Abstract
Background
Rhinovirus, the most common cause of upper respiratory tract infections, has been implicated in asthma exacerbations and possibly asthma deaths. Although the method of transmission of rhinoviruses is disputed, several studies have demonstrated that aerosol transmission is a likely method of transmission among adults. As a first step in studies of possible airborne rhinovirus transmission, we developed methods to detect aerosolized rhinovirus by extending existing technology for detecting infectious agents in nasal specimens.
Methods
We aerosolized rhinovirus in a small aerosol chamber. Experiments were conducted with decreasing concentrations of rhinovirus. To determine the effect of UV irradiation on detection of rhinoviral aerosols, we also conducted experiments in which we exposed aerosols to a UV dose of 684 mJ/m2. Aerosols were collected on Teflon filters and rhinovirus recovered in Qiagen AVL buffer using the Qiagen QIAamp Viral RNA Kit (Qiagen Corp., Valencia, California) followed by semi-nested RT-PCR and detection by gel electrophoresis.
Results
We obtained positive results from filter samples that had collected at least 1.3 TCID50 of aerosolized rhinovirus. Ultraviolet irradiation of airborne virus at doses much greater than those used in upper-room UV germicidal irradiation applications did not inhibit subsequent detection with the RT-PCR assay.
Conclusion
The air sampling and extraction methodology developed in this study should be applicable to the detection of rhinovirus and other airborne viruses in the indoor air of offices and schools. This method, however, cannot distinguish UV inactivated virus from infectious viral particles.
Background
Rhinoviruses have been associated with 40% to 65% of "common colds" studied by methods of virus culture [1,2]. With the use of reverse transcriptase – polymerase chain reaction (RT-PCR), rhinovirus has been shown to be associated with 53% of all colds throughout the year and up to 92% of colds during the autumn outbreak period [3]. Rhinviruses have been implicated in the exacerbation of asthma symptoms [4] and may even play a role in asthma related deaths [5]. Due to their role in the exacerbation of asthma symptoms, rhinoviruses are a major cause of pediatric hospitalizations [6].
Although rhinoviruses have been thought to be transmitted via large droplets or contact [7], studies in the 1980's demonstrated that fomites are not an effective means of transmitting rhinovirus infections [8-10]. While direct inoculation of respiratory secretions has been shown to transmit infection, this is not a likely route of transmission in older children and adults as survival on hands is brief [9]. Airborne transmission of infection was clearly demonstrated in one experimental study on adults [8]. However, in the absence of epidemiological studies designed to conclusively identify modes of transmission, the role of airborne transmission in viral respiratory infection remains controversial.
An impediment to future studies of airborne transmission of respiratory infections is the lack of established methods for the detection of airborne respiratory microorganisms appropriate for epidemiological studies. Several laboratory studies have been conducted in which aerosolized respiratory viruses have been collected and quantified with culture based methods [11-13]. These studies were conducted to determine the effects of temperature and relative humidity on the infectivity of respiratory viruses, but their methods are not suitable for the extremely low concentrations of virus in field experiments. PCR based assays have been employed to detect airborne respiratory viruses in a number of small studies conducted in hospital rooms of infected patients [14-17] and in pig houses [18].
To determine if airborne microorganisms can be inactivated, and therefore, prevent infection transmission, studies during the 1940's–1970's used ultraviolet germicidal irradation (UVGI) to inactivated airborne microorganisms under experimental conditions [19-22] and reduced airborne infections in a number of settings, such as hospitals [23], military housing [24], and classrooms [25]. However, UVGI was not effective against the highly contagious measles virus when children had extensive contact in buses and crowded tenements [26,27]. Thus, it fell into disfavor and has not been extensively employed nor has it been studied using molecular methods of detecting its efficacy.
Because we anticipate future studies of UVGI will use PCR based detection methods, it is important to determine how PCR assays react to UVGI damaged genetic material. In DNA, UVGI predominately produces damage in the form of cyclobutylpyrimidine dimers and (6–4) photoproducts. These photoproducts disrupt the replication of the genetic material and, therefore, inhibit the replication. There is limited information regarding the effect of UVGI on RNA. There are conflicting studies on viruses with RNA genomes that have attempted to determine if PCR assays are inhibited by UVGI. Two studies involving hepatitis A virus (single-stranded positive sense RNA genome) and rotavirus (double-stranded RNA genome), showed their PCR assays to be inhibited by UV inactivation of the viruses [28,29]. Conversely, Ma et al showed no effect of UV on a PCR assay when they irradiated poliovirus (single-stranded positive sense RNA genome) [30]. In this study, we present an extremely sensitive PCR based assay to detect airborne rhinovirus and demonstrate the effect of ultraviolet irradiation on the assay.
Methods
Organism
We obtained rhinovirus 16 strain 11757 from ATCC (Manassas, Virginia) with a titer of 107.25 particles per 0.2 ml. The suspension was diluted with phosphate buffered saline (PBS) to create four different concentrations to be used in the experiment.
Aerosolization and chamber description
We generated a rhinoviral aerosol with a 6-jet Collison nebulizer (model CN-38, BGI, Waltham, MA), which generates 2-μm mass-median diameter (MMD) droplets when run at 20 psig of air pressure [31]. The nebulizing solution consists of PBS, diluted virus, and 0.05 g/l uranine. We used the uranine as a fluorescent tracer, so that some filters could be used to determine the percentage of actual collection versus theoretical collection. The aerosol was generated in a specially designed aerosolization chamber that had been used in studies of UV sensitivity of Serratia marcescens and bacilli Calmette-Guérin (Figure 1) [32].
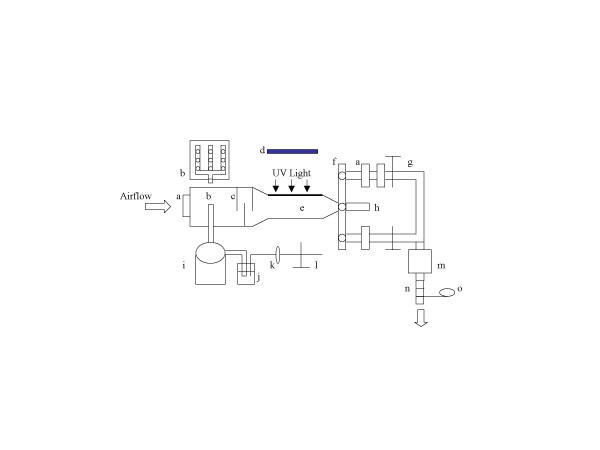
Figure 1.
Schematic diagram of the experimental chamber. a: Teflon filter; b: manifold; c: baffles; d: UV fixture; e: UV exposure chamber; f: sampling slide; g: air valve; h: port with the same features as the port below; i: drying jar; j: Collison nebulizer; k: pressure regulator; l: air valve; m: vacuum pump; n: rotameter; o: temperature and RH sensor
The chamber consists of four parts: a drying jar, a 165 mm square duct where the aerosol first enters the chamber, a UV exposure chamber, and sampling ports. We attached a Collison nebulizer to the drying jar, where the aerosol is given time to dry, before entering the square duct through a 9-port manifold, which creates a uniformly distributed aerosol. The aerosol next enters the UV exposure chamber. The UV exposure chamber is comprised of a fused quartz UV exposure window (279 × 254 mm), on which a UV fixture containing six 36-watt UV lamps (Lumalier, Memphis, Tennessee) sits. We inserted two types of screens beneath the UV fixture to adjust the level of irradiance. Downstream of the exposure chamber a slide contains three ports. One port is used to collect the aerosol sample with our sampling filters (Teflo 2.0 μm pore, Pall Gelman, Ann Arbor, Michigan) mounted in filter cassettes (Millipore Corp., Bedford, Massachusetts) while the other ports can be loaded with two Teflo filters in series, to allow a bypass while changing sampling filters. Two filters were used in the bypass to ensure that no aerosol escaped the chamber. The multiple ports allow changing of filters without interrupting the aerosol flow. Makeup air passes through a Teflon filter prior to entering the chamber to prevent environmental microorganisms from entering the chamber.
Filtered air was pumped through the chamber at a rate of 8.5 l/min monitored by a rotameter. To provide additional protection, the experimental chamber was housed within a Class II, Type A biosafety cabinet. Although temperature and %RH were not controlled, they were measured with continuous monitors (HOBO H08-004-02 four channel data loggers, Onset Computer Corp., Bourne, Massachusetts) downstream of the sampling filters.
Aerosol procedures
Airflow through the chamber was maintained at 8.5 l/min for 10 minutes. We then began the aerosolization of the rhinoviral solution and allowed the aerosol to flow through the bypass filter for 20 minutes. Air samples were then collected for 10 minutes. For sampling runs that included testing for effects of UV exposure, the UV radiation was turned on and allowed to warm up for 10 minutes while airflow was directed through the bypass filter. An air sample was then collected for 10 minutes. Three replicates of UV exposed and nonexposed particle samples were collected during each sampling run. Aerosols were exposed to an irradiance level of 30 mW/m2 for 22.8 s for a UV dose of 684 mJ/m2.
Spiking experiments
Prior to the aerosol experiments, we spiked filters with progressively more dilute viral suspensions to determine the limit of detection of our extraction and semi-nested RT-PCR assay. For spiking, 1 μl methanol was placed on the filter as a wetting agent followed by 1 μl of diluted virus. Filters were and allowed to dry prior to extraction. We also spiked one filter prior to each aerosol chamber run as a positive control. To test the stability of virus on the filters, a test was conducted in which spiked filters were used to collect air samples for one week in our laboratory, which has single pass air and is therefore an unlikely source of rhinovirus.
Filter extraction
We placed the filters face down in a 60 mm petri dish using sterile forceps. We then added 560 μl of prepared AVL buffer containing carrier RNA (Qiagen QIAamp Viral RNA Mini Kit) and 140 μl phosphate buffered saline (PBS). After placing the lids on the dishes, the dishes were secured to an orbital tabletop shaker (VWR Scientific Products, Thorofare, New Jersey) and rotated for 20 minutes at 360 RPM. After shaking, we pipeted the eluted material from the petri dish into a sterile microfuge tube.
RNA extraction
We extracted the RNA from the filter samples with the Qiagen QIAamp Viral RNA Mini Kit following manufacturer's protocol. The use of the Qiagen QIAamp Viral RNA Mini Kit resulted in 80 μl of purified viral RNA.
Semi-nested RT-PCR amplification
Following extraction, we first amplified the picornavirus RNA using primers OL26 (5'-GCA CTT CTG TTT CCC C-3') [33] and OL27 (5'-CGG ACA CCC AAA GTA G-3') [33] using the Invitrogen SuperScript One-Step RT-PCR with PLATINUM Taq Kit (Invitrogen Corp., Carlsbad, California). The PCR mixture contained 2 × reaction buffer (0.4 mM of each dNTP, 2.4 mM MgSO4), 1 μl of each primer (0.2 μM), 1 μl RT/PLATINUM Taq Mix and 10 μl of the RNA template in a final volume of 50 μl. We conducted the PCR reaction in a MJ Research (Watertown, Massachusetts) PTC-200 Peltier Thermal Cycler programmed with an initial cDNA synthesis and pre-denaturation step of 50°C for 30 minutes and 94°C for 2 minutes followed by then 36 cycles of denature, annealing and extension (15 s at 94°C, 30 s at 55°C, 30 s at 68°C).
We then amplified the PCR products from the RT-PCR step using primers OL26 (5'-GCA CTT CTG TTT CCC C-3') and JWA-1b (5'-CAT TCA GGG GCC GGA GGA-3') [34]. The PCR mixture contained 2 × reaction buffer (0.4 mM of each dNTP, 2.4 mM MgSO4), 1 μl of each primer (0.2 μM), 0.25 μl AmpliTaq (Applied Biosystems, Foster City, California) and 10 μl of RT-PCR product in a final volume of 50 μl. We conducted the PCR reaction for 36 cycles of denature, annealing and extension (15 s at 94°C, 30 s at 55°C, 30 s at 68°C), followed by a final extension of 4 min at 72°C.
Gel electrophoresis
Amplified products were detected by electrophoresis analysis on 2% agarose gel containing 8 μl of ethidium bromide per 200 ml of gel followed by examination under UV light.
PCR products
The OL26 and OL27 primer pair generates a 388 base pair amplicon from a region of the 5' noncoding region of the picornavirus genome. The semi-nested primer pair of OL26 and JWA-1b generates an amplicon of ~292 base pairs from within the original amplicon. The 292 bp amplicon does not differentiate between rhinovirus and enterovirus [34].
Uranine analysis
Uranine was used as a fluorescent tracer, which allowed us to determine a percent yield for each sampling run. To determine the amount of aerosol collected on each filter, uranine analysis was conducted on three filters per sampling run, one each from the beginning, middle and end of each run. The filters used for uranine analysis were removed from cassette holders and placed into a 100 mm test tube with sterile forceps. We added 5 ml of PBS to each tube and sonicated for an hour with vortexing every 15 minutes. We also collected samples of the nebulizing fluid before and after nebulization. Filter samples and nebulizing fluid samples were diluted with 0.01 N NaOH and analyzed with a spectrofluorometer (excitation 485 nm and emission 535 nm) (Tecan Corp. Spectrafluor Plus, Mannedorf, Switzerland). We averaged the results of the three filters to calculate an estimated percent yield for the sampling run.
Data analysis
The amount of uranine nebulized during each sampling run was determined by measuring the amount of fluid consumed and by measuring the fluorescence of nebulizing fluid before and after each sampling run. Using the total aerosolization time during the sampling run, we determined the amount of uranine that theoretically would be on each filter if we collected 100% of the aerosol generated during the 10-minute exposure of each filter. Using the filter samples that were analyzed for uranine, we determined the actual amount of uranine collected per sample. We averaged the three filter samples per run and divided by the total amount of uranine consumed to determine a percent yield (observed/expected) for the sampling run.
We computed the expected number of TCID50 on each filter sample from the known TCID50 and fluid volume added to the nebulizer, the amount of nebulizing fluid consumed per minute, and the sampling time. The expected TCID50 was adjusted for the yield for the uranine samples.
Results
We were able to obtain from the dilution gradient filter spiking experiments a positive band from a 1:500,000 dilution of virus, which translates into a detected amount of approximately 0.77 TCID50 or 0.39 PFU's per filter. The stability tests, in which, filters spiked with 25 TCID50/filter were used to collect air samples for 40 hours did not display any difference in band strength compared to control filters. Figure 2 displays the results from the aerosol experiments with the lowest concentration of virus detected.
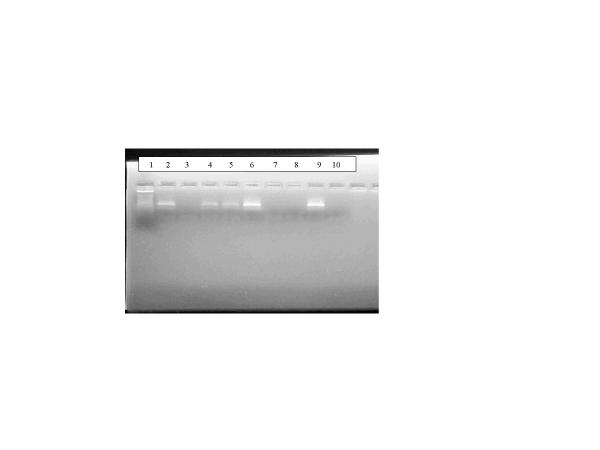
Figure 2.
PCR analysis of experimental contaminated air samples from VA Run 13 Lane 1: Marker; Lane 2:Nebulizer solution before run; Lane 3: Nebulizer solution after run; Lane 4: Filter sample; Lane 5: Filter sample; Lane 6: Spiked filter; Lane 7: Filter sample; Lane 8: Blank filter; Lane 9: Positive control; Lane 10: Negative control.
Table 1 summarizes results of the aerosol experiments, including the expected TCID50 captured on each filter during each sampling run and the number of filters that produced positive RT-PCR results. The limit of detection of our semi-nested RT-PCR assay with primers OL26, OL27 and JWA-1b is in the range of 1.3 TCID50/filter, yielding a clearly visible 292 bp band. No signal was detected from samples with 0.5 TCID50/filter. The percent yields for each sampling run, were low, ranging from 4% to 10%. Temperature and relative humidity were recorded in the air downstream of the chamber. The average temperature inside the chamber was very consistent, ranging from 26.4°C to 27°C and the %RH ranged from 24.9 to 50.7.
Table 1.
Detection of Experimental Rhinovirus 16 Aerosols by Semi-nested RT-PCR of Filter Samples
| Nebulizing Solution | ||||||
| Run Number | TCID50 per ml | Total ml Nebulized | Fractional Sampling Time per Filtera | Yieldb | Predicted TCID50 per Filterc | Positive Filters/Total |
| VA Run 12 | 16.1 | 15.2 | 0.136 | 0.041 | 1.4 | 3/3 |
| VA Run 17 | 10.1 | 24.2 | 0.085 | 0.103 | 2.1 | 3/3 |
| VA Run 13 | 6.5 | 15.0 | 0.142 | 0.093 | 1.3 | 2/3 |
| VA Run 14 | 3.3 | 16.7 | 0.119 | 0.072 | 0.5 | 0/3 |
aSampling time of each filter (10 minutes) divided by total run time. bFluorescence yield (observed/expected) for sampling run. cPredicted = TCID50 per ml * Total ml nebulized * Fractional sampling time per filter * Yield.
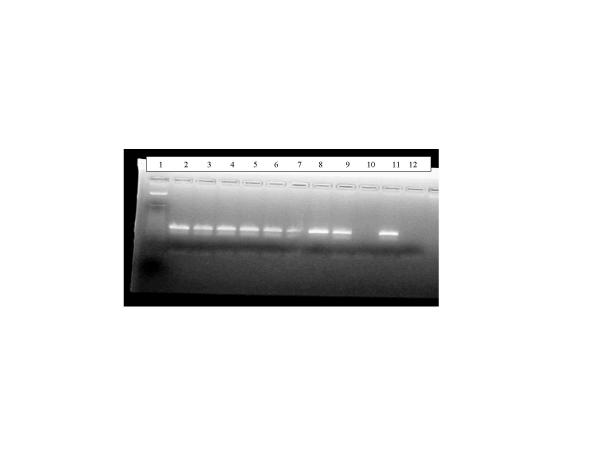
Figure 3 shows the results from test of the effect of UV exposure on aerosolized rhinovirus. Lanes 5 and 6 contain PCR products from filters containing UV exposed aerosol. No difference was detected between the UV exposed and nonexposed filter samples.
Figure 3.
PCR analysis of experimental contaminated air samples from VA Run 17 Lane 1: Marker; Lane 2:Nebulizer solution before run; Lane 3: Nebulizer solution after run; Lane 4: Filter without UV exposure; Lane 5: UV exposed filter; Lane 6: UV exposed filter; Lane 7: Filter without UV exposure; Lane 8: Spiked filter Lane 9: Filter without UV exposure; Lane 10: Blank filter; Lane 11: Positive control; Lane 12: Negative control.
In order to determine if the uranine in the viral suspension was inhibiting the extraction and detection assay, two otherwise identical runs were conducted – one with and one without uranine. Each sampling run was conducted with nebulizing solution containing 10.1 TCID50 per ml. Results of the two runs were identical, demonstrating that the uranine was not inhibitory to the extraction or RT-PCR assay.
Discussion
The majority of rhinoviral transmission among adults is thought to occur via airborne infectious droplet nuclei [8-10]. This study was conducted to assess the ability to detect airborne rhinovirus on air sampling filters and documents the first successful attempt to detect airborne rhinovirus using RT-PCR amplification-based techniques. These techniques allow for a highly sensitive method to conduct air sampling for rhinovirus. This assay also is capable of detecting aerosolized enterovirus, although future studies are necessary to confirm the limit of detection for enteroviral aerosols
Our limit of detection for the collection, extraction and amplification of aerosolized rhinovirus was 1.3 TCID50. The spiking experiments we conducted had a limit of detection of approximately 0.8 TCID50 or 60% of what was detected when the virus was aerosolized. These results can be interpreted in several ways. One possibility is that the virus may be damaged during the process of aerosolization in the Collison nebulizer, and that the RNA from the damaged virus cannot be amplified. This explanation would account for the results shown in Figure 2 where the nebulization solution prior to aerosolization yielded a positive result while after aerosolization we obtained a negative result. Another possible explanation is that it is easier to extract the virus from a dried liquid droplet, as was done in the spiking experiments, than from an aerosol spread uniformly over the filter surface.
We were unable to recover the majority of aerosol that was expelled into the chamber. The main reason for the low yields appears to be the result of large droplets falling out in the drying jar prior to entry into the exposure chamber. Because the yields were the same regardless of the UV status, the low yields should not affect our results.
Previous studies have shown that UV inhibits PCR assays in single and double stranded RNA viruses,[28,29] while our RT-PCR method did not detect a difference between UV exposed and nonexposed rhinoviral aerosols. The most likely reason for this is a matter of dose, although it is possible that characteristics of the particular viruses and PCR methods could also explain the results. Neither of the previous studies were conducted on aerosolized virus and the dose of UV was several orders of magnitude greater. This study, did not determine whether the rhinovirus aerosol was capable of infection after exposure to the high UV dose used in these experiments. It is possible that the UV irradiation damaged other regions of the viral RNA and not the region that our primers amplified, or that the amount of damage required to prevent RT-PCR amplification is greater than the amount of damage sufficient to prevent infection. This suggests possible future studies to investigate the relationship between UV damage, infectiousness, and detection by RT-PCR of various regions of the rhinovirus genome.
The limit of detection of our assay must be put into context with the amount of virus shed by an infectious individual. A study of rhinoviral shedding detected a titer of at least 1,600 TCID50 per ml of nasopharngeal wash in 17 out of 26 infected volunteers [35]. So it is clear that infected people can produce a large amount of virus, but it is unknown how much of that virus becomes airborne through coughs, sneezes, respiration and talking.
Two studies have attempted to determine the infectious dose 50% (ID50) of rhinovirus for humans [36,37]. The ID50 for microorganisms depend on at least four criteria: (1) the strain of the organism, (2) the route of transmission, (3) the method of inoculation, and (4) host immunity. In a study published in 1966, investigators from the U.S. Army Biological Laboratories and the National Institute of Allergy and Infectious Disease, conducted a series of tests designed to determine if respiratory viral agents were transmitted via the aerosol route. Using rhinovirus type 15, they inoculated volunteers in the nose with both nasal drops and an aerosol (particle size 0.3 to 2.5 μm). The authors estimated a human ID50 of 0.032 TCID50 for nasal drops and 0.68 TCID50 for aerosols [36]. In a study conducted in 1984, researchers inoculated volunteers with rhinovirus type 16 with droplets of viral suspension in several locations. The authors determined the ID50 for nasal drops was 0.28 TCID50 [37]. If we assume that the infectious dose is acquired through an aerosol, the aerosol data from 1966 suggests that our assay can detect an amount approximately equal to twice the ID50 in humans. But, if the spiking experiment is a better estimate of actual detection limit due to the inactivation by the nebulizer, then our detection limit is approximately equal to the ID50.
Caution must be taken when interpreting results from PCR-based air samples. Nucleic acid detection methods, such as the one presented here, are capable of detecting very small quantities of microorganisms, but these techniques cannot determine if the microorganism is infectious at the time of detection.
Conclusions
The filter collection and PCR-based detection methods developed here offers some clear advantages over the use of more traditional methods such as impinger collection and cell culture based detection. The use of filter collection allows for a much longer sample time, while impingers cannot be used for longer than 15–20 minutes. As was demonstrated here, filters can be used to collect at least 40-hour samples without reduced ability to detect virus on the filter. Cell culture methods, while useful in determining the amount of infectious virus, could not be used to detect the very low levels of infectious virus present in room air because collection of viable virus over long sampling times is not currently feasible. Therefore, the use of filter collection and PCR based detection is the only method presently available to detect low concentrations of respiratory virus in air. However, at UV doses much higher than typically used in upper room UV applications, this method was unable to distinguish between UV irradiated aerosols and nonexposed aerosols.
Abbreviations
PBS – Phosphate buffered saline
MMD – Mass-median diameter
RT-PCR – Reverse-transcriptase polymerase chain reaction
TCID50 – Tissue culture infectious dose 50%
UVGI – Ultraviolet germicidal irradiation
Competing interests
None declared
Authors' contributions
TM participated in the study design, the modification of the chamber, and performed the aerosol experiments and molecular assays. SJ participated in development of the molecular assays and study design. SR participated in the design of the study and modification of the chamber. DM participated in study design and coordination of the study.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by National Institute of Occupational Safety and Health, Grant number OH03694 and by National Institute of Environmental Health Sciences Center Grant 2P30ES00002. Dr. Myatt was supported by the National Institute Health's Training Program in Environmental Health Sciences number 2T32ES07155.
Contributor Information
Theodore A Myatt, Email: tmyatt@hsph.harvard.edu.
Sebastian L Johnston, Email: s.johnston@ic.ac.uk.
Stephen Rudnick, Email: srudnick@hsph.harvard.edu.
Donald K Milton, Email: dmilton@hsph.harvard.edu.
References
- Gwaltney J. The Jeremiah Metzger Lecture: climatology and the common cold. Trans Am Clin Climatol Assoc. 1984;96:159–175. [PMC free article] [PubMed] [Google Scholar]
- Hayden F, Kaiser D, Albrecht J. Intranasal recombinant alfa-2b interferon treatment of naturally occurring common colds. Anitmicrob Agents Chemother. 1988;32:224–230. doi: 10.1128/aac.32.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–42. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos N, Johnston S. The rhinovirus – not such an innocent? QJM. 2001;94:1–3. doi: 10.1093/qjmed/94.1.1. [DOI] [PubMed] [Google Scholar]
- O'Sullivan S, Cormican L, Faul JL, Ichinohe S, Johnston SL, Burke CM, Poulter LW. Activated, cytotoxic CD8(+) T lymphocytes contribute to the pathology of asthma death. Am J Respir Crit Care Med. 2001;164:560–4. doi: 10.1164/ajrccm.164.4.2102018. [DOI] [PubMed] [Google Scholar]
- Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O'Toole S, Myint SH, Tyrrell DA, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ. 1995;310:1225–9. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwaltney JM J, Moskalski PB, Hendley JO. Hand-to-hand transmission of rhinovirus colds. Ann Intern Med. 1978;88:463–7. doi: 10.7326/0003-4819-88-4-463. [DOI] [PubMed] [Google Scholar]
- Dick EC, Jennings LC, Mink KA, Wartgow CD, Inhorn SL. Aerosol transmission of rhinovirus colds. J Infect Dis. 1987;156:442–8. doi: 10.1093/infdis/156.3.442. [DOI] [PubMed] [Google Scholar]
- Jennings LC, Dick EC, Mink KA, Wartgow CD, Inhorn SL. Near disappearance of rhinovirus along a fomite transmission chain. J Infect Dis. 1988;158:888–92. doi: 10.1093/infdis/158.4.888. [DOI] [PubMed] [Google Scholar]
- Meschievitz CK, Schultz SB, Dick EC. A model for obtaining predictable natural transmission of rhinoviruses in human volunteers. J Infect Dis. 1984;150:195–201. doi: 10.1093/infdis/150.2.195. [DOI] [PubMed] [Google Scholar]
- Karim YG, Ijaz MK, Sattar SA, Johnson-Lussenburg CM. Effect of relative humidity on the airborne survival of rhinovirus-14. Can J Microbiol. 1985;31:1058–61. doi: 10.1139/m85-199. [DOI] [PubMed] [Google Scholar]
- Ijaz MK, Brunner AH, Sattar SA, Nair RC, Johnson-Lussenburg CM. Survival characteristics of airborne human coronavirus 229E. J Gen Virol. 1985;66:2743–8. doi: 10.1099/0022-1317-66-12-2743. [DOI] [PubMed] [Google Scholar]
- Ijaz MK, Karim YG, Sattar SA, Johnson-Lussenburg CM. Development of methods to study the survival of airborne viruses. J Virol Methods. 1987;18:2–3. doi: 10.1016/0166-0934(87)90114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aintablian N, Walpita P, Sawyer MH. Detection of Bordetella pertussis and respiratory synctial virus in air samples from hospital rooms. Infect Control Hosp Epidemiol. 1998;19:918–23. doi: 10.1086/647764. [DOI] [PubMed] [Google Scholar]
- Mastorides S, Oehler R, Greene J, Sinnott J, Kranik M, Sandin R. The detection of airborne Mycobacterium tuberculosis using Micropore membrane air sampling and polymerase chain reaction. Chest. 1999;115:19–25. doi: 10.1378/chest.115.1.19. [DOI] [PubMed] [Google Scholar]
- McCluskey R, Sandin R, Greene J. Detection of airborne cytomegalovirus in hospital rooms of immunocompromised patients. Journal of Virological Methods. 1996;56:115–118. doi: 10.1016/0166-0934(95)01955-3. [DOI] [PubMed] [Google Scholar]
- Sawyer MH, Chamberlin CJ, Wu YN, Aintablian N, Wallace MR. Detection of Varicella-Zoster Virus DNA in air samples from hospital rooms. The Journal of Infectious Diseases. 1994;169:91–94. doi: 10.1093/infdis/169.1.91. [DOI] [PubMed] [Google Scholar]
- Stark KD, Nicolet J, Frey J. Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Appl Environ Microbiol. 1998;64:543–8. doi: 10.1128/aem.64.2.543-548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D, Jones W, Newman C. Ultraviolet light inactivation and photoreactivation in the Mycobacteria. Infect Immun. 1971;4:318–319. doi: 10.1128/iai.4.3.318-319.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R, Knight M, Middlebrook G. Ultraviolet susceptibility of BCG and virulent tubercule bacilli. Am Rev Res Dis. 1976;113:413–418. doi: 10.1164/arrd.1976.113.4.413. [DOI] [PubMed] [Google Scholar]
- Edward D, Elford W, Laidlaw P. Studies of air-borne virus infections. J Hyg (Lond) 1943;43:1–15. doi: 10.1017/s0022172400035828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. Inactivation of airborne viruses by ultraviolet irradiation. Appl Microbiol. 1964;12:418–420. doi: 10.1128/am.12.5.418-420.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean R. General discussion: the mechanism of spread of asian influenza. Presented at the International Conference of Asian Influenza, Bethesda, MD. American Review of Respiratory Disease. 1961;83:36–38. [Google Scholar]
- Willmon T, Hollaender L, Langmuir A. Studies of the control of acute respiratory diseases among naval recruits I. A review of a four-year experience with ultraviolet irradiation and dust supressive measures, 1943–1947. Am J Hyg. 1948;48:227–232. doi: 10.1093/oxfordjournals.aje.a119237. [DOI] [PubMed] [Google Scholar]
- Wells W, Holla W. Ventilation in the flow of measles and chickenpox through a community: progress report, January 1, 1946 to June 14, 1949 – Airborne Infection Study, Westchester County Department of Health. Journal of the American Medical Association. 1950;142:1337–1344. doi: 10.1001/jama.1950.02910350007004. [DOI] [PubMed] [Google Scholar]
- Perkins J, Bahlke A, Silverman H. Effect of ultra-violet irradiation on classrooms on spread of measles in large rural central schools. Am J Public Health. 1947;37:529–537. [PMC free article] [PubMed] [Google Scholar]
- Council M. Air disinfection with ultraviolet irradiation: Its effect on illness in school-age children. London, Her Majesty's Stationary Office. 1954.
- Wang C-H, Tschen S-Y, Flehmig B. Antigenicity of hepatitis A virus after ultra-violet inactivation. Vaccine. 1995;13:835–840. doi: 10.1016/0264-410X(94)00054-Q. [DOI] [PubMed] [Google Scholar]
- Ojeh C, Cusack T, Yolken R. Evaulation of the effects of disinfectants on rotovirus RNA and infectivity by the polymerase chain reaction and cell-culture methods. Molecular and Celluar Probes. 1995;9:341–346. doi: 10.1016/s0890-8508(95)91652-0. [DOI] [PubMed] [Google Scholar]
- Ma J-F, Straub T, Pepper I, Gerba C. Cell culture and PCR Determination of poliovirus inactivation by disinfectants. Appl Environ Microbiol. 1994;60:4203–4206. doi: 10.1128/aem.60.11.4203-4206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May K. The Collison nebulizer: description, performance, and application. J Aerosol Sci. 1973;4:235. doi: 10.1016/0021-8502(73)90006-2. [DOI] [Google Scholar]
- Ko G, First M, Burge H. Influence of relative humidity on particle size and UV sensitivity of Serratia marcescens and BCG aerosols. Tuber Lung Dis. 2000;80:217–228. doi: 10.1054/tuld.2000.0249. [DOI] [PubMed] [Google Scholar]
- Gama R, Horsnell P, Hughes P, North C, Bruce C, Al-Nakib W, Stanway G. Amplification of rhinovirus specific nucleic acids from clinical samples using polymerase chain reaction. J Med Virol. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- Johnston SL, Sanderson G, Pattemore PK, Smith S, Bardin PG, Bruce CB, Lambden PR, Tyrrell DA, Holgate ST. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J Clin Microbiol. 1993;31:111–7. doi: 10.1128/jcm.31.1.111-117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RG, Jr, Cate TR, Gerone PJ, Couch RB. Quantitative rhinovirus shedding patterns in volunteers. Am Rev Respir Dis. 1966;94:159–67. doi: 10.1164/arrd.1966.94.2.159. [DOI] [PubMed] [Google Scholar]
- Couch R, Cate T, Douglas R, Gerone PJ, Knight V. Effect of route of inoculation on experimental respiratory viral disease in volunteers and evidence for airborne transmission. Bacteriol Rev. 1966;30:517–531. doi: 10.1128/br.30.3.517-529.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio DJ, Meschievitz CK, Peterson JA, Dick CR, Dick EC. Short-duration exposure and the transmission of rhinoviral colds. J Infect Dis. 1984;150:189–94. doi: 10.1093/infdis/150.2.189. [DOI] [PubMed] [Google Scholar]