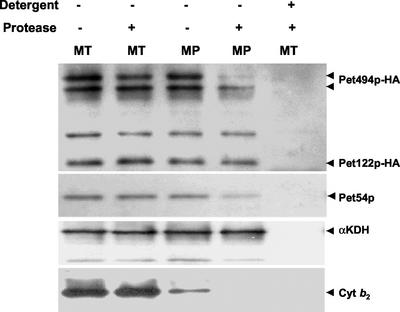
Protease protection experiments were used to determine the submitochondrial location of the three proteins of the COX3 mRNA-specific activator. Mitochondria were prepared from a yeast strain (CAB30) expressing both Pet494p-HA and Pet122p-HA from their chromosomal genes. Proteinase K treatment of detergent-solubilized mitochondria eliminated detectable Pet54p, Pet122p-HA, and Pet494p-HA, showing that none of these proteins are protected by a stable protein complex (Figure 2). However, these three proteins were protected from proteinase K when the mitochondria were intact. Protease treatment of mitoplasts, whose outer membranes were ruptured by osmotic shock, had relatively little effect on either Pet122p-HA or Pet54p, indicating that they are completely within the inner membrane (Figure 2). However, protease treatment of mitoplasts destroyed almost completely the full-length Pet494p-HA, while leaving a shorter form undegraded (Figure 2). This result suggests that the full-length integral membrane protein Pet494p is partially exposed on the outer surface of the inner membrane, probably at the N terminus. However, the HA-epitope was inserted in-frame with the PET494 gene at codon 120 (see MATERIALS AND METHODS), because changes in the C terminus of Pet494p lead to respiratory defects (our unpublished data), making interpretation of the topology difficult. Pet122p was HA tagged at its C terminus, which remains protected in mitoplasts. The soluble intermembrane space protein cytochrome b2 was largely lost during mitoplasting, whereas the soluble matrix protein α-ketoglutarate dehydrogenase remained completely protected (Figure 2).
Figure 2.
Submitochondrial localization of the COX3 mRNA translational activator proteins Pet54p, Pet122p, and Pet494p. Mitochondria were isolated from yeast a strain expressing HA-tagged form of Pet122p-HA and Pet494p-HA (CAB30). Purified mitochondria (MT) and mitoplasts (MP) were subjected to digestion with proteinase K (100 μg/ml) in presence (+) and in absence (−) of 1% octyl-glucoside, a nonionic detergent. Treated samples were analyzed by Western blotting by using anti-HA-HRP to detect Pet122p-HA and Pet494p-HA. Filters were stripped and reprobed with antisera against Pet54p, the soluble intermembrane space protein cytochrome b2 (Cyt b2) and the soluble matrix marker α-ketoglutarate dehydrogenase (αKDH).
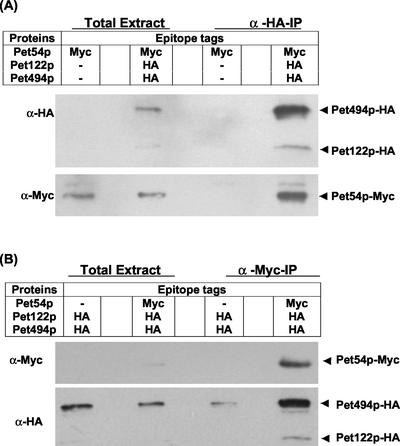
Two-hybrid analysis had previously suggested that Pet54p, Pet122p, and Pet494p interact to form a COX3 mRNA-specific activator complex (Brown et al., 1994). To confirm these interactions, we constructed a strain (SN25) expressing Pet122p-HA, Pet494p-HA, and Pet54p-Myc from their chromosomal loci and carried out coimmune precipitation experiments. Mitochondria purified from this strain were treated with the cross-linker DSP before solubilization with 1% digitonin (see MATERIALS AND METHODS). These soluble extracts were incubated with anti-HA antibody coupled to beads, and the resulting immunoprecipitates were examined by Western analyses by using anti-Myc, and anti-HA, antibodies (see MATERIALS AND METHODS). As shown in Figure 3A, Pet54p-Myc was coimmuneprecipitated with Pet122p-HA and Pet494p-HA, but was not precipitated from control extracts of mitochondria lacking HA tags on Pet122p and Pet494p (Figure 3A). In a converse experiment in which anti-Myc antibody coupled to agarose beads was used to precipitate Pet54p-Myc, specific coimmune precipitation of Pet122p-HA and Pet494p-HA was also observed (Figure 3B).
Figure 3.
Coimmune precipitation of the COX3 mRNA-specific activator proteins Pet54p, Pet122p, and Pet494p. Purified mitochondria containing the indicated epitope-tagged proteins were treated with 1 mM DSP, a membrane-permeable cross-linker at 25°C for 30 min followed by solubilization with 1% digitonin at 4°C for 30 min (see MATERIALS AND METHODS). After a clarifying spin, solubilized extracts (from 400 μg of mitochondria) were incubated with anti-HA affinity matrix (A) or with anti-Myc-agarose conjugate (B) for 4 h at 4°C under gentle shaking conditions and immunoprecipitates were analyzed by Western blotting with anti-HA-HRP and anti-Myc-HRP (see MATERIALS AND METHODS). Solubilized mitochondrial proteins (20 μg) were loaded in total extract lanes. Strains used were SN25, SN28, and CAB30 (Table 1).
Interactions between COX1 and COX3 mRNA-specific Translational Activator Proteins
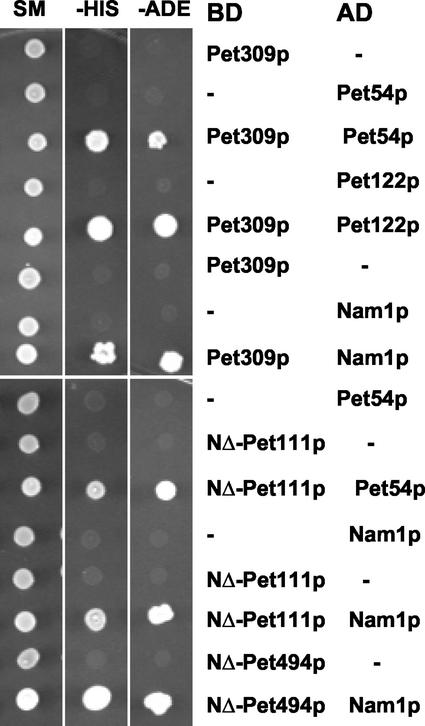
We first sought evidence for interactions of the COX1 mRNA-specific translational activator Pet309p with the COX3 translational activators Pet54p, Pet122p, and Pet494p by using the yeast two-hybrid system. Appropriate plasmid pairs were transformed into test strain PJ69-4a (James et al., 1996), and activation of the HIS3 and ADE2 reporter genes was monitored. Cells containing Pet309p fused to a DNA binding domain of Gal4p alone, and Pet54p or Pet122p fused to an activating domain of Gal4p alone did not express the reporters (Figure 4). However, reporter expression was observed when the Pet309p fusion protein was expressed with either the Pet54p or the Pet122p fusion protein (Figure 4). No two-hybrid interactions were detected between Pet309p and Pet494p, nor were any self-interactions detected in our experiments (our unpublished data).
Figure 4.
Identification of interactions by use of the yeast two-hybrid system. Plasmid pairs were transformed into host strain PJ69-4a and double transformants were selected on appropriate minimal media. All plasmids encoding transcriptional activation domain (AD) of Gal4p carried LEU2 selection marker, and vectors encoding DNA binding domain (BD) of Gal4p carried either TRP1 (pGBT9-based plasmids) or URA3 (pGBDU-C1) (see MATERIALS AND METHODS). Transformants were grown in liquid minimal medium at 30°C. Then 3 μl of each culture (0.2 OD600), was spotted on plates containing medium selecting for plasmid markers (SM), the HIS3 reporter (−His) (SC-histidine + 2.5 mM 3-aminotriazole), and the ADE2 reporter (−Ade) (SC-adenine). Plates were incubated at 30°C for 4 d. The symbol (−) indicates empty vector used in control experiment. In each case, empty vector corresponded to the plasmid carrying the experimental hybrid protein (see MATERIALS AND METHODS).
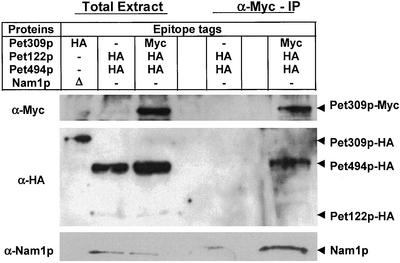
We further explored these interactions by coimmune precipitation. We constructed a strain (SN32) expressing Pet122p-HA, Pet494p-HA, and Pet309p-Myc from chromosomal genes. Mitochondria from this strain were incubated with 2 mM DTME before solubilization with 1% digitonin (see MATERIALS AND METHODS). The soluble extract was precipitated with anti-Myc-agarose and the precipitate analyzed by a Western probed with anti-HA antibody. Roughly 5% of the Pet309p-Myc was precipitated, and Pet494p-HA coimmuneprecipitated with approximately equal efficiency (Figure 5). The yield of coimmuneprecipitated Pet494p-HA was approximately fivefold lower when the cross-linker was omitted (our unpublished data). Taken together with the two-hybrid results, the coimmune precipitations strongly suggest an association between the COX1 and COX3 mRNA-specific translational activators.
Figure 5.
COX3 activator protein Pet494p, and Nam1p coimmuneprecipitate with the COX1 activator Pet309p. Purified mitochondria from a strain containing Pet122p-HA, Pet494p-HA, and Pet309p-Myc (SN32), and a strain containing Pet122p-HA and Pet494p-HA (CAB30) were incubated with the cross-linker 2 mM DTME at 25°C for 1 h, before solubilization with 1% digitonin at 4°C for 30 min (see MATERIALS AND METHODS). Clarified mitochondrial extracts (from 500 μg of mitochondria in each case) were incubated with anti-Myc-agarose conjugate at 4°C for 4 h (see MATERIALS AND METHODS). Immunoprecipitates were analyzed by Western blotting with anti-Nam1p, anti-HA, and anti-Myc antibodies. Solubilized proteins from 50 μg of mitochondria were loaded in total extract lane. To establish the identity of Nam1p, 50 μg of total mitochondrial protein from a nam1Δ strain, which also contains Pet309p-HA (SN24), was probed.
Interactions between COX2 and the COX3 mRNA-specific Activators
We tested for interactions of the COX2 mRNA-specific activator Pet111p with the COX3 activators Pet54p, Pet122p, and Pet494p by two-hybrid and coimmune precipitation experiments. An N-terminally truncated derivative of Pet111p fused to a DNA binding domain failed to activate reporter gene expression, but the combination of the Pet111p fusion and a Pet54p-activating domain fusion did, indicating physical interaction between Pet111p and Pet54p (Figure 4). Cells expressing the Pet111p fusion and an N-terminally truncated Pet494p-activating domain fusion (expressed from pNGB39 (Brown et al., 1994)) were able to activate only the HIS3 reporter weakly (our unpublished data). However, Pet111p did not exhibit two-hybrid interactions with Pet122p or Pet309p (our unpublished data). A fusion protein containing full-length Pet111p fused either to the activating or DNA binding domains of Gal4p failed to exhibit any interactions, including self-interactions.
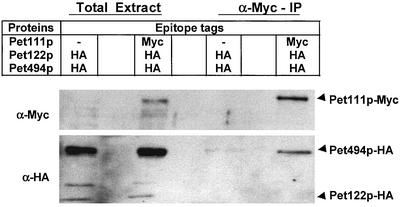
Coimmune precipitation experiments failed to confirm the interaction between Pet111p and Pet54p, but did confirm the interaction between Pet111p and Pet494p. Purified mitochondria from a strain (SN33) expressing Pet122p-HA, Pet494p-HA, and Pet111p-Myc from their chromosomal loci were treated with DTME, followed by solubilization with 0.5% Triton X-100. The solubilized extracts were incubated with anti-Myc-agarose conjugate and the immunoprecipitates were analyzed by Western blots. More than 10% of the Pet111p-Myc was precipitated from these extracts, whereas <0.5% of Pet494p-HA was coimmuneprecipitated (Figure 6). Significantly less Pet494p was present in the control precipitates from mitochondrial extracts containing unmodified Pet111p. In absence of the cross-linker, the yield of the Pet494p-HA in the coimmuneprecipitates was three- to fivefold lower. Coimmune precipitation of Pet54p and Pet122p-HA with Pet111p-Myc was not observed, nor were any interactions observed between the COX2 activator Pet111p and the COX1 activator Pet309p in the two-hybrid system or in coimmune precipitation experiments (our unpublished data).
Figure 6.
COX3 activator Pet494p coimmuneprecipitates with the COX2 activator protein Pet111p. Purified mitochondria containing the indicated epitope-tagged proteins were treated with the cross-linker 2 mM DTME at 25°C for 1 h, followed by solubilization with 0.5% Triton X-100 at 4°C for 30 min. The clarified extracts (from 350 μg of mitochondria in each case) were incubated with the anti-Myc-agarose conjugate for 4 h at 4°C under gentle shaking conditions and immunoprecipitates were analyzed by Western blotting (see MATERIALS AND METHODS). Anti-HA-HRP and anti-Myc-HRP antibodies were used for probing the Western blots. Solubilized proteins from 20 μg of mitochondria were loaded in total extract lanes. Strains were CAB30 and SN33.
Nam1p Interacts with Translational Activators Pet309p, Pet111p, and Pet494p
Nam1p/Mtf2p is a soluble matrix protein required for normal mitochondrial mRNA metabolism (Lisowsky and Michaelis, 1989; Wallis et al., 1994). Nam1p has been proposed to facilitate movement of mitochondrial transcripts to their sites of translation on the inner membrane (Wallis et al., 1994; Bryan et al., 2002) and interacts with the mitochondrial RNA polymerase (Rodeheffer et al., 2001). We therefore tested whether Nam1p interacts with translational activator proteins. First, two-hybrid interactions between Nam1p and the COX1, the COX2, and the COX3 activators were examined in pairwise manner (Figure 4). Cells expressing Nam1p-Gal4AD (from pACT-NAM1) or Pet309p-Gal4BD (from pSN21) alone were unable grow in absence of histidine and adenine. However, cells expressing Pet309p-Gal4BD and Nam1p-Gal4AD simultaneously were able to confer histidine and adenine prototrophy, indicating interaction between Nam1p and Pet309p. Nam1p exhibited clear two-hybrid interactions with Pet309p and with the N-terminally truncated forms of Pet111p and Pet494p (Figure 4). No interactions were observed between Nam1p and either Pet54p or Pet122p (our unpublished data).
To test for coimmune precipitation of Nam1p with translational activators, we used an anti-Nam1p polyclonal serum (Wallis et al., 1994) to probe Western blots. This serum recognized a 51-kDa protein in extracts of wild-type mitochondria that was absent in mitochondrial extracts from a nam1 deletion strain (Figure 5). Coimmuneprecipitation of Nam1p was observed with the COX1 translational activator Pet309p-Myc (Figure 5), but not with the other translational activator proteins tested.
DISCUSSION
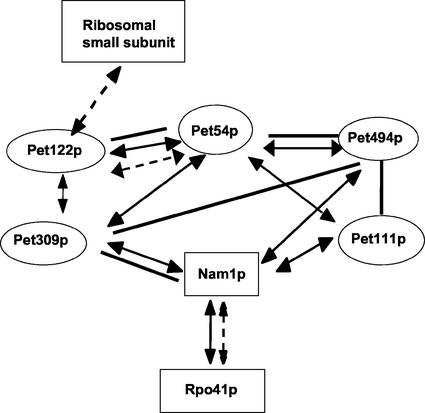
Our results demonstrate associations among nuclearly encoded mRNA-specific translational activator proteins, located on the matrix side of the inner membrane, that control the synthesis of the mitochondrially coded core subunits of S. cerevisiae cytochrome c oxidase. These interactions, as well as others established previously, are summarized in Figure 7. Because untranslated regions of the mitochondrial COX2 and COX3 mRNAs, which contain the targets of their translational activators, play a role in targeting translation for efficient cytochrome c oxidase assembly (Sanchirico et al., 1998), our present findings suggest that these interacting activators could be organized on the surface of the inner membrane such that synthesis of Cox1p, Cox2p, and Cox3p would be colocalized in a way that facilitates assembly of the enzymatic core of the cytochrome c oxidase complex.
Figure 7.
Summary of interactions among proteins involved in mitochondrial gene expression. Thick bars indicate coimmuneprecipitation, double-headed arrows connected by solid lines indicate two-hybrid interactions, and double-headed arrows connected by broken lines indicate genetic/functional interaction. Functional interactions between mitochondrial small subunit ribosomal proteins and Pet122p (McMullin et al., 1990; Haffter et al., 1991; Haffter and Fox, 1992) and interactions between of Nam1p and Rpo41p (mitochondrial RNA polymerase) (Rodeheffer et al., 2001) were reported previously. Two-hybrid and genetic interactions among Pet54p, Pet122p, and Pet494p were reported previously (Brown et al., 1994; Wiesenberger et al., 1995).
Our data do not distinguish whether interactions among the translational activators are transient and dynamic, or strong enough to form stable complexes in vivo. We were not able to detect stable complexes in detergent-solubilized extracts by using blue native gel electrophoresis (Schägger, 1995; our unpublished data). In any event, their physical association with each other is not required for their activity, because deletion of genes coding for one of the activators does not disrupt the mRNA-specific functions of the others (Fox, 1996a).
Why translation of the mitochondrially coded COX1, COX2, and COX3 mRNAs should be dependent on distinct activators, as opposed to a single “cytochrome c oxidase-specific activator” remains an open question. One possible rationalization is that mRNA specificity prevents competition among the three mRNAs for a single activator protein, where small differences in affinities could produce large differences in relative rates of synthesis. Interestingly, the amino acid sequences of translational activator proteins have diverged rapidly during fungal evolution, whereas the mRNA-specific relationships between activator protein homologues and target mRNAs have been conserved in those cases studied (Coffin et al., 1997; Costanzo et al., 2000). These orthologous functional relationships have been conserved despite the fact that the activator dependence of mitochondrial mRNAs can be altered experimentally in S. cerevisiae, without eliminating respiratory complex formation, by in vivo expression of chimeric mRNAs containing 5′-UTLs and coding sequences derived from different respiratory complex genes (Müller et al., 1984; Costanzo and Fox, 1986, 1988; Poutre and Fox, 1987; Rödel and Fox, 1987; Mulero and Fox, 1993b; Manthey and McEwen, 1995). Thus, although the interactions among translational activators for the core cytochrome c oxidase subunits are likely to confer a selective advantage, they are not absolutely required to form the enzyme.
PET494 expression levels (Marykwas and Fox, 1989) suggest that a diploid cell growing on nonfermentable carbon sources contains roughly 50–100 COX3-specific translational activators (Fox, 1996b). The expression of PET122 and PET111 seems to be comparably low (our unpublished data). These facts, taken together with the interactions reported herein suggest the possibility that there are a limited number of foci on the inner membrane where synthesis of the core subunits of cytochrome c oxidase is initiated. Interestingly, a genetic interaction between PET111 and COX18, which is required to translocate the Cox2p C-tail through the inner membrane, suggests that the levels of the COX2 translational activator and a Cox2p translocator are roughly comparable (Saracco and Fox, 2002). How the cytoplasmically synthesized subunits of cytochrome c oxidase would be targeted to such foci is an interesting question in view of the fact that published data (Vestweber and Schatz, 1988) suggest that there must be at least 10,000 sites per cell where cytoplasmically synthesized precursor proteins can be imported into mitochondria.
Interesting parallels can be drawn between the mRNA-specific translational activators of yeast mitochondria and bacterial type III secretion chaperones. These chaperones are typically specific for a single substrate and in at least some cases seem to function as translational activators with a role in targeting translation to the translocation apparatus, at least for flagellar assembly (Karlinsey et al., 2000; Aldridge and Hughes, 2001). Although there is no detectable homology between the mitochondrial proteins and any type III secretion components, the mechanisms used in yeast mitochondria are likely to have their origins in bacterial systems.
A substantial fraction of mammalian mitochondrial ribosomes are tightly associated with the inner membrane (Liu and Spremulli, 2000). However, it remains unknown whether or how translation of mammalian mitochondrial mRNAs is localized on the inner membrane. A mechanism to achieve this in animals must be at least somewhat different from that of yeast because animal mitochondrial mRNAs lack 5′-UTLs (Attardi and Schatz, 1988), which contain the recognition sites for yeast activators. An mRNA-specific mammalian system analogous or homologous to that of yeast would have to recognize RNA sites embedded in the coding sequences.
The interactions we observed between Nam1p/Mtf2p and translational activators suggest a direct link between transcription and translation within the mitochondrial matrix (Figure 7). Nam1p is a soluble matrix protein (Wallis et al., 1994) encoded by a gene originally identified as a high-copy suppressor of mitochondrial intron splicing defects and by temperature-sensitive mutations affecting mRNA levels (Lisowsky and Michaelis, 1989; Wallis et al., 1994). Nam1p is required to stabilize intron-containing mRNAs and has been proposed to “convey” mitochondrially coded mRNAs to the translation apparatus (Wallis et al., 1994) in a pathway involving the inner membrane protein Sls1p (Bryan et al., 2002). Importantly, Nam1p was recently found to interact with the amino terminal domain of the core subunit of mitochondrial RNA polymerase, Rpo41p, both genetically and in a two-hybrid test (Rodeheffer et al., 2001). Taken together, these data suggest the interesting possibility that mitochondrial transcriptional machinery is coupled to the membrane-bound translational activation system.
ACKNOWLEDGMENTS
We thank M.C. Costanzo for pPET494HA and N.G. Brown for pNGB67, pNGB68, pNGB69, and pNGB70S. We also thank G.S. Shadel for pACT-NAM1, N. Bonnefoy for anti-Nam1p antibody, and B. Glick for anti-α-ketoglutarate dehydrogenase. This study was supported by National Institutes of Health research grant GM-29362 (to T.D.F.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0490. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0490.
REFERENCES
- Aldridge P, Hughes KT. How and when are substrates selected for type III secretion? Trends Microbiol. 2001;9:209–214. doi: 10.1016/s0966-842x(01)02014-5. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular Interactions in Development: A Practical Approach. Oxford: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- Botstein D, Falco SC, Stewart SE, Brennan M, Scherer S, Stinchcomb DT, Struhl K, Davis RW. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brown NG, Costanzo MC, Fox TD. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C-T, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Coffin JW, Dhillon R, Ritzel RG, Nargang FE. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr Genet. 1997;32:273–280. doi: 10.1007/s002940050277. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Bonnefoy N, Williams EH, Clark-Walker GD, Fox TD. Highly diverged homologs of Saccharomyces cerevisiae mitochondrial mRNA-specific translational activators have orthologous functions in other budding yeasts. Genetics. 2000;154:999–1012. doi: 10.1093/genetics/154.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986;6:3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc Natl Acad Sci USA. 1988;85:2677–2681. doi: 10.1073/pnas.85.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Mueller PP, Strick CA, Fox TD. Primary structure of wild-type and mutant alleles of the PET494 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986;202:294–301. doi: 10.1007/BF00331654. [DOI] [PubMed] [Google Scholar]
- Fox TD. Genetics of mitochondrial translation. In: Hershey JWB, Matthews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996a. pp. 733–758. [Google Scholar]
- Fox TD. Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia. 1996b;52:1130–1135. doi: 10.1007/BF01952112. [DOI] [PubMed] [Google Scholar]
- Glick BS. Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae. Methods Enzymol. 1995;260:224–231. doi: 10.1016/0076-6879(95)60140-6. [DOI] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Green-Willms NS, Butler CA, Dunstan HM, Fox TD. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene. COX2. J Biol Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- Green-Willms NS, Fox TD, Costanzo MC. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′-untranslated leaders. Mol Cell Biol. 1998;18:1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Fox TD. Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and. PET127. Mol Gen Genet. 1992;235:64–73. doi: 10.1007/BF00286182. [DOI] [PubMed] [Google Scholar]
- Haffter P, McMullin TW, Fox TD. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991;127:319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of amino- and carboxy-termini, and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Fox TD. Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol Cell Biol. 1999;19:6598–6607. doi: 10.1128/mcb.19.10.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE, Lonner J, Brown KL, Hughes KT. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- Lisowsky T, Michaelis G. Mutations in the genes for mitochondrial RNA polymerase and a second mitochondrial transcription factor of Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:125–128. doi: 10.1007/BF00261167. [DOI] [PubMed] [Google Scholar]
- Lithgow T, Cuezva JM, Silver PA. Highways for protein delivery to the mitochondria. Trends Biochem Sci. 1997;22:110–113. doi: 10.1016/s0968-0004(97)01007-4. [DOI] [PubMed] [Google Scholar]
- Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Biol Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey GM, Przybyla-Zawislak BD, McEwen JE. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur J Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marykwas DL, Fox TD. Control of the Saccharomyces cerevisiae regulatory gene PET494: transcriptional repression by glucose and translational induction by oxygen. Mol Cell Biol. 1989;9:484–491. doi: 10.1128/mcb.9.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin TW, Fox TD. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem. 1993;268:11737–11741. [PubMed] [Google Scholar]
- McMullin TW, Haffter P, Fox TD. A novel small subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990;10:4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis U, Körte A, Rödel G. Association of cytochrome b translational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol Gen Genet. 1991;230:177–185. doi: 10.1007/BF00290666. [DOI] [PubMed] [Google Scholar]
- Mulero JJ, Fox TD. Alteration of the Saccharomyces cerevisiae COX2 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993a;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Fox TD. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993b;133:509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller PP, Reif MK, Zonghou S, Sengstag C, Mason TL, Fox TD. A nuclear mutation that post-transcriptionally blocks accumulation of a yeast mitochondrial gene product can be suppressed by a mitochondrial gene rearrangement. J Mol Biol. 1984;175:431–452. doi: 10.1016/0022-2836(84)90178-5. [DOI] [PubMed] [Google Scholar]
- Palacios IM, Johnston DS. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- Pon L, Schatz G. Biogenesis of yeast mitochondria. In: Broach JR, Pringle JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Vol. 1. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1991. pp. 333–406. [Google Scholar]
- Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel G, Fox TD. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5′-untranslated leader. Mol Gen Genet. 1987;206:45–50. doi: 10.1007/BF00326534. [DOI] [PubMed] [Google Scholar]
- Sanchirico ME, Fox TD, Mason TL. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail, and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell. 2002;13:1122–1131. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 1995;260:190–202. doi: 10.1016/0076-6879(95)60137-6. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D, Schatz G. A chimeric mitochondrial precursor protein with internal disulfide bridges blocks import of authentic precursors into mitochondria and allows quantitation of import sites. J Cell Biol. 1988;107:2037–2043. doi: 10.1083/jcb.107.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis MG, Groudinsky O, Slonimski PP, Dujardin G. The NAM1 protein (NAM1p), which is selectively required for cox1, cytb and atp6 transcript processing/stabilization, is located in the yeast mitochondrial matrix. Eur J Biochem. 1994;222:27–32. doi: 10.1111/j.1432-1033.1994.tb18837.x. [DOI] [PubMed] [Google Scholar]
- Wiesenberger G, Costanzo MC, Fox TD. Analysis of the Saccharomyces cerevisiae mitochondrial COX3 mRNA 5′-untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995;15:3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoladek T, Vaduva G, Hunter LA, Boguta M, Go BD, Martin NC, Hopper AK. Mutations altering the mitochondrial-cytoplasmic distribution of Mod5p implicate the actin cytoskeleton and mRNA 3′ ends and/or protein synthesis in mitochondrial delivery. Mol Cell Biol. 1995;15:6884–6894. doi: 10.1128/mcb.15.12.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]