Abstract
Background
In order to contribute to the establishment of a complete map of transcribed regions of the human genome, we constructed a testicular cDNA library for the cynomolgus monkey, and attempted to find novel transcripts for identification of their human homologues.
Result
The full-insert sequences of 512 cDNA clones were determined. Ultimately we found 302 non-redundant cDNAs carrying open reading frames of 300 bp-length or longer. Among them, 89 cDNAs were found not to be annotated previously in the Ensembl human database. After searching against the Ensembl mouse database, we also found 69 putative coding sequences have no homologous cDNAs in the annotated human and mouse genome sequences in Ensembl.
We subsequently designed a DNA microarray including 396 non-redundant cDNAs (with and without open reading frames) to examine the expression of the full-sequenced genes. With the testicular probe and a mixture of probes of 10 other tissues, 316 of 332 effective spots showed intense hybridized signals and 75 cDNAs were shown to be expressed very highly in the cynomolgus monkey testis, but not ubiquitously.
Conclusions
In this report, we determined 302 full-insert sequences of cynomolgus monkey cDNAs with enough length of open reading frames to discover novel transcripts as human homologues. Among 302 cDNA sequences, human homologues of 89 cDNAs have not been predicted in the annotated human genome sequence in the Ensembl. Additionally, we identified 75 dominantly expressed genes in testis among the full-sequenced clones by using a DNA microarray. Our cDNA clones and analytical results will be valuable resources for future functional genomic studies.
Background
Progress in genome biology has revealed the complete genome sequences of many non-mammalian species, such as yeast, nematodes, and the fruit fly. In addition, the much larger and more complicated genome sequences of the mouse and the human will soon be made completely available. However, decoding the genome sequences, especially the human sequence will be a long process. In order to achieve a comprehensive understanding of how an organism is established by its genome sequence, we must identify the structure, function, and interaction of as many genes as possible. First, we should accumulate and compile many types of evidence from computational and empirical data. The immediate challenge is establishing a complete map of transcribed regions in the human genome. Current comprehensive studies predicting protein-coding genes from the human genome [1,2] mainly employ three sources of information: empirical evidence provided by expressed sequence tags (ESTs) and cDNAs, nucleotide and protein sequence similarity to those of known genes, and statistical probability calculated by computer algorithms (ab initio prediction). All of these sources more or less lead to false-positive or false-negative types of errors. EST and cDNA sequences usually contain sequences that are not actually transcribed in vivo, i.e. artifacts arising from splicing intermediates, genomic DNA contamination, and transcription from nongenic regions [3,4]. Moreover, rarely expressed genes that may represent only a small portion of all transcripts cannot be easily represented in cDNA libraries. Predictions based on nucleotide and protein sequence similarities to those of other gene families and organisms might misassign pseudo genes, and cannot identify evolutionarily diverged genes that have no sequence similarity to known genes. Ab initio prediction works well for some organisms, such as yeast, nematodes, and the fruit fly. However, the human genome makes ab initio prediction of protein-coding genes difficult because it generally consists of small exons separated by long introns. Ultimately, in order to make a complete catalog of human genes, it will be necessary to gather undiscovered evidence from experiments and discard spurious evidence.
Our strategy for finding novel genes is to perform cDNA analysis using an organism closely related to humans, the cynomolgus monkey (Macaca fascicularis). In previous studies, we accumulated a number of 5'-end sequences of many clones derived from the oligo-capped cDNA libraries of the brain with high mRNA complexity, and determined approximately 1,500 full insert sequences of the clones whose 5'-end sequences showed no significant similarity to sequences in the public databases [5,6]. This method allowed us to identify many novel transcripts in the human genome sequence. Using fresh cynomolgus monkey tissues makes it possible to isolate rarely expressed genes, because mRNAs are so fragile that considerable portions of them degenerate during the usual construction of a cDNA library for humans. As an advantage of using cynomolgus monkey, evolutionary inspection can also provide information on gene function. If there are genes that evolved rapidly after the divergence of humans and cynomolgus monkeys, the function of the proteins and parts of the proteins might be important for human evolution. Moreover, biomedical interest in nonhuman primate genomes has been increased rapidly [7], especially in macaques, which also have been a material as transgenic primates [8], and thus genomic analysis of macaques will be important after the completion of human genome sequencing. In this study, we analyzed the cDNA library of the cynomolgus monkey testis. Analysis of testicular cDNA libraries has high potential for finding novel genes [9,10], because the testis is an organ in which transcripts have high complexity and where important biological processes, such as cell differentiation and meiosis, occur. The genes expressed in the testis are also important for medical, evolutionary, and developmental research. It is ironic that one of the most attractive tissues for biology expresses a number of undiscovered genes. We anticipated that analysis of the testicular cDNA library would lead to the discovery of novel genes that would facilitate post-genomic studies to attempt to unravel the complex genomes of higher organisms. Further, we conducted an expression analysis of our full-sequenced cDNAs with cDNA microarray. DNA microarrays are a versatile tool for evaluating gene expression and sequence variation [11]. We used a cDNA microarray, to determine whether our putative genes were actually transcribed in cynomolgus monkey tissues and whether they were expressed dominantly in the cynomolgus monkey testis.
Results
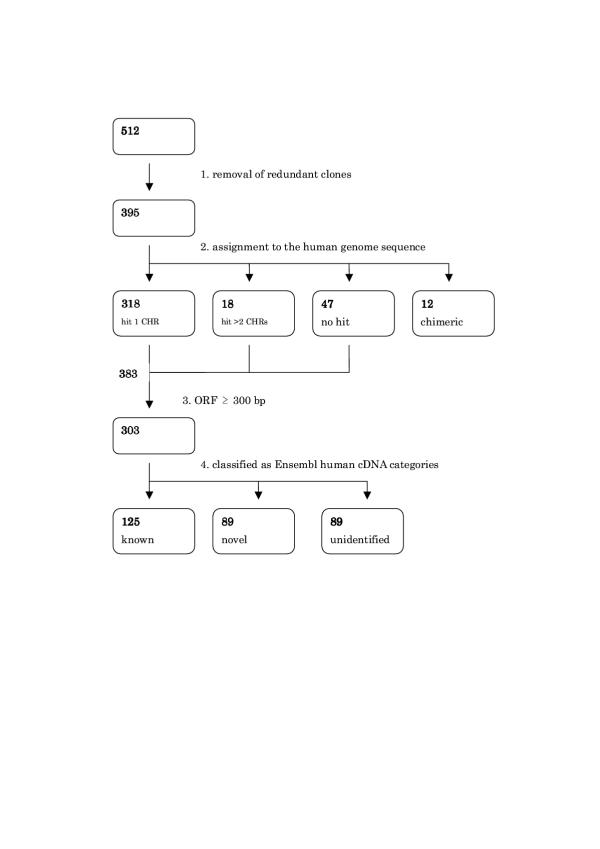
We constructed the cDNA library derived from the cynomolgus monkey testis (library name: QtsA) by the oligo-capping method. The 5'-ends of 10,426 clones isolated from the library were sequenced and yielded 5,381 clusters of sequences (the redundancy rate was 1.94). To classify these cynomolgus monkey cDNAs and find their human homologues, we performed the BLAST search [12] to human RefSeq databases [13]. The 5'-end sequences of 6151 clones were found to have high similarity to 2321 human RefSeq genes with a cut-off value of e-60. The results showed that most of frequently occurring genes in cDNA library, QtsA were those specifically expressed in testis and sperm. Breakdown of the numerically represented genes is shown in Table 1. The clones whose 5'-end sequences had no homologies to the sequences in the nr and EST databases in the Genbank and had the possibility of being a certain length of ORF were subjected to full-insert sequencing. The entire sequences of 512 clones were determined as a result, but the total number of non-redundant transcripts was smaller because we could not completely exclude the 5'-truncated clones of the same transcripts at the stage of 5'-end sequences. Further, we might obtain some alternatively spliced transcripts from the same gene. In such cases, we used the longest transcripts in this study. Ultimately, we obtained a total of 394 non-redundant full insert cDNA sequences (Figure 1). After masking the common repetitive elements in the Repbase Update database [14], we assigned all cDNA sequences to the human genome draft sequence by using the BLAST program. With 85% or greater sequence identity and 50% or greater overlap of cDNA sequence length as criteria, 12 clones were deduced to be chimeric because they could be divided into two regions, of which DNA sequences showed homology to sequences of different human chromosomes. Sequences of 317 cDNAs had only one homologous region in the human genome sequences, while 18 cDNA sequences had homology to more than two human chromosomal regions. The remaining 47 had no homologous sequences in the human genome based on the above criteria. The average nucleotide length of all full-sequenced clones was 2016 bp. Of the 382 non-chimeric sequences, 302 carried a putative CDS (coding sequence) longer than or equal to 300 bp. In order to determine how many human homologues of our full-sequenced cDNAs have been annotated from the human genome sequences, a search was made for 302 putative CDSs to the Ensembl human database (Release 5.28.1) [15], which comprised 29,076 putative transcribed sequences classified as 'known' and 'novel' genes (BLAST cut-off value was e-60, and the coverage was ≥ 50% of each putative CDS length). Genes classified as 'known' in Ensembl are more reliable and have valid cDNA and/or evolutionary evidence, whereas 'novel' genes lack credible sources of expression and are sometimes supported by only ab initio methods and ESTs. As a result, 124 and 89 putative CDSs had human homologous sequences in the known category and novel category, respectively. The other 89 putative CDS had no homologous sequences in Emsembl human database based on these criteria. We also searched 302 cDNA sequences against the Ensembl mouse database (Release 7.3b.2), in which 28,097 putative transcribed sequences were annotated, (cut-off: e-30, coverage: ≥ 50% of ORF length), resulting that 74 and 67 cDNAs had homologous mouse sequences predicted as Ensembl 'known' and 'novel' genes, respectively. Finally, 69 putative CDSs have no homologous sequences in the annotated mammalian genome sequences in Ensembl. The putative functions of 302 hypothetical proteins were predicted by searching against the InterPro database [16]. A list of their name and other information on the 302 cDNAs is provided in the supplementary table (additional file 1). We then constructed the putative human transcribed sequences corresponding to the 302 cynomolgus monkey cDNA sequences carrying enough length of ORFs by using the human genome draft sequences (see Materials and methods). The result showed that 117 putative human transcribed sequences, including 55 'known' and 48 'novel' genes in Ensembl had almost identical genomic structure to those of cynomolgus monkeys. We tested how many exons of 48 'novel' and 12 'unidentified' putative transcribed sequences can be predicted by the ab initio program, GENSCAN [17]. In total, 240 (53%) and 79 (17%) of 455 exons were correctly and partially predicted by GENSCAN, respectively, however, 136 exons (30%) were unpredictable. The list of putative human transcribed sequences is presented in Table 2 and their sequences are provided in the supplementary data (additional file 2), but the sequences have not been registered in the public database because they were not actually sequenced. We also compared the nucleotide and protein sequence similarity of 117 putative transcribed sequences between humans and cynomolgus monkeys. Amino acid sequence identity, nucleotide sequence identity for CDS, synonymous substitution per synonymous site, and non-synonymous substitution per non-synonymous site are presented in Table 2.
Table 1.
The list of 10 most frequently represented genes in the library QtsA.
| Accession | No. of clones | Gene name (Gene symbol) |
| NM_002762 | 318 | Protamine 2 (PRM2) |
| NM_004645 | 121 | Coilin (COIL) |
| NM_004362 | 108 | Calmegin (CLGN) |
| NM_005425 | 105 | Transition protein 2 (TNP2) |
| NM_004396 | 95 | DEAD/H box polypeptide 5 (DDX5) |
| NM_017769 | 83 | Hypothetical protein FLJ20333 (FLJ20333) |
| NM_030941 | 80 | Exonuclease NEF-sp (LOC81691) |
| NM_001402 | 77 | Eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) |
| NM_021009 | 76 | Ubiquitin C (UBC) |
| NM_004724 | 63 | ZW10 (Drosohplila) homolog (ZW10) |
Figure 1.
Flow of full-sequencing analysis of unidentified clones. 1) The 512 full-sequenced clones were reduced to 394 because slightly different 5'-end sequences could be derived from the same transcripts. 2) 394 non-redundant clones were assigned to the human genome sequence. 3) 302 of 383 non-chimeric clones carried ORFs longer than 300 bp. 4) 302 putative genes were classified as 'known' and 'novel' categories of Ensembl human cDNA sequences. *CHR: Chromosome
Table 2.
The list of 117 putative human transcribed sequences.
| Referenced macaca cDNAa |
Ensembl statusb |
length | CDS length: start..end (bp) | aa identity (%) | nt identity of CDS (%) |
Kac | Ksd |
| QtsA-10152 | novel | 1789 | 413AA: 42..1283 | 96.1 | 97.6 | 0.019 | 0.039 |
| QtsA-10154 | known | 2010 | 502AA: 377..1885 | 98.2 | 98.3 | 0.009 | 0.032 |
| QtsA-10162 | novel | 2444 | 718AA: 72..2228 | 96.5 | 97.6 | 0.017 | 0.040 |
| QtsA-10245 | known | 2598 | 752AA: 298..2556 | 94.4 | 96.1 | 0.027 | 0.078 |
| QtsA-10439 | novel | 2566 | 538AA: 271..1887 | 88.6 | 93.7 | 0.059 | 0.076 |
| QtsA-10472 | known | 2159 | 418AA: 76..1332 | 95.5 | 96.7 | 0.025 | 0.062 |
| QtsA-10491 | known | 2439 | 346AA: 1322..2362 | 98.0 | 98.6 | 0.009 | 0.027 |
| QtsA-10636 | novel | 2627 | 440AA: 57..1379 | 100.0 | 100.0 | 0.000 | 0.000 |
| QtsA-10679 | known | 2415 | 523AA: 726..2297 | 96.0 | 96.8 | 0.021 | 0.061 |
| QtsA-10739 | novel | 1880 | 231AA: 141..836 | 93.4 | 97.0 | 0.034 | 0.022 |
| QtsA-10833 | known | 2234 | 673AA: 89..2110 | 92.2 | 95.2 | 0.037 | 0.077 |
| QtsA-10891 | novel | 2049 | 343AA: 2..1033 | 93.2 | 95.2 | 0.038 | 0.084 |
| QtsA-10947 | known | 2132 | 540AA: 112..1734 | 93.2 | 94.4 | 0.033 | 0.126 |
| QtsA-10963 | known | 1924 | 462AA: 433..1821 | 98.9 | 98.6 | 0.005 | 0.039 |
| QtsA-11068 | unidentified | 2299 | 594AA: 405..2189 | 91.8 | 94.9 | 0.042 | 0.080 |
| QtsA-11127 | known | 2084 | 550AA: 54..1706 | 89.8 | 95.5 | 0.053 | 0.034 |
| QtsA-11181 | known | 3414 | 566AA: 84..1784 | 99.8 | 98.9 | 0.001 | 0.036 |
| QtsA-11319 | unidentified | 1559 | 104AA: 168..482 | 100.0 | 100.0 | 0.000 | 0.000 |
| QtsA-11379 | known | 2805 | 690AA: 106..2178 | 98.8 | 98.2 | 0.005 | 0.050 |
| QtsA-11535 | known | 2116 | 474AA: 304..1728 | 98.3 | 98.0 | 0.008 | 0.057 |
| QtsA-11567 | unidentified | 1376 | 376AA: 200..1330 | 96.0 | 97.7 | 0.020 | 0.034 |
| QtsA-11570 | unidentified | 2437 | 117AA: 1902..2255 | 90.6 | 95.5 | 0.046 | 0.042 |
| QtsA-11661 | novel | 2228 | 588AA: 227..1993 | 97.6 | 98.0 | 0.013 | 0.038 |
| QtsA-11670 | unidentified | 1785 | 325AA: 413..1390 | 99.7 | 99.2 | 0.002 | 0.028 |
| QtsA-11842 | known | 2173 | 225AA: 142..819 | 100.0 | 100.0 | 0.000 | 0.000 |
| QtsA-12007 | novel | 2316 | 724AA: 28..2202 | 96.6 | 97.1 | 0.017 | 0.053 |
| QtsA-12095 | novel | 710 | 231AA: 16..711 | 94.4 | 96.8 | 0.030 | 0.039 |
| QtsA-12142 | known | 1731 | 404AA: 395..1609 | 94.1 | 96.3 | 0.027 | 0.060 |
| QtsA-12155 | known | 1305 | 329AA: 252..1241 | 95.5 | 97.7 | 0.024 | 0.034 |
| QtsA-12190 | unidentified | 1962 | 600AA: 21..1823 | 94.0 | 96.6 | 0.030 | 0.044 |
| QtsA-12219 | known | 2480 | 793AA: 18..2399 | 100.0 | 100.0 | 0.000 | 0.000 |
| QtsA-12282 | novel | 2270 | 700AA: 93..2195 | 97.1 | 97.9 | 0.013 | 0.036 |
| QtsA-12354 | known | 2082 | 674AA: 5..2029 | 84.4 | 90.3 | 0.095 | 0.119 |
| QtsA-12362 | novel | 2177 | 695AA: 91..2178 | 93.3 | 95.9 | 0.034 | 0.068 |
| QtsA-12457 | novel | 2405 | 689AA: 105..2174 | 94.9 | 96.5 | 0.026 | 0.059 |
| QtsA-12579 | novel | 1499 | 491AA: 8..1483 | 93.1 | 94.9 | 0.034 | 0.103 |
| QtsA-12649 | novel | 2114 | 634AA: 33..1937 | 97.6 | 98.2 | 0.012 | 0.038 |
| QtsA-12757 | known | 1280 | 278AA: 201..1037 | 98.6 | 97.7 | 0.008 | 0.055 |
| QtsA-12767 | novel | 2100 | 622AA: 51..1919 | 98.6 | 98.0 | 0.007 | 0.060 |
| QtsA-12769 | known | 1530 | 395AA: 75..1262 | 92.2 | 95.8 | 0.038 | 0.052 |
| QtsA-12850 | known | 2825 | 854AA: 262..2826 | 97.5 | 97.7 | 0.011 | 0.053 |
| QtsA-13222 | known | 2806 | 2806 873AA: 68..2689 | 92.7 | 95.0 | 0.038 | 0.085 |
| QtsA-13252 | novel | 2229 | 669AA: 65..2074 | 95.7 | 96.9 | 0.021 | 0.059 |
| QtsA-13272 | novel | 1833 | 207AA: 184..807 | 98.1 | 98.6 | 0.010 | 0.033 |
| QtsA-13343 | novel | 1960 | 131AA: 171..566 | 91.6 | 96.2 | 0.041 | 0.026 |
| QtsA-13392 | unidentified | 1761 | 438AA: 313..1629 | 98.9 | 98.9 | 0.005 | 0.022 |
| QtsA-13406 | known | 1855 | 266AA: 930..1730 | 92.0 | 96.2 | 0.039 | 0.040 |
| QtsA-13432 | known | 1718 | 428AA: 360..1646 | 97.7 | 98.1 | 0.011 | 0.038 |
| QtsA-13460 | known | 1492 | 427AA: 26..1309 | 95.0 | 97.2 | 0.023 | 0.035 |
| QtsA-13672 | novel | 1824 | 363AA: 734..1825 | 95.3 | 96.9 | 0.023 | 0.046 |
| QtsA-13918 | known | 1730 | 537AA: 120..1733 | 97.2 | 98.1 | 0.012 | 0.047 |
| QtsA-13925 | novel | 1678 | 515AA: 114..1661 | 92.0 | 95.4 | 0.041 | 0.061 |
| QtsA-14022 | novel | 1653 | 517AA: 102..1655 | 96.3 | 96.8 | 0.020 | 0.064 |
| QtsA-14166 | novel | 1121 | 293AA: 177..1058 | 89.5 | 94.7 | 0.052 | 0.060 |
| QtsA-14245 | known | 1784 | 531AA: 5..1600 | 96.6 | 97.2 | 0.017 | 0.056 |
| QtsA-14351 | novel | 2938 | 839AA: 225..2744 | 91.8 | 96.0 | 0.041 | 0.046 |
| QtsA-14618 | known | 1273 | 363AA: 57..1148 | 94.5 | 97.0 | 0.027 | 0.037 |
| QtsA-14653 | known | 996 | 150AA: 405..857 | 100.0 | 98.7 | 0.000 | 0.049 |
| QtsA-14746 | known | 2049 | 528AA: 17..1603 | 96.8 | 97.2 | 0.016 | 0.060 |
| QtsA-14752 | known | 904 | 235AA: 184..891 | 97.5 | 97.0 | 0.012 | 0.078 |
| QtsA-14816 | unidentified | 2515 | 134AA: 242..646 | 98.4 | 99.2 | 0.008 | 0.013 |
| QtsA-14824 | known | 2965 | 891AA: 151..2826 | 95.7 | 97.5 | 0.020 | 0.036 |
| QtsA-14970 | known | 1282 | 168AA: 639..1145 | 100.0 | 99.2 | 0.000 | 0.017 |
| QtsA-15013 | novel | 2349 | 303AA: 364..1275 | 90.1 | 95.3 | 0.045 | 0.047 |
| QtsA-15139 | novel | 2487 | 740AA: 75..2297 | 96.4 | 96.9 | 0.017 | 0.061 |
| QtsA-15186 | known | 2181 | 588AA: 97..1863 | 93.1 | 96.4 | 0.034 | 0.044 |
| QtsA-15224 | novel | 2089 | 290AA: 336..1208 | 96.1 | 96.8 | 0.021 | 0.074 |
| QtsA-15268 | novel | 1808 | 396AA: 203..1393 | 96.2 | 96.4 | 0.018 | 0.092 |
| QtsA-15315 | known | 1284 | 344AA: 217..1251 | 88.8 | 94.0 | 0.054 | 0.073 |
| QtsA-15384 | known | 2169 | 565AA: 213..1910 | 95.9 | 96.9 | 0.019 | 0.062 |
| QtsA-15676 | novel | 2153 | 581AA: 184..1929 | 92.1 | 95.8 | 0.039 | 0.068 |
| QtsA-15696 | novel | 1856 | 563AA: 144..1835 | 93.1 | 96.5 | 0.034 | 0.046 |
| QtsA-15812 | novel | 2293 | 576AA: 491..2221 | 96.4 | 97.2 | 0.019 | 0.052 |
| QtsA-15844 | known | 2569 | 653AA: 174..2135 | 95.6 | 96.1 | 0.023 | 0.086 |
| QtsA-15875 | known | 2327 | 654AA: 357..2321 | 96.6 | 97.7 | 0.017 | 0.038 |
| QtsA-16005 | known | 1987 | 518AA: 433..1989 | 100.0 | 99.4 | 0.000 | 0.021 |
| QtsA-16015 | known | 2389 | 671AA: 345..2360 | 98.1 | 97.7 | 0.009 | 0.054 |
| QtsA-16028 | known | 1624 | 447AA: 23..1366 | 99.8 | 97.7 | 0.001 | 0.082 |
| QtsA-16077 | known | 2307 | 571AA: 576..2291 | 99.7 | 98.7 | 0.002 | 0.047 |
| QtsA-16107 | known | 2039 | 432AA: 301..1599 | 100.0 | 98.8 | 0.000 | 0.034 |
| QtsA-16118 | known | 1396 | 415AA: 57..1304 | 97.6 | 96.7 | 0.012 | 0.096 |
| QtsA-16284 | novel | 1199 | 342AA: 31..1059 | 96.5 | 96.8 | 0.017 | 0.071 |
| QtsA-16373 | known | 2085 | 433AA: 619..1920 | 99.5 | 98.5 | 0.002 | 0.048 |
| QtsA-16429 | known | 1783 | 413AA: 42..1283 | 96.1 | 97.6 | 0.019 | 0.039 |
| QtsA-16453 | novel | 1757 | 185AA: 793..1350 | 87.3 | 93.5 | 0.066 | 0.065 |
| QtsA-16496 | novel | 1599 | 448AA: 72..1418 | 95.5 | 96.1 | 0.023 | 0.081 |
| QtsA-16602 | unidentified | 2482 | 263AA: 315..1106 | 96.6 | 97.1 | 0.015 | 0.079 |
| QtsA-16622 | novel | 1364 | 313AA: 291..1232 | 95.1 | 96.9 | 0.024 | 0.045 |
| QtsA-16678 | known | 2325 | 688AA: 91..2157 | 98.5 | 96.5 | 0.008 | 0.096 |
| QtsA-16765 | novel | 2501 | 415AA: 862..2109 | 98.1 | 97.4 | 0.010 | 0.068 |
| QtsA-16837 | known | 3268 | 586AA: 873..2633 | 99.3 | 98.5 | 0.004 | 0.041 |
| QtsA-17449 | known | 1858 | 506AA: 262..1782 | 90.9 | 95.4 | 0.044 | 0.053 |
| QtsA-17495 | novel | 1026 | 261AA: 62..847 | 96.2 | 97.3 | 0.018 | 0.068 |
| QtsA-17616 | known | 2471 | 617AA: 435..2288 | 98.4 | 98.2 | 0.008 | 0.044 |
| QtsA-18070 | novel | 1997 | 585AA: 134..1891 | 97.8 | 97.7 | 0.009 | 0.054 |
| QtsA-18134 | known | 1832 | 309AA: 592..1521 | 99.7 | 98.2 | 0.002 | 0.053 |
| QtsA-18363 | novel | 1807 | 315AA: 638..1585 | 95.9 | 97.5 | 0.020 | 0.040 |
| QtsA-18372 | novel | 972 | 128AA: 337..723 | 97.7 | 97.4 | 0.011 | 0.069 |
| QtsA-18427 | known | 2198 | 565AA: 416..2113 | 99.3 | 98.9 | 0.003 | 0.033 |
| QtsA-18831 | unidentified | 2133 | 555AA: 47..1714 | 92.1 | 95.1 | 0.041 | 0.082 |
| QtsA-18885 | known | 3250 | 642AA: 314..2242 | 96.6 | 95.8 | 0.017 | 0.102 |
| QtsA-19023 | novel | 2072 | 500AA: 84..1586 | 91.0 | 95.6 | 0.043 | 0.047 |
| QtsA-19036 | novel | 955 | 214AA: 313..957 | 100.0 | 99.5 | 0.000 | 0.014 |
| QtsA-19380 | unidentified | 2158 | 412AA: 625..1863 | 98.1 | 97.3 | 0.009 | 0.071 |
| QtsA-19788 | novel | 1080 | 295AA: 116..1003 | 98.6 | 98.3 | 0.006 | 0.040 |
| QtsA-19856 | known | 2055 | 352AA: 497..1555 | 98.9 | 98.4 | 0.005 | 0.039 |
| QtsA-19961 | known | 1025 | 283AA: 62..913 | 100.0 | 98.0 | 0.000 | 0.069 |
| QtsA-20273 | novel | 1783 | 420AA: 79..1341 | 92.7 | 96.1 | 0.039 | 0.029 |
| QtsA-20302 | known | 2889 | 882AA: 87..2735 | 94.6 | 97.2 | 0.026 | 0.042 |
| QtsA-20424 | unidentified | 2056 | 505AA: 147..1664 | 99.2 | 98.4 | 0.005 | 0.041 |
| QtsA-20433 | novel | 1981 | 559AA: 73..1752 | 94.8 | 96.5 | 0.027 | 0.057 |
| QtsA-20664 | known | 2396 | 616AA: 231..2081 | 97.1 | 96.2 | 0.015 | 0.095 |
| QtsA-20987 | known | 3090 | 561AA: 636..2321 | 97.9 | 97.7 | 0.011 | 0.053 |
| QtsA-21536 | novel | 1409 | 350AA: 134..1186 | 92.3 | 95.7 | 0.042 | 0.052 |
| QtsA-21565 | novel | 1810 | 367AA: 276..1379 | 94.2 | 95.6 | 0.028 | 0.093 |
| QtsA-21583 | novel | 2640 | 761AA: 260..2545 | 90.4 | 95.0 | 0.046 | 0.060 |
| QtsA-21585 | known | 2252 | 202AA: 38..646 | 91.8 | 94.5 | 0.045 | 0.085 |
a) Cynomolgus monkey cDNA sequence that was used to deduce putative human transcribed sequence. b) Classification of human transcribed sequence in the Ensembl human database. c) Synonymous substitution rate per synonymous site between human and cynomolgus monkey genes. d) Non-synonymous substitution rate per non-synonymous site between human and cynomolgus monkey genes.
In order to investigate the expression pattern of the testicular full-sequenced cDNAs, we designed a DNA microarray containing approximately 400 spots of cDNA, full-sequenced samples and controls. Fifty clones carrying common repetitive elements and 12 clones deduced to be chimeric were excluded from further analysis, although they were spotted on the slides. Ultimately, 332 spots were used for quantification of gene expression. First, we investigated whether the putative genes were transcribed in a ubiquitous manner or had a tissue-specific pattern of expression especially in the testis. RNA pools from the testis of the cynomolgus monkey and the mixture of equal amounts of RNA from 10 other cynomolgus monkey tissues (brain, heart, skin, liver, spleen, renal, pancreas, stomach, small intestine, and large intestine) were independently labeled and co-hybridized to the DNA microarray. When the signal intensity of the testicular probe is greater than that of the mixed probe, the gene was concluded to be over-expressed in the testis, or to be transcribed in the testis and a few other tissues, but not ubiquitously. When the intensity of both signals was equal, the gene was concluded to be expressed in a ubiquitous manner. When the signal intensity of the testicular probe was lower than that of the mixed probe, the gene was concluded to be mainly transcribed in non-testicular tissues. We calculated the ratio of the testicular probe intensity to the mixed probe intensity and the ratio was normalized by using the beta-actin cDNA spot. A total of 316 (95%) of the 332 effective spots showed an intense and reproducible signal with either the testicular RNA probe or the mixed RNA probes or both. The signals of 75 spots were four fold or more intense with the testicular probe, and human homologues of the 15 genes among 75 cDNAs had been registered in the RefSeq database (Table 3). Eight of the 15 RefSeq genes were reported to be expressed exclusively or dominantly in the human testis in the literature and the databases: TSGA10, expressed during spermatogenesis [18]; ACTL7B, an intronless gene strongly expressed in the testis and weakly expressed in the prostate [19]; SOX30, Sry-related transcriptional factor specifically expressed in the testis [20]; and five NYD-SP genes, functionally anonymous but highly expressed in the testis in other DNA microarray experiments [21]. The other seven genes had ORFs of hypothetical proteins and were deduced from only the cDNA sequence evidence. Four of the cDNA clones were derived from human testis, and the other three cDNAs were from brain, placenta, or teratocarcinoma (Table 3). The results indicated that the remaining 60 clones that have no human RefSeq homologues are expressed exclusively or dominantly in the cynomolgus monkey testis.
Table 3.
The list of genes that were highly expressed in a testis and had human RefSeq homologues
| Macaca clone | Human RefSeq | Description | Ratioa | Expression (Reference) |
| QtsA-10833 | NM_032559 | kinesin protein (LOC84643) | 8.7 | derived from testis |
| QtsA-13647 | NM_025244 | testis specific, 10 (TSGA10) | 8.6 | testis specific [18] |
| QtsA-16118 | NM_006686 | actin-like 7B (ACTL7B) | 8.5 | testis and prostate [19] |
| QtsA-14409 | NM_018418 | hypothetical protein (HSD-3.1) | 7.8 | derived from testis |
| QtsA-13567 | NM_033122 | testis development protein NYD-SP26 (NYD-SP26) | 7.5 | testis |
| QtsA-14035 | NM_033123 | testis-development related NYD-SP27 (NYD-SP27) | 7.2 | testis |
| QtsA-11842 | NM_032130 | hypothetical protein DKFZp434J0113 (DKFZP434J0113) | 6.9 | derived from testis |
| QtsA-15256 | NM_032126 | hypothetical protein DKFZp564J047 (DKFZP564J047) | 6.6 | derived from brain |
| QtsA-14560 | NM_032599 | testes development-related NYD-SP18 (NYD-SP18) | 6.6 | testis |
| QtsA-12850 | NM_019038 | hypothetical protein (FLJ11045) | 6.4 | derived from placenta |
| QtsA-15384 | NM_030672 | hypothetical protein FLJ10312 (FLJ10312) | 5.1 | derived from teratocarcinoma |
| QtsA-10245 | NM_007017 | SRY (sex determining regionY)-box 30 (SOX30) | 5.0 | testis specific [20] |
| QtsA-18012 | NM_032596 | testes development-related NYD-SP22 (NYD-SP22) | 5.0 | testis |
| QtsA-14618 | NM_032598 | testes development-related NYD-SP20 (NYD-SP20) | 4.7 | testis |
| QtsA-19865 | NM_033364 | AAT1-alpha (AAT1) kinesin-like 6 (mitotic centromere-associated kinesin) | 4.5 | derived from testis |
| QtsA-16015 | NM_006845 | (KNSL6) | 4.1 | thymus and testis [21] |
a) The ratio of signal intensity of testicular probe to mixed probe.
Discussion
In this study we analyzed a cDNA library derived from a cynomolgus monkey testis. Although most of the human genome sequence has been determined, many unidentified genes remain, and a complete catalog of protein-coding genes is desired. Sequence similarity search of our full-sequenced cDNAs to the human draft genome sequence resulted in the assignment of 347 cDNA sequences to at least one human chromosome, indicating that most genes in the cynomolgus monkey have homologous regions in the human genome. The primary objective of this analysis was to find genes that have not been experimentally identified in the human genome. Among the 302 cDNAs carrying enough length of ORFs ( = 300 bp), we succeeded in identifying 89 putative genes that have no counterparts in the Ensembl 29,076-gene set. Another 89 genes that had highly similar sequences to Ensembl 'novel' genes were discovered in our full-sequenced cDNAs. The latter 89 genes strongly support the existence of predicted 'novel' cDNA sequences, which are relatively less accurate.
Many genes expressed in the testis cause male infertility in humans [22]. Since it is estimated that up to 11% of all genes in the fruit fly might lead to male sterility [23], in view of the complexity of the human genome, at least 4000 genes might be responsible for male infertility in humans and there must be many as yet unidentified genes that are related to male fertility. Functional analysis of 75 genes found to be highly expressed in the cynomolgus monkey testis may contribute such a medical interest about male infertility. A DNA microarray analysis is an appropriate method not only of annotating the pattern of expression of our full-length cDNAs, but of demonstrating that our strategy for finding novel gene works well. In the first set of the DNA microarray experiment, among the 199 genes that displayed two fold or more higher expression with the testicular probes than with the mixed probes, 67 (34%) were classified as the Ensembl 'known' genes, whereas among the 45 genes that showed ubiquitous pattern of expression (signal intensities within 1.5 fold of each other with both probes), 23 (51%) were classified as Ensembl 'known' genes. This finding indicated that the probability that transcripts overexpressed in testis are derived from unidentified novel genes is significantly higher than that of ubiquitous transcripts (p = 0.028: Fisher's exact test).
Evolutionary inspection is also important, especially for gene analysis of the testis, because genetic diversity in the male reproductive system is quite large, even among closely related species. Many reproductive proteins have evolved rapidly at the molecular level [24,25]. We compared 117 sequences of cynomolgus monkey cDNA and the corresponding human genome sequences described above, and use of the cDNA microarray revealed that 79 of the 117 cDNAs were overexpressed in testis ( = 2.0 fold in testis) and 15 were ubiquitously expressed (within 1.5 fold of each other). We estimated the sequence divergence of putative coding sequences between humans and cynomolgus monkeys and found that the average non-synonymous nucleotide divergence of testis-dominantly expressed genes (0.024) was significantly greater than that of ubiquitously expressed genes (0.012; p value < 0.01), whereas divergence in synonymous sites were not different significantly (testis-dominant genes: 0.54, ubiquitous genes: 0.51). This finding is also highly consistent with a report that the proteins of genes expressed in a tissue-specific manner evolve an average of twice as fast as those that are ubiquitously expressed [26].
Although a number of full and partial sequences of human genes have been deposited in the public databases, many of the genes in the human genome have not yet to be discovered experimentally. Most of the undiscovered genes may be expressed very seldom or their expression may be restricted to certain tissues and developmental stages. The complete human genome will be available in 2003, and a search of the entire genome for novel genes by oligonucleotide-based microarray analysis is designed; i.e. an attempt to predict all candidate human genes from the human genome and experimentally confirm the transcript status of the predicted regions as well as the entire region by using a oligo-nucleotide-based microarray [27,28]. However, it is difficult to overcome the problem of rarely or temporarily expressed genes for practical reasons. The transcriptional and genomic approaches will compensate for each other's blind spots, and many tissues, developmental stages, and other organisms should become experimental subjects for finding undiscovered genes to complete the human gene catalog.
Materials and Methods
cDNA library from cynomolgus monkey testis
A 15-year-old male cynomolgus monkey was used as the source of the testis, and a 1-year-old and 21-year-old female cynomolgus monkeys were used for other RNA samples. The monkeys were cared for and handled according to guidelines established by the Institutional Animal Care and Use Committee of the National Institute of Infectious Diseases (NIID) of Japan and the standard operating procedures for monkeys at the Tsukuba Primate Center, NIID, Tsukuba, Ibaraki, Japan. Tissues were excised in accordance with all guidelines in the Laboratory Biosafety Manual, World Health Organization, and were carried out at the P3 facility for monkeys of the Tsukuba Primate Center, NIID. Immediately after collection, the tissues were frozen with liquid nitrogen and used for RNA extraction. Oligo-capped cDNA libraries were constructed according to the method described previously [29,30].
DNA sequencing
The 5'-end sequences of the clones were sequenced using ABI 3700 sequencer (Applied Biosystems), and categorized using DYNACLUST (DYNACOM), based on a BLAST search against the GenBank database. The entire sequences of clones were determined by the primer walking method. Cycle sequencing was performed with an ABI PRISM BigDye Terminator Sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Computational analyses
The Sim4 program was used to align each cynomolgus monkey cDNA sequence with the human genome sequence [31]. Whenever Sim4 failed to align cynomolgus monkey cDNA sequence with human genome DNA sequence, comparison by BLAST program was executed, and the alignment was corrected manually. In the intron sequences, GT at the 5' splice site and AG at the 3' site (GT-AG pattern), and the GC-AG pattern were regarded as conserved splice sites, and corresponding human genome regions were concatenated to construct a hypothetical human transcribed sequence. 117 Cynomolgus monkey cDNA sequences and the putative human transcribed sequences were aligned by using the ClustalW program [32]. Synonymous substitution per synonymous site and non-synonymous substitution per non-synonymous site were estimated by the method of Li [33].
cDNA microarray
An aliquot of the same DNA preparation used in the 5'-end-sequencing reactions provided material for the PCRs. Inserts were amplified by PCR using 5'-CTTCTGCTCTAAAAGCTGCG-3' as a forward primer and 5'-CGACCTGCAGCTCGAGCACA-3' as a reverse primer, in a volume of 100 μl. Successful amplification was confirmed by agarose gel electrophoresis. When the first PCR failed to amplify enough products, the first PCR products were amplified again. Four hundred cDNA clones were amplified and samples of approximately 300 μg /ml DNA in 2 × Solution-T reagent (Takara Bio) were printed on duplicate glass-slides with a GMS 417 arrayer (Genetic MicroSystems). The testicular RNA was obtained from only one 15-year-old male cynomolgus monkey, and the other RNA was a mixture of RNA obtained from 10 tissues (brain, heart, skin, liver, spleen, renal, pancreas, stomach, small intestine, and large intestine) of two cynomolgus monkeys, a 1-year-old female and a 21-year-old female. RNA was isolated with Trizol (Life Technologies) and purified with Oligo-Tex (Takara Bio). Both 0.7 μg mRNA probes were labeled with Cy3- and Cy5- dioxynucleotide (Pharmacia) and co-hybridized to DNA spots. The amount of RNA from each tissue was 0.07 μg in the mixed RNA probe. After the hybridization and washing procedure, slides were scanned with ScanArray (GSI Inc.). Several experiments were conducted, and the duplicated spots on the slides, where the most intense signals were obtained, were processed to measure the transcriptional status. When the relative intensity of Cy3/Cy5 signals of duplicated spots differed more than 1.5 times compared to that of the corresponding spots in duplicate, the spots were not processed for the subsequent analyses. Finally, the ratio of signal intensities of Cy3 (the testicular probe) and Cy5 (the mixed probe) was obtained from average value of duplicated spots and normalized by dividing by the ratio of the beta-action spots as a control.
List of abbreviations
EST: expressed sequence tag.
CDS: coding sequence.
ORF: open reading frame.
Authors' contributions
NO was involved in design of the study, construction of cDNA library, in silico analysis, expression analysis with DNA microarray and preparation of the manuscript. M. Hida and SS performed construction of cDNA libraries and analysis of 5'-end sequence analysis. JK participated in the design and implementation of the study, contributed to writing and revising the manuscript. RT and M. Hirata participated in the sequencing of cDNAs and in-silico analyis of cDNA sequences. YS and M. Hirai participated in the design and implementation of the study on microarray, and obtained funding for the study. KT contributed to obtaining the tissues for cDNA libraries and total RNA from cynomolgus monkeys. KH was involved in the design and implementation of the study, writing and editing the manuscript and obtained funding for the study.
All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
Acknowledgements
This study was supported in part by the Health Science Research grant for the Human Genome Program from the Ministry of Health, Labor and Welfare of Japan.
Contributor Information
Naoki Osada, Email: osada@nih.go.jp.
Munetomo Hida, Email: ss77233@ims.u-tokyo.ac.jp.
Jun Kusuda, Email: jkusuda@nih.go.jp.
Reiko Tanuma, Email: rtanuma@nih.go.jp.
Makoto Hirata, Email: mhirata@nih.go.jp.
Yumiko Suto, Email: suto@k.u-tokyo.ac.jp.
Momoki Hirai, Email: mhirai@k.u-tokyo.ac.jp.
Keiji Terao, Email: terao@nih.go.jp.
Sumio Sugano, Email: ssugano@ims.u-tokyo.ac.jp.
Katsuyuki Hashimoto, Email: khashi@nih.go.jp.
References
- International Human Genome Sequencing Consortium: Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 milliion nucleotides of cDNA sequence. Nature. 1995;377:3–17. [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Osada N, Hida M, Kusuda J, Tanuma R, Iseki K, Hirata M, Suto Y, Hirai M, Terao K, Suzuki Y, et al. Assignment of 118 novel cDNAs of cynomolgus monkey brain to human chromosomes. Gene. 2001;275:31–37. doi: 10.1016/S0378-1119(01)00665-5. [DOI] [PubMed] [Google Scholar]
- Osada N, Hida M, Kusuda J, Tanuma R, Hirata M, Hirai M, Terao K, Suzuki Y, Sugano S, Hashimoto K. Prediction of unidentified human genes on the basis of sequence similarity to novel cDNAs from cynomolgus monkey brain. Genome Biol. 2002;3:research0006.1–0006.5. doi: 10.1186/gb-2001-3-1-research0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Dejong PJ. Biomedical applications and studies of molecular evolution: A proporsal for a primate genomic library resource. Genome Res. 2002;12:673–678. doi: 10.1101/gr.250102. [DOI] [PubMed] [Google Scholar]
- Chan AWS, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- Andrews J, Bouffard CG, Cheadle C, Lu Jining., Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster tesis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.GR1547R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooedewier MO, Warren PV. Gene expression microarrays and the integration of biological knowledge.: Trends Biotecnol. 2001;19:412–415. doi: 10.1016/S0167-7799(01)01735-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;9:418–420. doi: 10.1016/S0168-9525(00)02093-X. [DOI] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nuc Acid Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Modarressi MH, Cameron J, Taylor KE, Wolfe J. Identification and characterisation of a novel gene, TSGA10, expressed in testis. Gene. 2001;262:249–255. doi: 10.1016/S0378-1119(00)00519-9. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Mull J, Helbling LA, Gill S, Leyne M, Robbins CM, Pinkett HW, Makalowska I, Maayan C, Blumenfeld A, et al. Cloning, mapping, and expression of two novel actin genes, actin-like-7A (ACTL7A) and actin-like-7B (ACTL7B), from the familial dystonomia candidate region on 9q31. Genomics. 1999;58:302–309. doi: 10.1006/geno.1999.5848. [DOI] [PubMed] [Google Scholar]
- Osaki E, Nishina Y, Inazawa J, Copeland NG, Gilbert DJ, Jenkins NA, Ohsugi M, Tezuka T, Yoshida M, Semba K. Identification of a novel Sry-related gene and its germ cell-specific expression. Nuc Acid Res. 1999;27:2503–2510. doi: 10.1093/nar/27.12.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiahao S, Zuomin ZH, Jianmin L. Preparation of human testicular cDNA microarray and initial research of gene expression library related to spermatogenesis. In: Chan HS, editor. In Epithelial Cell Biology-A primer. Beijing; 2000. pp. 274–277. [Google Scholar]
- Okabe M, Ikawa M, Ashkenas J. Male infertility and the genetics of spermatogenesis. Am J Hum Genet. 1998;62:1274–1281. doi: 10.1086/301895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein JHP, Hochstenbach R, Pearson PL. Towards an understanding of the genetics of human make infertility: lessons from flies. Trends Genet. 2000;16:565–572. doi: 10.1016/S0168-9525(00)02140-5. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the decent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving make reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Schadt EE, Armour CD, He YD, Garrett-Engele P, McDonagh PD, Loerch PM, Leonardson A, Lum PY, Cavet G, et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SPA, Gingeras TA. Large-Scale Transcriptional Activity in Chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5' untranslated region of human mRNA using "Oligo-Capped" cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Hida M, Suzuki Y, Sugano S, Hashimoto K, Terao K, Hayasaka I, Hirai M. Construction and preliminary characterization of full length enriched cDNA libraries for nonhuman primates. Primate Res. 2000;16:95–110. [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.