Abstract
Inflammation or infection down-regulate the activity and expression of cytochrome P450 (P450) enzymes involved in hepatic drug clearance, possibly altering drug effectiveness and leading to toxicity. The regulation of UDP-glucuronosyltransferases (UGTs) in inflammation and infection is less well characterized. To determine the response of hepatic and renal UGTs during inflammation and infection, mice were administered either saline or 1 mg/kg LPS (16 hours), or Citrobacter rodentium by oral gavage (6 days). Hepatic mRNA expression of UGT1A1, 1A9, and 2B5 were similarly down-regulated after LPS exposure and C. rodentium infection, whereas UGT1A2 and 1A6 mRNAs were unchanged. Effects of C. rodentium infection did not require a functional Toll-like receptor 4 (TLR4). Conversely, renal UGT isoforms were relatively unaffected, except for UGT2B5 induction after LPS treatment. Regulation of UGTs during the inflammatory response exhibits similarities and differences with regulation of P450s, and may be cytokine-mediated.
Abbreviations: UGT, uridine diphosphate glucuronosyltransferase; P450, cytochrome P450; IL, interleukin; TNFα, tumor necrosis factor alpha; LPS, lipopolysaccharide; RT-PCR, reverse transcription-polymerase chain reaction; GAPDH, glyceraldehyde phosphate dehydrogenase
INTRODUCTION
UDP-glucuronosyltransferases (UGT) reside in the endoplasmic reticulum and catalyze the biotransformation of a variety of substrates (Wells et al., 2004). Addition of glucuronic acid by UGTs generally increases the water solubility of compounds for subsequent excretion in bile or urine. UGTs have multiple constitutive and inducible isoforms with overlapping substrate specificities, and are expressed in both hepatic and extrahepatic tissues (Wells et al., 2004).
Inflammation and infection can alter drug metabolism in humans and rodents, resulting in significantly increased or decreased drug efficacy and potential toxicity. Both cytochrome P450 (P450) expression and metabolic activities in liver and extrahepatic tissues can be down-regulated during inflammation (Morgan, 2001; Renton, 2004). However, little is known regarding UGT activities during the inflammatory response. Because hepatic P450 metabolism is significantly suppressed during inflammation, hepatic glucuronidation may be similarly affected, and contribute to alterations in drug efficacy.
Injection of bacterial lipopolysaccharide (LPS) is a widely used model of inflammation, and is well characterized regarding its effects on basal and inducible hepatic P450 expression. Many in vivo effects of LPS-induced inflammation are due to proinflammatory cytokines, and these cytokines mimic the effects of LPS when injected in vivo or incubated with hepatocytes (Morgan, 2001; Renton, 2004). In contrast to P450s, very little data exist on UGT regulation during LPS-induced inflammation. Significant decreases in hepatic UGT activities were reported after LPS administration in rats and isolated perfused mouse liver (Chen et al., 1992; Banhegyi et al., 1995), but no effect on UGTs was found in mouse hepatocytes (Banhegyi et al., 1995). Glucuronidation was unchanged after injection of individual cytokines in rats (Chen et al., 1992), yet was inhibited in isolated pig hepatocytes after cytokine administration (Monshouwer et al., 1996). Even less information exists regarding UGT regulation during live bacterial infection. Different UGT isoforms may be differentially modulated with differences in magnitude, time course, and effect, depending on the type of inflammatory or infectious agent administered. Here, we investigated mouse hepatic and renal UGT mRNA expression in the LPS inflammation model, and in a model of live bacterial infection, Citrobacter rodentium.
MATERIALS AND METHODS
Chemicals, Animals and Treatments
Unless otherwise specified, all chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Adult female 129S1/SvImJ (Sv129) and C3H/HeOuJ (HeOu) mice were obtained from Jackson Laboratory (Bar Harbor, ME), and acclimatized to the animal facility for 1 week. In one experiment, Sv129 mice (7 weeks old) were provided rodent chow and water ad libitum. E .coli lipopolysaccharide (LPS), serotype 0127:B8 was dissolved in sterile 0.9% saline and Sv129 mice were injected intraperitoneally with either 1 mg/kg LPS or saline (n = 5). At 16 hours, livers and kidneys were collected and stored at −80°C until RNA or microsome preparation. In a second experiment, five-week-old HeOu mice were infected with 2.0 × 108 CFU of C. rodentium in saline by gavage as described in the companion manuscript (Richardson et al., 2006). Control mice were administered saline by gavage the day after the infected group, and were pair-fed: i.e. they received the same amount of food eaten by the infected mice on the previous day. Mice were sacrificed 6 days after administration of saline or bacteria (n = 6), and livers and kidneys were collected. The Institutional Animal Care and Use Committee of Emory University approved all procedures.
Preparation of Total RNA and Microsomes
Total liver and kidney RNA was prepared using RNA-Bee isolation reagent (Tel-test, Friendswood, TX). RNA concentration was verified spectrophotometrically at 260 nm, and RNA purity and integrity was confirmed by formaldehyde-agarose gel electrophoresis and visualization with ethidium bromide. Liver microsomes were prepared by differential centrifugation (Haugen and Coon, 1976), and protein concentrations were determined using bovine serum albumin (Lowry et al., 1951).
Primer Sequences
Primers for mouse UGTs and glyceraldehyde phosphate dehydrogenase (GAPDH) were designed using the Primer Select software program (DNASTAR, Inc., Madison, WI). All primers were submitted to the National Center for Biotechnological Information (NCBI) for nucleotide comparison by the basic local alignment search tool (BLASTn; Altschul et al., 1990). For UGT1A family members, the 5′-variable region was used as a target sequence to guarantee specific amplification of an individual isoform over similar ones within the same family. For UGT2B5, the entire sequence was used for primer design. Oligonucleotides with a high degree of similarity (>80%) to other mouse UGT mRNAs were eliminated. Primers were custom-synthesized by MWG Biotech, Inc. (High Point. NC), diluted to 100 μM in deionized water, and stored at −80°C. Primer sequences are as follows (UGT isoform, forward 5′-3′, reverse 3′-5′, annealing temperature [°C]): UGT1A1, ccagcagaaggggcacgaagttg, tgaccacgcgcagcagaaaagaat, 56.9; UGT1A2, tgccccttcgaggaatctcagg, ctcccgcacaacatccctcatg, 59.4; UGT1A6, ctggctgatggtggctgactg, actgaggcccaaagcactaggaa, 56.9; UGT1A9, cagagggcatgaggttgtggta, tcctgcagtgtgaaaatgttagtt, 53.2; UGT2B5, accccagcaactttaggacacaat, tgatagatcgcctcgtagacacca, 54.7. GAPDH primers were also designed for use at annealing temperatures corresponding to the various primer sets.
Quantitative reverse transcriptase PCR (real-time PCR)
Purified total RNA was reverse-transcribed using the SuperScript First-Strand Synthesis System for RT-PCR kit (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocol. Real-time RTPCR was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Bedford, MA) and SyBr Green reagent, to determine UGT mRNA expression in mouse tissues by the method of Livak and Schmittgen (2001) as previously described (Richardson and Morgan, 2005). Each primer set yielded a single PCR product of expected size by agarose gel electrophoresis, and specificity was monitored using product melting curves in each reaction well.
Western Immunoblotting
Hepatic UGT microsomal proteins were measured by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blotting, as previously described in Richardson and Morgan (2005). Anti-peptide antibodies to rat UGT1A and UGT2B1 were generously provided by Dr. Shin-ichi Ikushiro (University of Hyogo, Hyogo, Japan). Antibody to the common region of rat UGT1A isozymes was diluted 1:5000, whereas UGT2B1 antibody was diluted 1:20000. The goat anti-rabbit secondary antibody was diluted 1:10000. All assays were performed within a linear range and band intensity was measured by laser densitometry.
Statistical Analysis
Control and experimental groups were compared by Student’s t-test (p<0.05).
RESULTS AND DISCUSSION
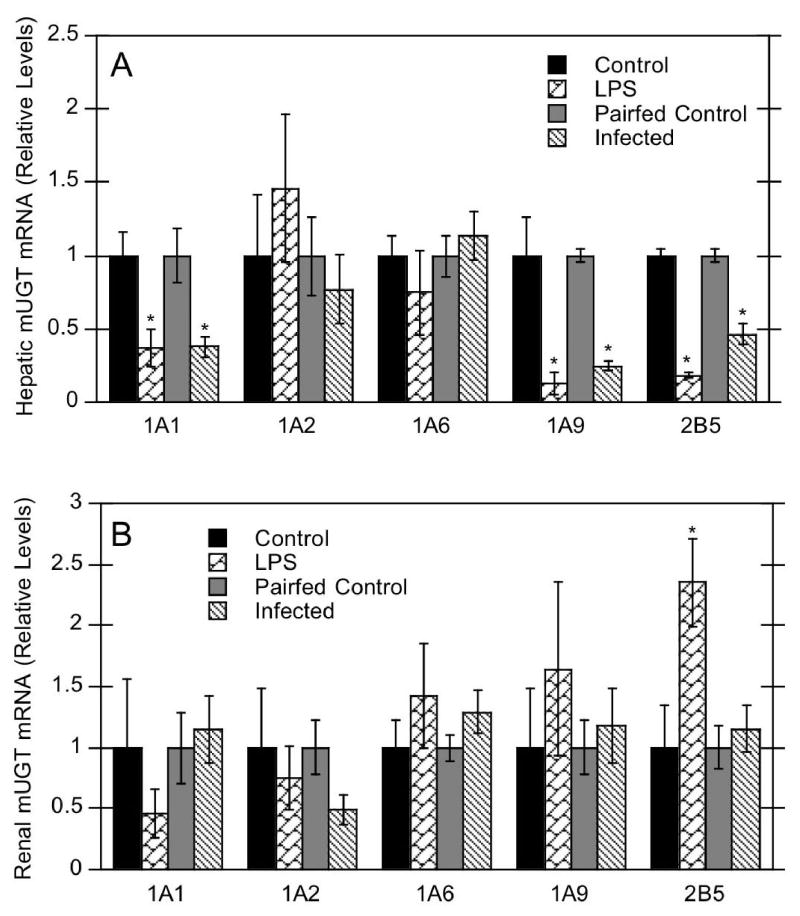
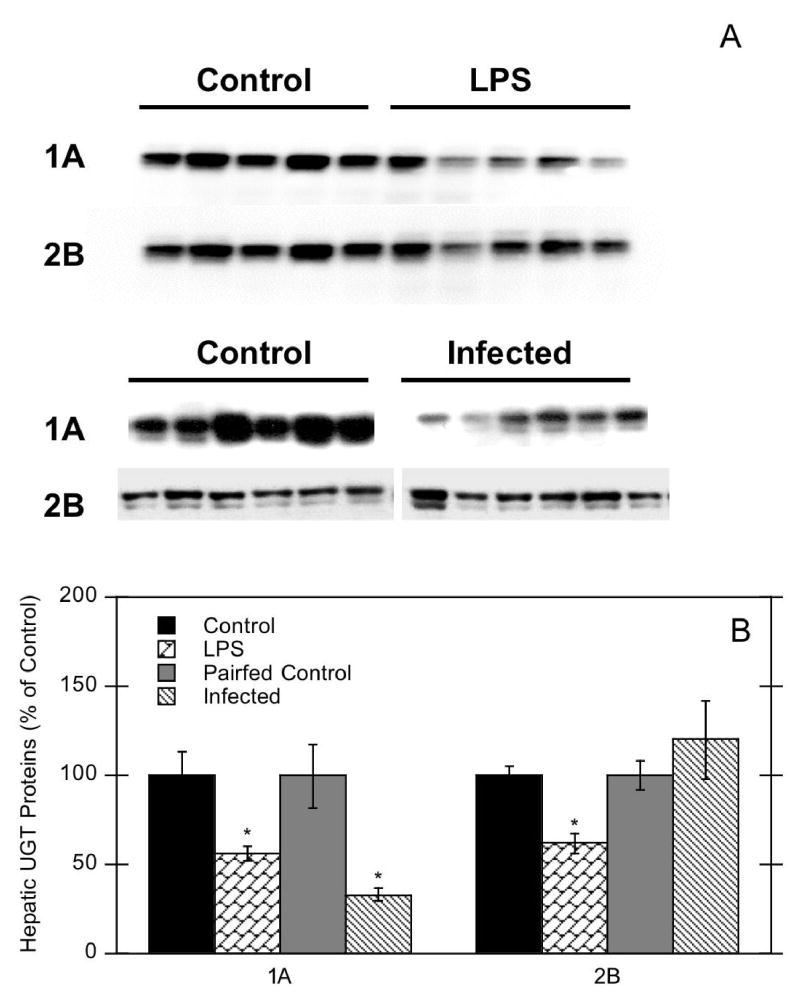
LPS-induced inflammation has been well studied with respect to its effects on hepatic P450s, but very little data exist on UGT expression during inflammation. As a live bacterial infection model, Citrobacter rodentium is the rodent equivalent of human enteropathogenic E. coli and a model of experimental colitis (Wales et al., 2005). Therefore, we investigated mouse hepatic and renal UGT mRNA expression in C. rodentium infection as well as in the well-characterized LPS model. LPS administration significantly decreased mouse hepatic UGT1A1 (37% of control), 1A9 (13% of control), and 2B5 mRNAs (18% of control) (Fig. 1A). Similar responses were observed in mice infected with C. rodentium, with suppression of UGT1A1 (38% of control), 1A9 (25% of control), and 2B5 (46% of control) mRNAs (Fig. 1A). In contrast, UGT1A2 and 1A6 mRNAs were not affected during either LPS-induced inflammation or bacterial infection. Effects of LPS on hepatic UGT proteins were consistent with effects on mRNA expression. LPS exposure significantly down-regulated hepatic UGT1A and 2B proteins to 50–60% of control. However, only UGT1A proteins were decreased in C. rodentium infected mice (to 33% of control) (Fig. 2).
Figure 1.
Effect of LPS administration or C. rodentium infection on hepatic (A) and renal (B) mRNA expression of UGT isoforms. Mice were treated with either saline or 1 mg/kg LPS, or treated with saline or C. rodentium. Hepatic and renal mRNA expression was determined, and values are expressed relative to the appropriate control group. Values represent means ±S.E.M. for each group (n = 5 or 6), and designations denote significant differences (p<0.05) from respective control (*).
Figure 2.
Effect of LPS exposure or C. rodentium infection on mouse hepatic UGT proteins. Mice were treated with either saline or 1 mg/kg LPS, or treated with saline or C. rodentium. Western blot data of UGT proteins from control and LPS-treated mice, and mice infected with C. rodentium (A). Quantitative analysis of western blot data (B). Values represent means ±S.E.M. for each group (n = 5 or 6), and designations denote significant differences (p<0.05) from respective control (*).
Others have shown that hepatic UGT mRNAs are suppressed after exposure to an inflammatory stimulus. Strasser et al. (1998) reported down-regulation of hepatic UGT mRNAs from turpentine-treated rats, including UGT1A1 (16% of control) and UGT2B3 mRNAs (53% of control). In addition, Congiu et al. (2002) correlated decreased human UGT1A4, 2B4 and 2B7 mRNA levels from liver biopsies with increased inflammation. Moreover, changes in hepatic UGT protein and/or activities during inflammation are also documented. Chen et al. (1992) showed significantly decreased rat UGT activities toward 1-naphthol and p-nitrophenol (UGT1A6), and testosterone (UGT2B3) after administration of LPS (to 77-83% of control). Strasser et al. (1998) found glucuronidation of testosterone was decreased to 65% of control, whereas activity towards p-nitrophenol was unchanged in turpentine-treated rats.
Our data provide a more comprehensive analysis of hepatic UGT mRNA regulation after LPS exposure and bacterial infection (Fig. 1), and demonstrate that this regulation is isoform-specific. Furthermore, our hepatic protein data indicate significantly decreased UGT1A and 2B proteins in LPS-treated mice and UGT1A proteins in infected mice, comparable to mRNA effects. The lack of effect on UGT2B protein in infected mice may be the result of variability in individual protein samples (Fig. 2), as well as the simultaneous detection of multiple UGT2B proteins that may be differentially regulated.
Renal UGT mRNAs seem relatively unaffected by inflammation or infection, except UGT2B5, which was significantly increased after LPS administration (Fig. 1B). Decreased renal UGT1A2 in infected mice narrowly missed significance (p = 0.073). In contrast to a lack of effect on UGTs, renal CYP4A and 2E1 mRNAs are induced in rat kidney after injection of LPS or irritants (Sewer et al., 1997; Mitchell et al., 2001) and CYP4A in mouse kidney (Barclay et al., 1999). In addition, C. rodentium infection has variable effects on renal P450 mRNAs, depending on the mouse strain (Richardson et al., companion manuscript).
The effects on hepatic UGTs during inflammation may be cytokine dependent, similar to cytokine-mediated P450 suppression after LPS treatment. We observed increased hepatic expression of IL-1β, IL-6, TNFα, and acute phase protein mRNAs during both LPS-mediated inflammation (Richardson et al., 2005) and C. rodentium infection (Richardson et al., 2006, companion manuscript). Other studies lend support to the idea that effects on UGTs are cytokine dependent. In isolated pig hepatocytes, IL-1α and TNFα caused early inhibition of glucuronidation (>60% at 12 hr), whereas glucuronidation was inhibited 51% by IL-6 at 24 hr (Monshouwer et al., 1996). Moreover, treatment of rat hepatocytes with IL-6, but not IL-1β, suppressed expression of UGT1A1 and 2B3 mRNAs (Strasser et al., 1998). Gut-derived cytokines could contribute to the effects observed during C. rodentium infection. Further investigation of UGT down-regulation is necessary to determine the role of cytokines in these effects.
Other studies in our laboratory with hepatic and renal P450 expression during LPS-induced inflammation and C. rodentium infection found striking differences in P450 regulation for each model (Richardson and Morgan, 2005; Richardson et al., 2006, companion manuscript). To the contrary, in this study we observed similar regulation of UGT isoforms in both models, despite several notable experimental differences between them. The LPS model is essentially an acute exposure (16 hr), whereas the C. rodentium infection model could be considered a chronic exposure (6 days). Also, different mouse strains and routes of administration were used for each experiment. In order to determine whether the effects of C. rodentium infection on hepatic and renal UGTs were dependent on bacterial LPS and toll-like receptor 4 (TLR4) signaling, we investigated these effects in TLR4 mutant mice. Our data indicate that the effects of C. rodentium infection on UGT mRNAs are independent of TLR4 and bacterial LPS signaling, because the effects of C. rodentium in TLR4- mutant mice (C3H/HeJ) were similar to effects in HeOu mice (supplemental data). Therefore, it appears that LPS and C. rodentium infection achieve the same effects on UGT expression via different mechanisms.
In summary, these data indicate that two different models of inflammation, LPS exposure and C. rodentium infection, produce similar down-regulation of hepatic UGT isoforms. This could have important consequences for drug and bilirubin metabolism in disease states. In contrast to P450 enzymes and hepatic UGTs, renal UGT mRNAs were not significantly decreased during these exposures. Additionally, the down-regulation of hepatic UGT mRNAs and proteins by C. rodentium infection is independent of TLR4.
Supplementary Material
Acknowledgments
The authors thank Kimberly L. Pierce for technical assistance, Dr. Gary W. Miller (Emory University) for the use of equipment, and to Dr. Shin-ichi Ikushiro (University of Hyogo) for graciously providing rat UGT antibodies.
Footnotes
Reprint requests should be sent to: Dr. Edward T. Morgan, Department of Pharmacology, Emory University School of Medicine, 5119 O. Wayne Rollins Research Center, 1510 Clifton Road NE, Atlanta, GA 30322
The National Institutes of Health Grant GM46897 and DK072372 (ETM), AR002157 (MS), and AI056067-01 (DK) provided funding for this study.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Banhegyi G, Mucha I, Garzo T, Antoni F, Mandl J. Endotoxin inhibits glucuronidation in the liver. An effect mediated by intercellular communication. Biochem Pharmacol. 1995;49:65–8. doi: 10.1016/0006-2952(94)00389-4. [DOI] [PubMed] [Google Scholar]
- Barclay TB, Peters JM, Sewer MB, Ferrari L, Gonzalez FJ, Morgan ET. Modulation of cytochrome P-450 gene expression in endotoxemic mice is tissue specific and peroxisome proliferator-activated receptor-α dependent. J Pharmacol Exp Ther. 1999;290:1250–1257. [PubMed] [Google Scholar]
- Chen YL, Florentin I, Batt AM, Ferrari L, Giroud JP, Chauvelot-Moachon L. Effects of interleukin-6 on cytochrome P450-dependent mixed-function oxidases in the rat. Biochem Pharmacol. 1992;44:137–48. doi: 10.1016/0006-2952(92)90047-m. [DOI] [PubMed] [Google Scholar]
- Congiu M, Mashford ML, Slavin JL, Desmond PV. UDP glucuronosyltransferase mRNA levels in human liver disease. Drug Metab Dispos. 2002;30:129–34. doi: 10.1124/dmd.30.2.129. [DOI] [PubMed] [Google Scholar]
- Haugen DA, Coon MJ. Properties of electrophoretically homogeneous phenobarbital-inducible and beta-naphthoflavone-inducible forms of liver microsomal cytochrome P450. J Biol Chem. 1976;251:7929–7939. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mitchell SR, Sewer MB, Kardar SS, Morgan ET. Characterization of CYP4A induction in rat liver by inflammatory stimuli: Dependence on sex, strain, and inflammation-evoked hypophagia. Drug Metab Dispos. 2001;29:17–22. [PubMed] [Google Scholar]
- Monshouwer M, Witkamp RF, Nujmeijer SM, Van Amsterdam JG, Van Miert AS. Suppression of cytochrome P450- and UDP glucuronosyl transferase-dependent enzyme activities by proinflammatory cytokines and possible role of nitric oxide in primary cultures of pig hepatocytes. Toxicol Appl Pharmacol. 1996;137:237–44. doi: 10.1006/taap.1996.0077. [DOI] [PubMed] [Google Scholar]
- Morgan ET. Regulation of cytochrome P450 by inflammatory mediators: Why and how? Drug Metab Dispos. 2001;29:207–212. [PubMed] [Google Scholar]
- Renton KW. Cytochrome P450 regulation and drug biotransformation during inflammation and infection. Curr Drug Metab. 2004;5:235–243. doi: 10.2174/1389200043335559. [DOI] [PubMed] [Google Scholar]
- Richardson TA, Morgan ET. Hepatic cytochrome P450 gene regulation during endotoxin-induced inflammation in nuclear receptor knockout mice. J Pharmacol Exp Ther. 2005;314:703–709. doi: 10.1124/jpet.105.085456. [DOI] [PubMed] [Google Scholar]
- Richardson TA, Sherman M, Antonovic L, Kardar SS, Strobel HW, Kalman D and Morgan ET (2006) Hepatic and renal cytochrome P450 gene regulation during Citrobacter rodentium infection in wildtype and toll-like receptor 4 mutant mice. Drug Metab Dispos This Issue [DOI] [PMC free article] [PubMed]
- Sewer MB, Koop DR, Morgan ET. Differential inductive and suppressive effects of endotoxin and particulate irritants on hepatic and renal cytochrome P450 expression. J Pharmacol Exp Ther. 1997;280:1445–1454. [PubMed] [Google Scholar]
- Strasser SI, Mashford ML, Desmond PV. Regulation of uridine diphosphate glucuronosyltransferase during the acute-phase response. J Gastroenterol Hepatol. 1998;13:88–94. doi: 10.1111/j.1440-1746.1998.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Wales AD, Woodward MJ, Pearson GR. Attaching-effacing bacteria in animals. J Comp Pathol. 2005;132:1–26. doi: 10.1016/j.jcpa.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, Hansen AJ, Kessler FK, Kim PM, Chowdhury NR, Ritter JK. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab Dispos. 2004;32:281–90. doi: 10.1124/dmd.32.3.281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.