Abstract
Summary: Living systems operate under interactive selective pressures. Populations have the ability to anticipate the future by generating a repertoire of elements that cope with new selective pressures. If the repertoire of such elements were transcendental, natural selection could not operate because any one of them would be too rare. This is the problem that vertebrates faced in order to deal with a vast number of pathogens. The solution was to invent an immune system that underwent somatic evolution. This required a random repertoire that was generated somatically and divided the antigenic universe into combinatorials of determinants. As a result, it became virtually impossible for pathogens to escape recognition but the functioning of such a repertoire required two new regulatory mechanisms: 1) a somatic discriminator between Not-To-Be-Ridded (‘Self’) and To-Be-Ridded (‘Non-self’) antigens, and 2) a way to optimize the magnitude and choice of the class of the effector response. The principles governing this dual regulation are analyzed in the light of natural selection.
I.Introduction
A. ‘...doth protest too much’
To confess, I was very hesitant to accept this invitation because my major contribution to immunology has been the gifted students that I unleashed over the years on the community and that didn't seem like an appropriate qualification for this assignment. However, as I mulled over the goal, ‘to infuse the next generation with the excitement of immunology’, I recalled the ‘infusion of excitement’ that my teachers, no longer with us, had given to me as a heritage. As their faces flashed across my recall, Adams, Avery, Beadle, Delbruck, Ephrussi, Heidelberger, Kabat, Lwoff, Macleod, Monod, Ochoa, Pappenheimer, Racker, Salk, Spiegelman, Szilard and so many more to whom I owed guidance by close encounter, I felt an imperative to attempt this part of the assignment, fail or succeed, as a tribute to them. The other half of our mission, ‘a perspective on how our field has developed and where you think things are (or should be) going’ is a much less difficult task as it only requires a tasteful balance between a sufficiency of ‘self-esteem’ and an awareness of ones intellectual limitations. I imagine that it would be salutary for the aspiring generation to appreciate that science advances more as a social than as an individual endeavor and, all too often, historical or clairvoyant essays end up being no more than an assay of the common sense and good taste of the reader, rather than of the author. Further, the best science is ascetic, reflective and single-minded, a burden few wish to bear, but from which all hope to profit.
I am aware that the names of my mentors are, in large measure, unknowns to the next generation (how often have I tested this on my students!). Their contributions were the ingredients that were fermented as a database from which were distilled the principles that, upon aging by experience, became truths. Our students are given these truths as a foundation on which to build future progress. The creators of this legacy all too often pass into anonymity. What they leave for us is a rich heritage of knowledge and understanding, the two most precious products of scientific endeavor. There is no better defense of reason at this moment in time when so many members of our species seek answers by the use of machines of death hopefully made superior by the blessings, spiritual endorsement, mythologies and self-righteousness of the feuding almighties, Jehovah and Allah.
What I think of as my contribution to this heritage is a way of thinking about the immune system. I will illustrate it in this essay without philosophy, justification or analysis of methodology. I keep toying with the idea of writing a philosophical essay entitled ‘How not to think about the immune system!’ but my colleagues tell me that, as a matter of prudence, it would have to be published posthumously. One has only to consider the misdirected bandwagons engendered by idiotype networks, suppressor circuitry and transcendental repertoires, or the semantic folderol surrounding the terms ‘self’ and ‘nonself’, etc., to appreciate the need for such an essay.
B. Living things obey the laws of natural selection
Living organisms are understood by us only in terms of the concept referred to as ‘evolution’. Evolution is a historical process that has been recently described with great precision by delineating the meanings of its elements, replication, variation, and selection (1). When we analyze a given segment of biology, as I will try to do here, the mini-laws that I formulate are really the offspring of the law of natural selection. This ‘mother’ law is so probing that the mini-laws of the immune system take on a strong explicative character. My goal here is to illustrate these laws.
C. What started the wars between the DNAs?
In the beginning on God's Little Acre was only ‘messes and messes of firmament’ from which a replicating molecule arose by an improbable, yet inevitable, event given ‘infinite’ time. It was imperfect in copying itself, and it varied. These variants interacted with the ‘firmament’ to set up an interactive selection pressure that resulted in some of the variants being copied and others becoming extinct. The molecule, known to be a nucleic acid, was selected upon over evolutionary time to encode not only the information to copy itself, but also the information to construct the machinery and provide the driving energy needed to capture raw materials and integrate them into replicating structures, cells and organisms. These evolving structures are referred to as living, and the dominant nucleic acid encoding all of this is DNA. Today we take this process of evolution as a given.
Living organisms competing for resources quickly discovered that their neighbours are the best and cheapest source of building materials; as a consequence, some ‘chose’ a life of predation and some ‘chose’ a life of parasitism. These latter, in turn, set up the selection pressure for host mechanisms to protect against parasitism, and to this end, all living organisms have biodestructive and ridding mechanisms. Bacteria have ‘restriction’ enzymes to protect against viruses; insects have a vast array of toxic and lytic agents to rid bacteria and fungi. Plants have lectins and degrading enzymes; vertebrates have both ‘innate’ and ‘adaptive’ biodestructive mechanisms, the subject of this essay.
This war between the DNAs is by its nature a ‘no exit’ process. No one can become a complete victor. No protective mechanism can evolve to perfection as there would be no selection pressure to maintain it; no pathogen can evolve to destroy all of its hosts, as that would result in its own extinction. Consequently, a pseudo-steady state or a slowly evolving boundary is established between pathogen and host, so that both species survive and the cycles of replication, variation and selection never let up, generation after generation, the result being lineages of evolved forms.
A large variety of mechanisms of protection and of pathogenesis were invented by the warring DNAs, and this variety, in turn, led to the view that nature tinkers. In a sense it does; nevertheless, the huge variety of mechanisms are subject to a common law, that of natural selection, and I apologize for being so repetitive. By analogy from physics, there are many tinker toys that perform work — gasoline engines, steam engines, electric motors, metabolic engines, etc. — but, in doing so, they all obey the second law of thermodynamics, no work without waste. Frankly, were it not for the pleasure to be found in revealing these laws, science would be a bore. However, to appreciate and take pleasure in this baroque activity requires education and taste no different from that needed to appreciate the arts and the humanities.
D. The passage from germline to somatic evolution
The non-vertebrates invented all of the biodestructive and ridding effector mechanisms used by the vertebrate immune systems. The defense mechanisms of non-vertebrates differ from the immune system of vertebrates in the nature and behaviour of the recognitive sites that guide the biodestructive and ridding effector functions to their targets. For the non-vertebrates, the recognition of the target is and was a germline encoded and selected property. As germline evolution favors the rapidly replicating prokaryotic pathogens over the slower eukaryotes, the selection was either for a recognitive site of as broad a specificity as possible (e.g. a particle sensor) or for one targeting a structure on the pathogen that is of comparably slow variation (e.g. a carbohydrate). Why wasn't the selection for a seemingly ideal solution, a universal glue?
The use of a protective mechanism that is biodestructive and ridding requires that the potential targets be separated into those which, if attacked, would destroy the host and those which, if attacked, would protect the host from pathogens that would otherwise kill it. We will refer to the potential host targets which are Not-To-Be-Ridded as NTBR-antigens and the potential non-host targets which are To-Be-Ridded as TBR-antigens (2). Any effector mechanism that was guided to its target by a recognitive site that had the specificity of a universal glue would kill the host that it is supposed to protect. Consequently, we have a basic principle. Any protective mechanism that is biodestructive and ridding must make an NTBR—TBR discrimination. This discrimination is the responsibility of the recognitive site, henceforth referred to as a paratope.
Whether the selection is on the germline or the soma, the sole selection pressure on the specificity of the paratope is the necessity to make an NTBR—TBR discrimination. In the case of germline-selected (GS) paratopes, the NTBR-antigens are the totality of those of the species. Any mutation in the specificity of a GS-paratope that resulted in recognition of an NTBR-antigen of the species would be lethal for the individual resulting from a mating that expressed both entities. The result is that the GS-paratopes of the non-vertebrate defense mechanisms are blind to the NTBR-antigens of the species. This is why one can, as a general rule, successfully transplant tissues between individuals of a non-vertebrate species (those with GS-paratopes only).
As evolution progressed to result in vertebrates that encountered an increased and more varied pathogenic load, the germline evolution of the paratopes of the non-vertebrate defense mechanisms became inadequate, because too many pathogens escaped recognition. Many factors contributed to this; as a general picture, the vertebrates are long-lived, warm-blooded and occupy very varied water and land habitats, giving the pathogenic load a significant edge by including one that escaped recognition. Also, germline evolution to increase the size of the recognitive paratopic repertoire had reached a limit when the probability of mutating to recognize an NTBR-antigen of the species became too high (see Section IV.D).
This limit resulted in a crafty solution, namely, to somatically generate a large and random paratopic repertoire that is expressed on a one paratope—one cell basis. This repertoire divided the antigenic universe into epitopes that are combinatorially distributed on antigens, making it virtually impossible for a pathogen (TBR-antigen) to escape recognition. However, a somatically generated random repertoire created two new problems: 1) a somatic mechanism was required to sort this somatically generated random repertoire into anti-NTBR and anti-TBR, and 2) a mechanism became essential that would both regulate the magnitude of the response and couple the paratopic repertoire to the appropriate effector function, one effective in ridding the parasite.
The non-vertebrate defense mechanisms, being germlineselected, express their effector activities, ‘one paratope—one effector mechanism’. The vertebrate immune system possessing a somatically selected, large random repertoire expresses its effector activities on a ‘many paratopes—one effector mechanism’ basis. This somatically generated large random paratopic repertoire and its expression by coupling to the handful of germline encoded effector mechanisms can be described as undergoing somatic evolution. The rate of somatic evolution in an individual is comparable to that of the germline evolution of pathogens. Unfortunately, but inevitably, what is somatically ‘learned’ by and stored in the ‘memory’ of the individual is not passed on to succeeding generations, as is the case for germline evolution.
From this discussion it should be appreciated that the sole evolutionary selection pressure for the specificity of paratopes is the necessity to make the NTBR—TBR discrimination. Specificity is heuristically defined in terms of this discrimination as the probability that a change in amino acid sequence of an antibody resulting in a functionally different specificity is anti-NTBR (3-5). It can be loosely translated as the Specificity Index (SI), the probability that a given cell in an antigen-unselected population will be anti-NTBR (5, see Section IV.D).
E. Two classes of pathogen must be faced
There are enemies from within and from without that require different protective mechanisms. There is an asymmetry between intracellular and extracellular pathogens. The enemies from within, viruses and certain bacteria, must at some time be expressed from without, whereas the enemies from without need never be expressed from within. However, the need for a regulatory mechanism to make the NTBR—TBR discrimination is common to the effector activities that deal with both classes of pathogen, and this has forced the immune system to find a way to process extracellular pathogens to ligands recognizable by the regulatory mechanism. This became the role of special peptide presenting cells, referred to as antigenpresenting cells (APC).
A virus pathogenic for a cell must at some time leave it to infect another cell. If it did not leave, the virus would die with the cell it is killing. This process would not be true of non-cytopathogenic viruses, but in this case the host would not require a biodestructive and ridding effector response. The virus while intracellular makes the entire cell a TBR-antigen, or better still a unit of elimination (an eliminon). When the virus is expressed from without, it itself becomes an eliminon. The infected cell and the free virus are eliminated by different effector mechanisms. Intracellular pathogens are eliminated by the cell-mediated effector systems and extracellular pathogens by the humoral effector systems.
The cell-mediated or T-cell system must examine the cells of the organism to determine whether a pathogen lurks inside without lysing the cell to analyze its contents. The only solution was to examine the garbage. The T-cell system capitalized on a mechanism used early in evolution to rid intracellular waste. During protein synthesis a certain proportion of the nascent polypeptide is mis-folded or truncated and degraded to be ridded by the cell as peptides. This garbage disposal process was turned into a detection mechanism for intracellular pathogen. Intelligence agencies have learned from the immune system that a good way to gather information is to examine the garbage (see Section IV.C). The extracellular form of the virus is dealt with by the humoral or B-cell system as are all extracellular TBR-antigens or, to use a more meaningful vocabulary, TBR-eliminons.
F. Two pathways are required for a successful immune response
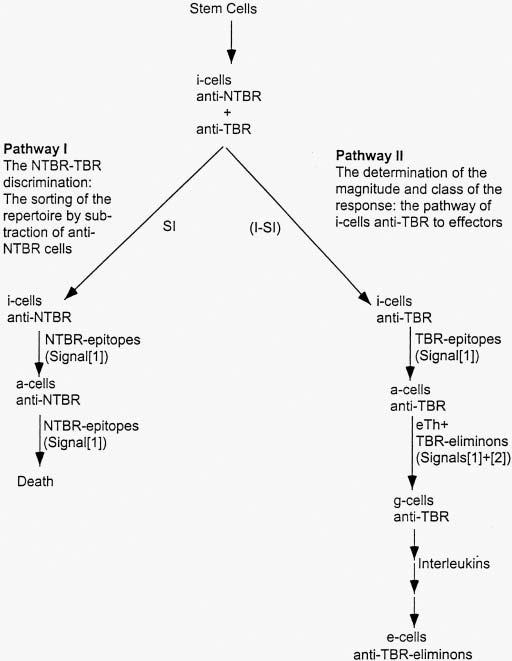
Upon encountering an antigen, the immune system must decide if it is NTBR or TBR (Fig. 1). The mechanism of Pathway I, the NTBR—TBR discrimination, is under spirited debate (6) even though a small number of principles limit how the para- topic repertoire can be sorted. Having decided that the antigen is TBR, Pathway II kicks in to regulate the magnitude and class of the response required to rid the TBR-antigen.
Fig. 1.
The pathways to an effective effector response (see list of Abbreviations).
II. The NTBR—TBR discrimination
A. The three laws of the NTBR—TBR discrimination (Pathway I)
A somatically generated random repertoire must be sorted by a somatic mechanism. Paratopes define epitopes, the ligand on antigens with which paratopes interact. Paratopes do not recognize antigens; they recognize epitopes. Cells express paratopes on a one paratope—one cell basis, the mechanism and parameters of which have been recently delineated (7; 8, click on ‘Ig haplotype exclusion’).
In order to sort this repertoire into anti-NTBR (to be inactivated) and anti-TBR (by definition the repertoire remaining after subtraction of anti-NTBR), there must exist a way for the individual to distinguish NTBR-epitopes from TBR-epitopes. NTBR-antigens express NTBR-epitopes only, but TBR-antigens may express either TBR-epitopes only or a combination of TBR- and NTBR-epitopes. This latter category, TBR-antigens that share epitopes with NTBR-antigens, is, in the end, the most serious challenge faced by any mechanism of the NTBR—TBR discrimination. The immune system must be able to determine if an epitope is NTBR or TBR, independent of the antigenic context in which it is found (i.e. the decision must operate epitope-by-epitope).
It might be recalled that, in contrast to the germline encoded mechanisms of non-vertebrates, for the somatic immune system of vertebrates, what is TBR for one individual is NTBR for another. One cannot, as a general rule, transplant tissues within a species from one individual to another. This means that TBR-antigens cannot be distinguished from NTBR-antigens by any physical or chemical property. The consequence was that evolution was forced to use the only criterion possible, namely the separation of NTBR and TBR during developmental time. All vertebrates with immune systems pass through a window of embryonic development when they are shielded from TBR-antigens by maternal mechanisms. During this period, all NTBR-antigens are present and no TBR—antigens. The interaction of the NTBR-epitopes with anti-NTBR results in the deletion of the latter, leaving by subtraction the anti-TBR repertoire. When this developmental window closes (around birth), and TBR-antigens appear on the scene, the anti-TBR repertoire is now regulated to respond by Pathway II. If I may be allowed to get ahead of myself, the decision between Pathway II and Pathway I is made by the sufficiency or insufficiency of effector T-helpers (eTh) specific for the antigen (Fig. 1).
This sorting of anti-NTBR from anti-TBR is a steady state process throughout life. As long as NTBR-antigen persists, the repertoire is purged of anti-NTBR, not to zero, but to some steady state acceptable level. Any NTBR-antigen that does not persist is eventually redefined as a TBR-antigen. No new NTBR-antigens appear after the developmental window closes.
In summary, the NTBR—TBR discrimination (Pathway I) is a product of two laws:
NTBR-antigens are prior and persistent. TBR-antigens are posterior and transient.
The somatically generated large random repertoire of paratopes must be sorted paratope-by-paratope by interactions epitope-by-epitope that subtract anti-NTBR and leave anti-TBR, a steady state lifelong process.
Anti-TBR does not and cannot define TBR-eliminons by whether or not they are noxious or dangerous. TBR-antigens are defined by the immune system as anything that is not NTBR. As anti-NTBR is expressed on a one paratope—one cell basis, the subtraction of the paratope from the repertoire involves the deletion of the cell.
The non-vertebrate defense mechanisms generate newborn cells as effectors. Interaction with the target triggers the biodestructive and ridding effector activity. This is possible because the recognitive site of the effector function is germlineselected. In the case of the vertebrate immune systems, which somatically generate large random repertoires, if the cells were born as effectors, it would not be possible to make an NTBR—TBR discrimination, as the newborn effectors anti- NTBR would kill the host. So evolution had to put an initial state cell, or i-cell, in the pathway to effectors. The i-cell expresses no effector activity on interacting with an epitope and it is the only stage at which the NTBR—TBR discrimination is or can be made. The i-cell has two pathways open to it upon encountering antigen, death (Pathway I) or activation (Path way II). This law is our third of the NTBR—TBR discrimination:
Cells of the immune system must be born as i-cells without effector activity that upon interaction with antigen have two pathways open to them as required by the NTBR—TBR discrimination.
B. The mechanism of the NTBR—TBR discrimination
An i-cell interacting with an epitope on an antigen cannot tell whether it is NTBR or TBR. It must be told. The signal to the i-cell (Signal[1]), via its antigen-receptor is the same whether it is NTBR or TBR. The i-cell receiving Signal[1] is converted to an anticipatory cell or a-cell, which is the stage with two pathways open to it, either death (Pathway I) or differentiation to effectors (Pathway II) (Fig. 1).
The death Pathway I is signaled epitope-by-epitope. However, the entry into expression of effector activity (Pathway II) is signaled antigen-by-antigen or, more precisely, eliminon-by-eliminon. In order to rid the pathogen (eliminon), the response to each of its epitopes that are recognized must be coherent (in the same class and of appropriate magnitude). The Signals[1] and [2] must be delivered to i- and a (anticipatory)-cells by epitope—specific interactions.
In summary, Pathway I, the NTBR—TBR discrimination, must be mediated epitope-by-epitope, whereas Pathway II, the determination of effector class, must be mediated eliminon-by-eliminon. This rule is very useful. Any proposal requiring that the NTBR—TBR discrimination (Pathway I) be made eliminon-by-eliminon (e.g. suppression, ‘danger’, ‘pathogenicity’, localization, mode of entry, etc.) should be viewed with circumspection. Any role they are found to play will be in the regulation and choice of effector class (Pathway II).
Referring to Fig. 1 , the two pathways (I or II) open to i-cells are distinguished by the insufficiency or sufficiency of effector T-helpers, a special class of regulatory cells. These cells are the source of an antigen-specific Signal[2] delivered to the a- cell by associative recognition of the eliminon. The somatic sorting of the repertoire is the responsibility of the eTh, which must be the end product of a pathway unique to it (see Section II.C).
The delivery of Signal[2] to an a-cell receiving Signal[1] converts it to an activated (or galvanized, g) g-cell responsive to interleukins that determine the class of the response. The a-cell is a required intermediate in order to insure that no cell can be activated that, in principle, could not have been inactivated (Pathway I).
Given that the somatically determined NTBR—TBR sorting of the somatically generated random immune repertoire is totally dependent on the sufficiency or insufficiency of eTh, several points arise:
What about antigens the response to which is eTh-independent? In this case, the specificity to which the response is directed must be absent from the NTBR-antigens of the mating pool (species). In other words, the paratope induced to expression in effectors must be germline-encoded and selected (9).
What happens if there is an error in the decision, Pathway I or Pathway II, and an anti-NTBR cell is activated? Once in Pathway II no discrimination between NTBR and TBR is possible. The g-cell anti-NTBR goes on to become an effector, the debilitating consequences of which depend on the frequency with which such errors occur (normally they must be infrequent) and on the NTBR-target (renewing or non-renewing).
How can the immune system deal with eliminons that share epitopes with NTBR-antigens?
There are several factors at play here, the most interesting being the competition between NTBR-antigens which can deliver Signal[1] only (obligatorily tolerigenic) and the TBR-antigen cross-reactive with NTBR, which activates by delivering both Signals ([1] and [2]). The NTBR-antigen competes to prevent the breaking of tolerance by the NTBR TBR> eliminon. If ‘suppression’, not ‘help’, were to regulate the NTBR—TBR discrimination, the response to the cross-reactive pathogen would be blocked, a lethal situation. However, to propose suppression as the sorting mechanism is at best an error in thinking.
We are not out of the woods yet! If helper cells arise as iTh and eTh is required to activate them to become effectors, where does the first or primer eTh come from?
C. Facing the ‘chicken and egg’ problem
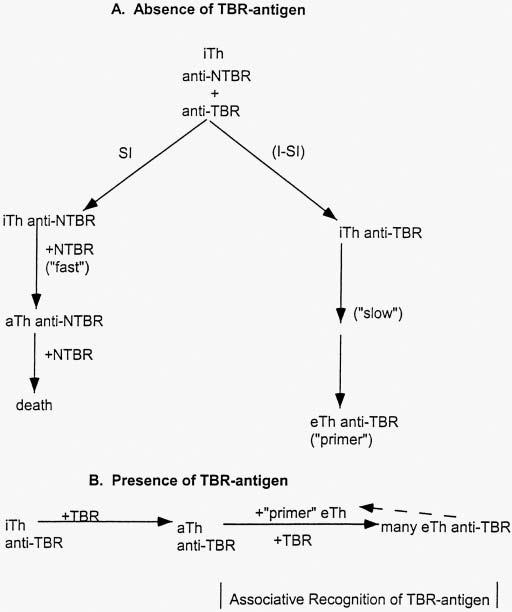
Whatever solution is envisaged, it must involve a pathway from iTh to primer eTh that is independent of an interaction with a TBR-eliminon and this requires a mechanism for the purging of iTh anti-NTBR and the accumulation of primer eTh anti-TBR. A precise model (Fig. 2) has been formulated (8, click on ‘Th-genesis’) based on an antigen-independent differentiation of iTh to eTh that is slow, and the rapid blockage of entry into this pathway upon interaction with antigen. The only way that an aTh can become an eTh is by associative recognition of the eliminon by an eTh delivering Signal[2]. The slow spontaneous differentiation of iTh to eTh is unique to this cell. As NTBR-antigens are prior and persistent, whereas TBR-antigens are posterior and transient, the iTh anti-NTBR are maintained at a threshold below the functional level, whereas eTh anti-TBR are maintained at a priming level. The TBR-eliminon is responded to ‘autocatalytically’ by a primer eTh-APC-aTh interaction resulting in the rapid pathway aTh→gTh→eTh. In sum then, the initiation of induction of effector activity (Pathway II) requires a special regulatory eTh-cell somatically selected to be specific for TBR-antigens that is derived by a TBR-antigen-indepen- dent pathway, iTh→eTh.
Fig. 2.
A, The TBR-antigen independent pathway to ‘primer’ eTh. B, The ‘autocatalytic’ induction of eTh initiated by ‘primer’ eTh in the presence of TBR-antigen.
An aside: The uncritical popularity of suppression as a determinant of the NTBR—TBR discrimination deserves comment. The reasons why it is unlikely to play this role has been discussed repeatedly by us (10-13). Here, we make an additional point. In order to determine the NTBR—TBR discrimination, suppressors (Ts) must themselves be sorted. This sorting can only be accomplished if, during the developmental window, Signal[1] (interaction with ligand) induces the conversion of iTs anti-NTBR to eTs anti-NTBR, epitope-byepitope, and iTs-cells anti-TBR that do not interact with ligand undergo an antigen-independent or default pathway to death. The process must be halted when the window closes and TBR-eliminons are encountered. The eTs anti-NTBR must then be self-renewing. As T-helpers are still required, that is, they remain necessary and sufficient, adding a double bluff control over the NTBR—TBR discrimination would be unselectable. Regulation by suppression plays its role in determining magnitude and class, and suppressors themselves, like all i-cells, must be regulated by eTh.
III. The regulation of effector class (Pathway II)
Evolution selects on the immune system at the level of the effector function. For it to be effective, the NTBR—TBR discrimination (Pathway I), the regulation of magnitude/class (Pathway II) and an effective destructive/ridding activity must operate. Central to this regulation is that the response to an eliminon be coherent, necessitating associative recognition of the eliminon by regulatory effectors and executive i-cells.
The Pathway II to effectors requires a set of sequential decisions. First, is the eliminon an intracellular or an extracellular pathogen? If intracellular, a cell-mediated response is indicated. If extracellular, a humoral response is indicated. Second, if cell-mediated, is the response to be lytic (an effector cytotoxic T-cell) or chemical (an effector inflammatory T-cell)? Third, if humoral, which isotype will be effective in arming an effector activity to rid the eliminon, immunoglobulin (Ig)M, IgG, IgA, IgE?.
Why do we need differential expression of the class of response? Why not respond by turning on all of the effector activities and let those that are effective rid the pathogen and those that are ineffective be wasted? The reason is that the ineffective classes/isotypes block the functioning of the effective classes/isotypes. Lighting up the immune response like a Christmas tree would be ineffective. Given that there is a steady state antigenic load and that effector activities entail a certain level of innocent bystander biodestruction, turning them on essentially constitutively would be debilitating.
What is the criterion that evolution had to use in order to select for the expression of the appropriate effector response? The criterion had to be based on an assay of ridding the pathogen. Effective classes rid the eliminon; ineffective classes do not.
What is there about a TBR-eliminon that the immune system can utilize to choose the class? There are two categories of choice. First, there is the stereotyped or germline-encoded recognition of pathogenicity (mode of entry, localization, inflammatory effects, etc.). This category was germline-selected based on an assay of the ability to rid the invader. For all we know, the regulation of class (Pathway II) could be entirely germline-selected, unlike the NTBR—TBR discrimination (Pathway I), which is entirely somatically selected. The coherence of the response of g-cells entering Pathway II is dependent on associative recognition of antigen (eliminon) by regulatory helper and suppressor cells that are told what to do by stereotyped decisions.
It is only because pathogens are so wily and escape germline-selected mechanisms so easily that one cannot help but be suspicious that a somatic mechanism must also be operative. What might such a mechanism entail? It would require a way to assay ineffective and effective ridding of the eliminon. Ineffective classes would accumulate along with the eliminon and have to be suppressed; effective classes would be ridded along with the eliminon and their production need only be controlled by the presence or absence of the eliminon.
The reader will notice the sharp change in style in this discussion of class regulation. Instead of an analysis of competing models compatible with the data, the analysis is a set of questions. Solving these questions is a real challenge; the next generation should be grateful that this generation left them something to do. There is no proper model for the regulation of class, the consequence of which is that much of the relevant data is cast in the wrong framework and difficult to extract from the information overload. We might console ourselves by pointing out that asking the right question is an important prerequisite to getting the right answer (i.e. the Socratic approach).
IV. The somatically selected immune repertoire
A somatically generated large and random repertoire is what distinguishes the immune system of vertebrates from the defense mechanism of non-vertebrates. The vertebrates hijacked the effector mechanisms of the non-vertebrates by coupling them to this somatically generated random repertoire (random with respect to the recognition of NTBR-epitopes). However, a somatically generated repertoire must be built on a germline-selected base. Before dealing with this point, it is important to realize that the immune system is modular in construct.
A. The Protecton is the unit of function
Animals ranging in size from a pygmy shrew (107 lymphocytes) to mice (108 lymphocytes) to humans (1012 lymphocytes) are equally protected per individual. This occurrence tells us that the immune system is constructed as an iterated minimum unit of function, which we have named a Protecton (3,14).
The Protecton is simply iterated proportionally to the size of the animal. Our best estimate of the size of the Protecton is 107 cells at an average density of 107 cells/mL. The mouse immune system is estimated to consist of 10 Protectons, whereas humans consist of 105 Protectons. The individual is protected per milliliter, not per individual. This concept has astounding consequences.
In summary then, the immune system is modular in construct. The modules referred to as Protectons are equivalent in function. The Protecton is the minimum sample of the immune system of an individual that possesses all of the protective properties of the whole. The Protecton is the unit upon which evolution selects, and its resultant parameters are also those of the immune system of the individual (8, click on ‘Protecton’).
Any discussion of the repertoire must be separated into that of the humoral (B-cell) and that of the cell-mediated (T-cell) systems.
B. The humoral immune system
Antibody functions in a concentration-dependent manner. On average, it must reach a threshold concentration in a short enough time to effectively protect against a pathogen. It requires that a minimum number of B-cells per ml respond initially to arrive at the threshold in the required time. For example, a reasonable estimate would be that antibody must reach 100ng/ml in 5days to protect against most pathogens. This process requires that 100 iB-cells per mL specific for the given pathogen respond initially. This response must be true for each and every milliliter of animal, which is another way of arriving at the concept of a Protecton. A Protecton of 107 cells cannot have a repertoire greater than 107, and if it is to be functional, must be considerably less.
At this point let us turn to the requirement for coupling antibody to effector function. The triggering of effector function by antibody requires that it be aggregated by interaction with the eliminon. The reason is that antibody which is bound to the eliminon, must trigger effector function in the face of a large excess of unbound Ig in tissue fluids. There are two competing selection pressures operating here. On the one hand, the level of reserve serum Ig must be maintained as high as possible to immediately protect against secondary encounter with as large a variety of pathogens as possible. Also, and not to be underestimated, the newborn is protected initially by maternally derived antibody. The newborn lives off the experience of the mother, until its own immune system is effective or mature. On the other hand, the unbound antibody competes to inhibit the bound antibody from interacting with the Fc receptors of the effector mechanism. From this point of view, the reserve serum Ig must be maintained as low as possible. Unbound antibody inhibits the interaction of the aggregated bound antibody with Fc receptors by what might be described as anticooperative unzipping. The optimum between these opposing selection pressures occurs when the concentration of fluid or unbound Ig is at the Kd of the Fc-FcR interaction, and this scenario is what is experimentally found.
The limiting selection pressure on the size of the repertoire is exerted by monomers (toxins and lytic enzymes) produced by pathogens. In order for a monomeric protein to aggregate antibody so that it can effectively trigger effector function, the monomer must be seen in at least 3 ways. In order for the majority of monomers to be seen in at least 3 ways, the average number of ways that the repertoire must see monomers can be calculated from the Poisson distribution. If the repertoire were to see monomers an average of 5 ways, then it would fail to see 12 out of 100 monomers in 3 ways (unacceptable), whereas an average of 10 ways would fail to see 3 out of 1000 monomers in 3 ways (acceptable). A reasonable estimate, then, would be that the repertoire recognizes on average 10 epitopes per antigen. What is required to deal with monomers (the limiting selection pressure) in the end also applies to polymers.
Before continuing, it might be helpful to point out that evolution is selecting upon the properties of the primary or virgin repertoire. Any individual who is not protected on initial encounter with a pathogen is unlikely to worry about a secondary encounter. Much of the phenomenology of the secondary encounter, like affinity maturation, is built into the mechanism of antigenic selection and requires nothing that would be unique to the secondary encounter.
Different species use different mechanisms to arrive at the primary repertoire but its final characteristics must be common to all given that each immune system must be acceptably protective. There are three categories of antibody based on where their subunits are encoded and then selected (Table 1).
Table 1.
Categories of antibody
| Germline |
Somatically |
|||
|---|---|---|---|---|
| Category | encoded | selected | encoded | selected |
| I | + | + | - | - |
| II | + | - | - | + |
| III | - | - | + | + |
Illustrating the antibody categories with mice or humans, the germline is selected upon to encode in VLVH pairs a specificity critical to the survival of the animal. The rules for this selection are identical to those used by the defense mechanisms of non-vertebrates. For Category I, the selecting agent must follow these rules:
be a polymer, as each VLVH pair selected in the germline must be able to trigger effector activity without collaboration from any other antibodies;
vary in structure sufficiently slowly to permit the germline to track it;
be able to induce effector T-helper independently (9).
Carbohydrates on pathogens are known candidates for this selection.
The germline then adds VLVH pairs by gene segment duplication and variation. A duplicated set of VLVH adds nothing to the repertoire. It must be selected upon to result in a new specificity that recognizes a determinant of the selecting agent (a pathogen) that is absent in the NTBR-antigens of the species. The selection is stepwise, adding one new VLVH at a time.
From sequence studies, there are roughly 50 VLVH pairs in Category I. The expression of these 50 VL and 50 VH gene segments permits complementation of subunits to yield 2450 (502-50) complements of random specificity that are germline-encoded but now somatically selected (Category II). Prior to the appearance of a somatic NTBR—TBR discrimination, Category II limited the size of the germline-encoded repertoire because the probability that a newly selected VLVH pair of Category I would generate an antispecies NTBR specificity by random complementation in Category II became too high. This feature provided the selection pressure for a somatic NTBR—TBR discrimination (Pathway I,Fig. 1).
We refer to this germline-encoded repertoire as the Stage I repertoire (Categories I + II). NVLVH pairs are germline-selected and (N2-N)VLVH pairs are somatically selected (N is estimated to be ∼50). The Stage I repertoire (Categories I+II) is the substrate for somatic hypermutation to result in a Stage II repertoire (Category III), which taking into account known parameters of a virgin iB-cell population can be estimated to be ∼20N2 or 5×104 specificities per Protecton (for detailed discussion see 3).
The Stage I germline encoded repertoire is expressed in high copy number (HCN), which can be estimated to be 102<HCN<103 copies per Protecton, the range dependent upon the functional role assumed for the NDN (‘CDR3’) region of the H-chain of Ig (7). The Stage II somatically encoded mutant repertoire is expressed in low copy number (LCN), which is roughly one copy per Protecton.
These two repertoires of approximately 5×104 total specificities see an average of 10 epitopes per eliminon. The repertoire of the B-Protecton divides the antigenic universe combinatorally into epitopes. A repertoire of 5×104 looking at antigens 10 epitopes at a time can distinguish 5×104C10= 1043 antigens but can effectively rid (because of the 3 or more rule) only 1013 eliminons, a reasonable number (see Section IV.D).
The HCN and LCN repertoires interact synergistically with eliminons. The HCN repertoire permits a rapid response to reach threshold concentration but being small would miss too many antigens. The LCN repertoire is sufficiently large to see most antigens but would respond too slowly. Acting together they resolve their respective limitations.
Thus far, by way of illustration only, mice and humans have been discussed. However, the principles should apply to other species. Consider birds as another example. They express a single germline-selected acceptor VLVH pair that is diversified by gene conversion from a set of donor V-gene segments. The donor gene segments may be viewed as having been selected in the germline for the specificities they encode. Single conversion events generate primarily the Category I specificities, whereas 2 or more conversions per accept or VLVH generate the Category II and Category III repertoires. This then results in an HCN and LCN repertoire. (see 15 for detailed discussion).
C. The cell-mediated immune system
The B-cell antigen receptor (BCR) and the T-cell antigen-receptor (TCR) are ‘isomers’, but do not be fooled. Looking alike does not imply functioning alike. They behave in quite different ways and are under distinctly different selection pressures.
When restrictive recognition of antigen was clearly established by Zinkernagel and Doherty, we, along with most immunologists, considered the phenomenon to show that the T-cell was concerned with cell-associated antigen. The antigen- specific receptor was visualized to recognize an major histocompatibility complex (MHC)-encoded restricting element(R) as a marker of cell-ness and antigen as cell-bound. However, thanks to a set of remarkable experiments by a large group of gifted experimentalists, it became clear that the restricting element was a presenter of intracellular peptide (P), and this changed the whole picture. The T-cell is not interested in cell-associated antigen but in intracellular antigen, a crucial difference. In hindsight, this association should have been predicted, and today we can only blush and maybe even learn something besides the need for humility.
The tendency was and still is to treat the TCR as a BCR by assuming that antigen interacts with the allele-specific determinants to create a new antigenic epitope seen by an Iglike TCR. In support of this view there was a collection of experiments interpreted to show that the TCR was indeed an Ig. In fact, some immunologists even produced data interpreted to show that the humoral response to viruses was also MHC-restricted. We never subscribed (16, p. 177; 17, 18) to this view that the TCR was an Ig or even that it recognized an interaction antigen. The TCR, unlike the BCR, had to be dually recognitive in its interaction with ligand. Why?
The recognition of allele-specific determinants on restricting elements is germline-selected. There is no way that a somatic process can determine what the alleles of the species are and therefore cannot distinguish allele-specific from shared determinants on R-elements. The recognition of antigen (now known to be peptide) had to be the consequence of a somatically generated repertoire. Two distinct selection pressures on the single TCR requires two distinct recognitive sites upon which each selection pressure operates. However, the original dual recognitive model was disproven by the discovery that the ligand for the TCR was [RP] and a new model was generated (19) referred to here as the Tritope Model.
The encoding of the recognition of allele-specific determinants is postulated to be in single Vα orβ gene segments. The 2V-gene loci act as a single [Vα+Vβ] pool. The reason for this assumption is that encoding in complemented VαVβ is ruled out, leaving only one choice. If VαVβ specified anti-R, then there could be only as many selectable VαVβ pairs as there are V-gene segments in the smallest locus. In mice there are roughly 20 Vβ and 80 Vα, permitting therefore only 20 VαVβ to be under germline selection for anti-R specificity. Clearly, given at least 2 Class I MHC loci (K and D) and 4 Class II loci (Aα+β and Eα+β) as well as > 10 alleles at each locus, 20 VαVβ pairs is insufficient to recognize all allele-specific determinants. Given 100 V-gene segments at the two loci acting as a single Vα+Vβ pool, a total of 100 allele-specific determinants are functional in mice.
The recognition of peptide (P) is somatically selected. The encoding of recognition of P is postulated to be encoded in the complementation between the (NDN)β regions of the β- subunit and the (NJ)α junctions of the α-subunit. As the Vα and Vβ gene segments encode anti-R, the only other source of amino acid variability is the (NDN)β and (NJ)α regions of the TCR. As only ∼5 amino acids in P contribute to the interaction and roughly 10 out of 20 possible replacements change the specificity in a significant way, the total size of this repertoire is ∼105. It is certainly less than 106 and greater than 104. An anti-P repertoire capped at ∼105 has many surprising consequences (4, 5, 19).
Unlike the repertoire of the B-Protecton, the repertoire of the T-Protecton has no germline-selected anti-P specificities. Its anti-P repertoire is somatically generated and random; it is the same size (105) for each family of TCRs restricted to a given allele of R. Given that a heterozygous mouse expresses a total of roughly 10 allele-specific determinants on its RI plus RII elements, then the anti-P repertoire restricted to a given allele would possess a copy number of 10 [107 ÷ (10 × 105)].
The TCR has 3 combining sites (Tritope). Positive selection in the thymus operates on one site (Vα or Vβ) to establish the restriction specificity, anti-R. The other subunit (Vβ or Vα)is entrained and expresses recognition of an allele-specific determinant which is responsible for alloreactivity, a phenomenon of no physiological importance but key to understanding the structure-function relationships of the TCR. The third site is the result of complementation (αβ) to specify anti-P. This structure derived from considerations of function has many consequences for the behavior of the TCR among them a requirement for a set of conformational changes in order to signal (19).
What about the allele-specific determinants? The evolutionary selection pressure is on the restricting element to bind peptide by anchor residues. As pathogens (viruses) can evade recognition by changing anchor residues, a selection pressure is established that drives polymorphism at the MHC loci. This fact leads to the postulate that the allele-specific determinants recognized by the [Vα+Vβ] pool have as their origin the anchoring sites for peptide.
Once a polygenic locus of V gene-segments is matched to a polymorphic set of R-elements, the system is locked in and becomes resistant to change. Two phenomena are explained:
alloreactivity is allele-specific and of much higher frequency than the restrictive recognition of any given eliminon;
xenoreactivity is also allele-specific and of high frequency.
For example, mouse will recognize human alleles and vice versa.
The anti-P repertoire poses a problem for the NTBR—TBR discrimination. The genome of most vertebrates encodes roughly 105 proteins. If these are expressed as peptides bound to restricting elements, it would appear that the total anti-P repertoire is also anti-NTBR, which poses a paradox. There seems to be only one solution. There is a sharp threshold of concentration (density of peptide presentation) above which the peptide is defined as NTBR and below which it is TBR. Reading such a threshold requires a conformational change in order to signal. In any case, once again, thresholds become a central property of responsiveness.
D. The meaning of specificity
Given that specificity of the paratope is being driven by the necessity to make an NTBR—TBR discrimination, its definition is best formulated in terms of that selection pressure. The formal definition would be that the degree of specificity, or SI, is the probability that a change in sequence of a given antibody that results in a new functional specificity will be anti-NTBR. SI varies between 0 and 1; SI = 1 is a universal glue, SI = 0 is ‘infinitely’ specific. However, any value of SI < 1/repertoire would be unselectable. Our best estimate for SI is ∼ 0.01.
The value of the Specificity Index was fixed early in evolution and, because a multigenic complementing system encodes the Stage I repertoire, SI, which has an average value, is very resistant to change. The Stage I repertoire could only have evolved stepwise by adding 1 VLVH at a time and selecting on it as a Category I antibody. In the absence of a somatically determined NTBR—TBR discrimination, germline evolution might have permitted as many as 20 VLVH to be selected in Category I before the probability of an anti-NTBR appearing in the Category II germline repertoire became prohibitive and required the appearance of a somatic NTBR—TBR discriminative mechanism. Table 2 illustrates this process.
Table 2.
Probability that addition in the germline of one anti-TBR will entrain a lethal anti-NTBR
| Adding one new VLVH at a time | Probability of adding one or more anti-NTBR by complementation |
|---|---|
| 20→21 | 0.33 |
| 30→31 | 0.45 |
| 40→41 | 0.55 |
| 50→51 | 0.63 |
Illustrative calculation: (20×2)×0.01 SI=0.4; (1 - e-0.4)=0.33.
Adding one new selected VLVH to 20 existent VLVH has a 0.33 probability of adding by complementation one or more anti-NTBR that would be lethal. This is the selection pressure for a somatic NTBR—TBR discrimination.
What structural property of paratopes is evolution selecting upon to determine the value of SI? In all likelihood the selection is on the size of the combining site. As SI increases, specificity decreases, and the size of the combining site decreases. If a paratope were of such small size that it responded to a single amino acid as an epitope, it could not distinguish NTBR-antigens from TBR-antigens (SI→1). If the paratope were so large that it viewed an entire antigen as an epitope, then it would be maximally specific; there would be no crossreactivity between antigens (SI→0). Evolution selected for a paratope of a size that distinguishes NTBR from TBR epitopes with sufficient precision to keep the frequency of debilitating autoimmunity acceptably low. It should be appreciated that the ability to distinguish an NTBR from a TBR epitope is equivalent to distinguishing any 2 TBR epitopes.
A repertoire of paratopes that recognizes an average of 10 epitopes per antigen (eliminon) would display a cross-reactivity between NTBR and TBR antigens calculatable by [SI+(1-SI)]10.At SI = 0.01 there is a 10% chance that a random TBR-antigen will express an NTBR-epitope (i.e. 1-0.9910).
What is the relationship between SI and the size of the repertoire? As SI increases, a smaller and smaller repertoire can recognize the antigenic universe but the ability to distinguish antigens decreases, until an NTBR—TBR discrimination is not possible. A repertoire of one is a universal glue. As SI decreases, a larger and larger repertoire is required to recognize the universe of antigens, until the size of the repertoire would equal the number of antigens in the universe. However, the time it would take a given antibody to reach a functional threshold concentration would quickly limit the size by making the repertoire non-functional.
The consequence of these considerations is that small paratopic repertoires can be used to effectively rid vast numbers of different eliminons by viewing them as combinatorial collections of epitopes. In order to be effective, paratopes must recognize shape, not chemistry. There is a price to be paid for this solution to recognition, which is the necessity to deal with TBR-antigens that share epitopes with NTBR-antigens. Thus, there is a limit to the ability of selection to reduce the frequency of autoimmunity.
The Specificity Index is tied into a set of parameters that describe the properties of the repertoire. Let us consider some illustrative empirical relationships. The parameters under discussion are the following:
SI = Specificity Index; varies 0<SI<1;
Z = relative size of the paratope; varies 0<Z<1;
E = average number of epitopes per eliminon seen by the repertoire;
R = size of the paratopic repertoire;
G = number of antigens distinguishable by R;
C = probability TBR shares epitopes with NTBR.
Consider a paratope characterized as having a relative size Z that varies between 0 and 1,0 = minimum, 1 = maximum. The paratope defines an epitope of a size that determines E, the total number of epitopes per eliminon, 102 being set as the maximum. Using Protecton theory (3) a set of descriptive (‘dead reckoning’) relationships can be formulated that tie these parameters together:
SI = 0.01 log E;
Z = 1 - 0.5 log E;
C = 1 - (1-SI)E.
From these relationships, Table 3 can be constructed.
Table 3.
Dead reckoning relationships
| SI | E | Z | C |
|---|---|---|---|
| 0 | 1 | 1 | 0 |
| 0.001 | 1.3 | 0.95 | 0.0013 |
| 0.005 | 3.2 | 0.75 | 0.016 |
| 0.01 | 10. | 0.5 | 0.096 |
| 0.016 | 40. | 0.2 | 0.474 |
| 0.02 | 100 | 0 | 0.867 |
As SI→0 (maximal degree of specificity), the size of the paratope is maximal (Z→1) and the eliminon behaves as a single epitope. Cross-reactivity between eliminons vanishes(i.e. C=0). As SI increases (degree of specificity decreases), the number of epitopes that constitute an eliminon increases, because the paratope is looking at a smaller and smaller area of the eliminon. At SI = 0.02 the maximal value of E (= 100) is reached. As an illustration, if the replacement of 15 out of 20 amino acids changed the epitope in a significant way, then the epitope would be defined by 2 amino acids (i.e. 15C2 = 105), clearly a limit. We estimate SI to be 0.01, and this estimate defines E = 10, Z = 0.5 and C = 0.096 an upper, yet acceptable level of cross-reactivity (sharing of NTBR-epitopes with TBR-antigens).
The relationship between the number of antigens (G) distinguishable by a repertoire of size R is given by
The primary repertoire calculated by Protecton theory is R = 4.53×104 at E = 10. This gives us the descriptive (‘empiric’) relationship between R and G atE=10,log R=0.1 log G+0.656.
Putting R together with the other parameters yields Table 4. At E = 10, A Repertoire Of 4.53×104 will distinguish 1040 antigens but given the 3 or more rule will fail to effectively rid 3 per 103 monomers, the key factor determining E. If ridding is saturated at 3 so that more is not any better, then a repertoire of 4.53×104 seeing, on average E = 10, will effectively rid 1.6×1013 eliminons (monomers). These ‘transcendental’ numbers tell us why pathogens are unlikely to escape recognition even with small random repertories. In general, they escape at the level of regulation of effector function.
Table 4.
The Specificity Index (SI) determines the size of the repertoire(R)
| SI | E | Z | C | G=RCE |
|---|---|---|---|---|
| 0 | 1 | 1 | 1040 | 1040 |
| 0.01 | 10 | 0.5 | 4.53×104 | 1040 |
| 0.02 | 100 | 0 | 1.49×102 | 1040 |
Much is made of the degeneracy and cross-reactivity properties of receptor—ligand interactions in the immune system. Viewed from the vantage point of natural selection, one can propose these simplifications:
two or more paratopes distinguishable by the experimenter that functionally interact with a single epitope are defined by the immune system as a single paratope (degeneracy of paratopes), and
two or more epitopes distinguishable by the experimenter that functionally react with a single paratope are defined by the immune system as a single epitope (cross-reactivity of epitopes).
The word ‘functionally’ needs delineation. The characteristics of the immune response are dominated by a selective pressure imposed by the fastest growing pathogens. As a reasonable estimate, a protective threshold concentration of antibody (or density of effectors) must be reached in roughly 5days. Therefore, the interaction between paratope and eliminon must be at a threshold of ‘avidity’ that can protect the individual when the paratope reaches the threshold concentration attainable in ∼5days (3).
From the point of view of the responding cell, a threshold of occupancy of its antigen-receptors determines signaling. A number of factors are integrated to determine occupancy, among which are affinity, concentration of ligand and density of receptors. In the end, the cell must count the number of signaling interactions (occupancy) and, if above a threshold, respond. The immune system, unlike the immunologist, has no way to assay degeneracy and cross-reactivity.
V. Why understand when you can cure without it?
Analyzing the immune system from the perspective of natural selection will seem to clinicians and biotechnologists to be a rather abstract activity. There is a certain truth in that view but two points should be made. First, this perspective keeps one focused on how the immune system does work, not on how it might work. Second, while history provides us with many examples of major biotechnological advances made before one had ‘understanding’, the converse, namely a ‘hit and miss’ approach, without understanding, remains a lottery ticket. It is possible to rationally program a search for understanding, and, if successful, it inevitably improves the chance of productively manipulating nature. It is not possible to design a rational program to ‘cure’ by ‘hit and miss.’ Students should be made aware of the importance of hypothesis-driven research. By way of example, evolution is selecting on the effector output to maintain the probability of debilitating autoimmunity acceptably low and of an effective ridding response acceptably high. Selection can never operate to perfection. What medicine is trying to do is alter these probabilities.
What has our analysis shown us? By definition, debilitating autoimmunity is the breaking of tolerance. This is not too useful a definition experimentally as the assay for breaking tolerance is debilitating autoimmunity. Tolerance cannot be assayed without breaking it. Tolerance is conceptual, defined as a state of unresponsiveness to NTBR-antigens as the result of a developmentally time-based somatic sorting process. If the theory is solid, then debilitating autoimmunity can be characterized.
There are two ways to break tolerance to NTBR-antigens(i.e. making them behave like TBR-antigens). First, tolerance can be broken by a TBR-antigen that shares epitopes with an NTBR-antigen. This mechanism results in autoimmunity to a specific target (e.g. the acetylcholine receptor, myelin basic protein, β-adrenergic receptors). The breaking of tolerance is the result of associative recognition of antigen by eTh-cells recognizing the TBR-epitopes and a-cells anti-NTBR rescued from the death pathway, recognizing the NTBR-epitopes. Most often this response would be seen as debilitating autoimmunity when the NTBR-target is non-renewing (e.g. an endocrine or neuronal cell). In most cases of self-renewing NTBR targets (e.g. fibroblasts, epithelial cells), the TBR-antigen is ridded and the autoimmune attack ceases without symptoms. Second, tolerance can be broken by interfering with the Signal[1]-driven death pathway for T-helpers so that iTh anti-NTBR accumulate and go down the antigen-independent pathway to effectors (eTh). This scenario does not apply to iB-cells and iTc-cells as the requirement for passage via the a-state assures that no cell can be activated that in principle could not have been inactivated. In other words, activation requires Signal[1] plus Signal[2].
Another way to break tolerance is by non-associative recognition of NTBR-antigen. If an eTh recognizes a surface component on an a-cell that is unrelated to the ligand recognized by the antigen-specific receptor, that a-cell will be activated whether it is anti-NTBR or anti-TBR. Whether tolerance is broken by interfering with Signal[1] to iTh or by non-associative delivery of Signal[2], the result is generalized autoimmunity. The targets attacked, and the consequences of the attack, are the result of a variety of factors dependent on the nature of the target. Experimental examples of generalized autoimmunity are ‘lupus’ in NZ mice or graft-vs.-host syndrome. A possible origin of naturally occurring generalized autoimmunity would be a non-cytopathogenic virus expressed in an a-cell that becomes the target of eTh. All other examples of immune system attack on host components are best defined as immunopathology, an innocent bystander consequence of the biodestructive and ridding effector activity itself.
Autoimmunity arises from abnormalities in the regulation of Pathway I; immunopathology arises from abnormalities in the regulation of Pathway II. When the NTBR—TBR discrimination breaks down (Pathway I), the most likely time would be prior to the end of the reproductive period of life. Evolutionary selection operates during this time to keep the frequency of debilitating autoimmunity sufficiently low. All too often, the term ‘autoimmunity’ is applied to chronic diseases with an onset well past the reproductive age. This kind of immune destruction should not be confused with the breakdown of the NTBR-TBR discrimination (Pathway I), which occurs in the prime of life and upon which evolution is selecting. More than likely the late onset ‘autoimmunity’ is of an origin best described as immunopathology.
What can we do about it? In principle, autoimmunity can be arrested by recreating in the adult individual the conditions of that unique embryonic window when no eTh and all NTBR-antigens were present. From what we know now, attempts to achieve this recreation in humans are unlikely to be successful. At best we can delete adult eTh activity and hope for the recreation of the developmental window when it regenerates. A much more likely approach is to manipulate Pathway II, the magnitude and class of the response. There is no doubt that the ability to shift in an antigen-specific manner from an effective to ineffective or, vice versa, from an ineffective to effective response will solve at the clinical level many autoimmune and immunopathology problems, not to mention optimize protection against infection. This example illustrates a well-known Murphy law. In one case we know everything and can do nothing; in the other case we know nothing but should be able to do everything.
VI. Coda: Extracting the postulates used to explain immune behavior
Throughout this manuscript, I have paused in order to state the law or postulate that was the basis for my argument. Here I will restate all of these laws and postulates so that they can be viewed as a unity.
Any protective mechanism that is biodestructive and ridding must make an NTBR—TBR discrimination.
NTBR-antigens are prior and persistent.TBR-antigens are posterior and transient.
The somatically generated large random repertoire of paratopes must be sorted paratope-by-paratope by interactions epitope × by-epitope that subtract anti-NTBR and leave anti-TBR, a steady state lifelong process.
Cells of the immune system must be born as i-cells without effector activity and upon interaction with antigen have 2 pathways open to them as required by the NTBR—TBR discrimination.
The NTBR—TBR discrimination must be mediated epitopeby-epitope, whereas the pathway to effectors must be mediated eliminon-by-eliminon.
The initiation of induction of effector activity requires a special regulatory cell (referred to as a T-helper) that is somatically selected to be devoid of anti-NTBR specificities. This population is by definition anti-TBR, the effector activity of which is derived by a TBR-antigen-independent pathway, iTh→eTh.
The immune system is modular in construct. The module referred to as a Protecton is the minimum sample of the immune system of an individual that possesses all of the protective properties of the whole.
The sole evolutionary selection pressure driving the specificity of the paratope is the necessity to make an NTBR—TBR discrimination.
Specificity defined by the NTBR—TBR discrimination, dictates the size of the repertoire (R), the average number of epitopes per eliminon seen by the repertoire, and the proportion of TBR-eliminons that share epitopes with NTBR-antigens.
The T-cell antigen-receptor (TCR), unlike the B-cell antigen-receptor (BCR), must be dually recognitive when mediating restrictive recognition of the eliminon. One combining site must engage the allele-specific determinant on the MHC-encoded restricting element, and the other combining site must engage the bound peptide. A third site responsible for alloreactivity is key to understanding the structure, but it is of no physiological importance.
Signaling of i-cells via ligand interactions with their antigen-specific receptors (TCR or BCR) requires a conformational change that results in self-complementation of the receptor, the transducer of the signal.
Triggering of effector activity by secreted Ig requires aggregation of the Ig by the eliminon. Monomeric eliminons must be seen in 3 or more ways to aggregate Ig sufficiently to effectively trigger effector activity.
The behavior of the immune system is sharply dependent on the reading of threshold concentrations and densities.
Acknowledgment
Supported by NIH Grant RR07716.
References
- 1.Hull DL, Langman RE, Glenn SS. A general account of selection: biology, immunology and behavior. Behav Brain Sci. 2001;24:511–528. [PubMed] [Google Scholar]
- 2.Cohn M, Langman RE. To be or not to be ridded? — That is the question addressed by the Associative Antigen Recognition model. Scand J Immunol. 2002;53:1–6. doi: 10.1046/j.1365-3083.2002.01059.x. [DOI] [PubMed] [Google Scholar]
- 3.Cohn M, Langman RE. The Protecton: the evolutionarily selected unit of humoral immunity. Immunol Rev. 1990;115:1–131. doi: 10.1111/j.1600-065x.1990.tb00783.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohn M. A new concept of immune specificity emerges from a consideration of the Self-Nonself discrimination. Cell Immunol. 1997;181:103–108. doi: 10.1006/cimm.1997.1212. [DOI] [PubMed] [Google Scholar]
- 5.Langman RE. The specificity of immunological reactions. Mol Immunol. 2000;37:555–561. doi: 10.1016/s0161-5890(00)00083-3. [DOI] [PubMed] [Google Scholar]
- 6.Langman RE, editor. Self-Nonself discrimnation revisited. Sem Immunol. 2000;12:159–344. doi: 10.1006/smim.2000.0227. [DOI] [PubMed] [Google Scholar]
- 7.Langman RE, Cohn M, editors. Haplotype exclusion at the Ig loci. Sem Immunol. 2002;14 doi: 10.1016/s1044-5323(02)00037-4. [DOI] [PubMed] [Google Scholar]
- 8.http://www.cig.salk.edu The Salk Institute website.
- 9.Cohn M. Some thoughts on the response to antigens that are effector T-helper independent (‘thymus-independence’) Scand J Immunol. 1997;46:565–571. doi: 10.1046/j.1365-3083.1997.d01-172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohn M. Conversations with Niels Kaj Jerne on immune regulation: Associative versus network recognition. Cell Immunol. 1981;61:425–436. doi: 10.1016/0008-8749(81)90390-7. [DOI] [PubMed] [Google Scholar]
- 11.Langman RE. The self-nonself discrimination is not regulated by suppression. Cell Immunol. 1987;108:214–219. doi: 10.1016/0008-8749(87)90205-x. [DOI] [PubMed] [Google Scholar]
- 12.Cohn M.The ground rules determining any solution to the problem of the self/nonself discrimination. In: The Tolerance Workshop. Proceedings of the EMBO Workshop on Tolerance; 1986 October 20-26; Basel, Switzerland. Basel: Basel Institute for Immunology, 1987: 3-35
- 13.Cohn M. The self-nonself discrimination: Reconstructing a cabbage from sauerkraut. Res Immunol. 1992;143:323–334. doi: 10.1016/s0923-2494(92)80132-5. [DOI] [PubMed] [Google Scholar]
- 14.Langman RE, Cohn M. The E-T (elephant- tadpole) paradox necessitates the concept of a unit of B-cell function: the Protecton. Mol Immunol. 1987;24:675–697. doi: 10.1016/0161-5890(87)90050-2. [DOI] [PubMed] [Google Scholar]
- 15.Langman RE, Cohn M, editors. 52nd Forum in Immunology: ‘The challenges of chickens and rabbits to immunology’. Res Immunol. 1993;144:421–495. [Google Scholar]
- 16.Cohn M, Langman RE, Geckeler W. Immunology 1980: Progress in Immunology. Academic Press; New York: 1980. Diversity 1980; pp. 153–201. [Google Scholar]
- 17.Cohn M. Pinch hitting for Hans Wigzell. Immunoglobulin Idiotypes and Their Expression. In: Janeway C, Sercarz EE, Wigzell H, editors. ICN-UCSA Symposium on Molecular and Cell Biology. Academic Press; New York: 1981. pp. 831–849. [Google Scholar]
- 18.Cohn M. The T-cell receptor mediating restrictive recognition of antigen. Cell. 1983;33:657–669. doi: 10.1016/0092-8674(83)90009-0. [DOI] [PubMed] [Google Scholar]
- 19.Langman RE, Cohn M. The Standard Model of T-cell receptor function: a critical reas- sessment. Scand J Immunol. 1999;49:570–577. doi: 10.1046/j.1365-3083.1999.00569.x. [DOI] [PubMed] [Google Scholar]