Abstract
Hydrophobic mismatch, which is the difference between the hydrophobic length of trans-membrane segments of a protein and the hydrophobic width of the surrounding lipid bilayer, is known to play a role in membrane protein function. We have performed molecular dynamics simulations of trans-membrane KALP peptides (sequence: GKK(LA)nLKKA) in phospholipid bilayers to investigate hydrophobic mismatch alleviation mechanisms. By varying systematically the length of the peptide (KALP15, KALP19, KALP23, KALP27, and KALP31) and the lipid hydrophobic length (DLPC, DMPC, and DPPC), a wide range of mismatch conditions were studied. Simulations of durations of 50–200 ns show that under positive mismatch, the system alleviates the mismatch predominantly by tilting the peptide and to a smaller extent by increased lipid ordering in the immediate vicinity of the peptide. Under negative mismatch, alleviation takes place by a combination of local bilayer bending and the snorkeling of the lysine residues of the peptide. Simulations performed at a higher peptide/lipid molar ratio (1:25) reveal slower dynamics of both the peptide and lipid relative to those at a lower peptide/lipid ratio (1:128). The lysine residues have favorable interactions with specific oxygen atoms of the phospholipid headgroups, indicating the preferred localization of these residues at the lipid/water interface.
INTRODUCTION
Membrane proteins constitute a significant fraction of all proteins, yet little is known about their structure and function when compared to cytosolic proteins. The primary reason for the lack of structural knowledge is the difficulty with which these highly hydrophobic structures are crystallized. In addition, membrane protein structure depends, to a certain extent, on the interactions of the protein with the surrounding lipids (1). Generally, membrane protein segments are arranged as bundles of α-helices or β-barrels such that the interactions between the polar residues and the hydrophobic lipid acyl chains are minimized. A typical lipid bilayer consists of a highly hydrophobic central region containing acyl chains and two flanking headgroup regions, which are more polar. The hydrophobic thickness, representing the thickness of the acyl chain region, can range from ∼20 Å for short lipids to ∼35 Å for longer lipids. For common lipids such as POPC or DPPC, the typical hydrophobic thickness is ∼25 Å. For straight or slightly tilted α-helical trans-membrane proteins embedded in lipid membranes, this thickness corresponds to ∼20 amino-acid residues. It has been proposed that it is energetically favorable for the membrane protein to match the hydrophobic thickness of the lipid bilayer with a similar length of hydrophobic domain of the peptide. When a hydrophobic mismatch exists, the peptide-lipid system undergoes compensatory adjustments to mitigate the energetically unfavorable mismatch in lengths. Such adjustments in structure or orientation of peptides or lipids could play important roles in protein activity (1–4). It is also observed that polar and charged protein residues tend to cluster on each side of the membrane-spanning hydrophobic segments, interacting with the membrane-water interfacial region. Experimental and computational studies on trans-membrane proteins such as the mechanosensitive channels have shown that changes in membrane structure, arising from hydrophobic mismatch, influence the protein function (5–13).
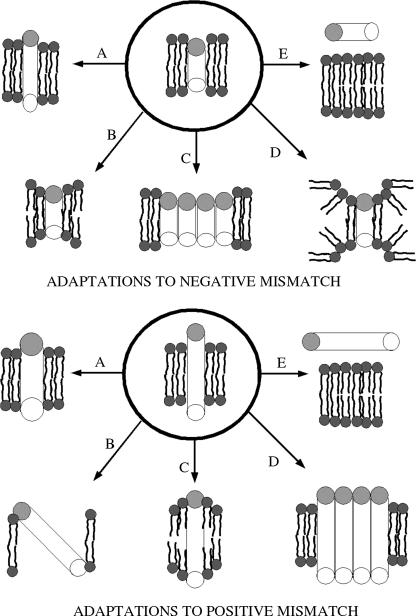
To investigate systematically complex membrane-protein interactions, simplified model membranes and peptides have been designed (14–22). The model lipid membranes typically constitute just one or two lipid species, rather than the complex mixture of lipids present in the cell membranes. The model proteins are typically single α-helical peptides with a large central hydrophobic region and flanking polar or charged residues (23–26). Peptides containing poly-leucine/alanine flanked by tryptophan or lysine have established themselves as preferred models of trans-membrane peptides (23,27–31). These peptides are called WALP or KALP peptides, depending on whether the flanking residues are tryptophans or lysines. To alleviate hydrophobic mismatch, trans-membrane peptides and lipids can undergo adaptations, such as those shown in Fig. 1, adapted from de Planque and Killian (3). For positive hydrophobic mismatch, i.e., a protein hydrophobic length that is greater than the thickness of the lipid hydrophobic region, one or more of the following adaptations can occur:
The α-helix can reduce its hydrophobic length by becoming a less tightly bound π-helix.
The peptide can tilt, reducing its exposure to polar groups.
The acyl chains near the peptide can order, increasing the local bilayer hydrophobic width.
The peptides can oligomerize or aggregate.
The peptide can be expelled from the bilayer.
In case of a negative hydrophobic mismatch, one or more of the following adaptations can occur:
The α-helix can increase its hydrophobic length by becoming a more tightly bound 310 helix.
The bilayer width near the peptide can decrease, by acyl-chain disordering.
The peptides may aggregate or oligomerize.
The peptide can induce nonlamellar phase formation.
The peptide can be expelled from the bilayer.
In addition to the hydrophobic mismatch, the flanking polar or charged residues of the peptide can also take on additional adaptations, due to their preferred position and orientation in the lipid/water interface. Tryptophan residues have been found to anchor themselves at the lipid water interfacial region (23,29). Although the positively charged lysines can be expected to prefer a similar position (32), the exact location of the lysine residues is difficult to determine experimentally (3).
FIGURE 1.
Possible adaptations to positive and negative mismatch conditions.
Molecular dynamics simulations are increasingly able to answer questions in structural biology that are difficult to answer experimentally (33,34). With the help of simulations, remarkable progress is being made in the understanding of protein folding, lipid bilayer behavior, and peptide-lipid interactions (35–41). Current state-of-the-art atomistic simulations allow the study of peptide binding to lipid/water interfaces (42), as well as trans-membrane peptide tilt behavior (43). The timescales of these simulations range from tens of nanoseconds to a few hundred nanoseconds. However, phenomena such as peptide insertion into lipids and peptide aggregation require longer timescales and are not accessible with the current computational resources. Short timescale (1.5 ns) simulations of WALP peptides at a peptide/lipid (P/L) ratio of 1:30 (44) have shown that peptides can tilt and affect local bilayer thickness and lipid ordering. Recent coarse-grained simulations by Venturoli et al. (45) and Nielsen et al. (46) have also shown that peptide tilt and lipid adjustments can lead to mismatch alleviation. Here, we use atomistic molecular dynamics simulations to study systematically the interactions of model trans-membrane KALP peptides (peptides with alternating alanine/leucine stretch flanked by lysine residues) with phosphatidylcholine bilayers. By systematically varying the extent of hydrophobic mismatch, we hope to understand the adaptations that the lipid/peptide systems undergo to relieve the unfavorable mismatch condition. We also seek to locate the preferred position of the lysine side chains in the lipid/water interface. Within the simulation timescale (10–200 ns), phenomena such as peptide expulsion or aggregation are not expected to occur. However, we hope to determine the extents of other adaptation mechanisms such as peptide tilt, local bilayer bending, peptide secondary change, etc., that can occur in our simulations.
METHODS
We use the GROMACS simulation tool for all our simulations (47). The united atom lipid parameters were adapted from the work of Berger and co-workers (48) and the peptides used the GROMOS force field. We performed a total of 33 simulations: three 100-ns simulations of pure lipid bilayers and 30 simulations of peptide/lipid systems of durations ranging from 50 to 200 ns. The peptide/lipid simulations were performed with a starting condition in which the peptide was in a vertical, trans-membrane state. This starting condition was used so that all the simulations started at an identical state. We considered three different lipid molecules (DLPC, DMPC, and DPPC) and five different peptides (KALP15, KALP19, KALP23, KALP27, and KALP31). The amino-acid sequences of all the peptides are given in Table 1. A DPPC bilayer was downloaded from http://moose.bio.ucalgary.ca/downloads, which contained 128 lipids (64 per leaflet) and 3655 water molecules. A DMPC lipid bilayer was created by removing the last two carbons of both tails of all the lipid molecules in the DPPC bilayer. A subsequent 20-ns equilibration simulation at 303 K yielded a DMPC bilayer with the correct interleaflet distance. In a similar fashion, a DLPC bilayer was constructed by removing the last two lipid carbon atoms from both tails from the DMPC bilayer. A subsequent 20-ns equilibration simulation at 298 K yielded a DLPC bilayer with the correct interleaflet distance. The DPPC, DMPC, and DLPC bilayers were each simulated for 100 ns at 323 K, 303 K, and 298 K, respectively, in an NPT ensemble. These temperatures were chosen to ensure that the lipids were in their fluid phases. A time step of 2 fs was used and the temperature was coupled semiisotropically to a Berendsen thermostat (49) at 1 atm with a coupling constant of 0.1 ps. The pressure was coupled semiisotropically to a Berendsen barostat with a coupling constant of 1 ps. Both the short-range electrostatic and van der Waals interactions used a short-range cutoff of 1.2 nm and the long-range electrostatics were calculated by the PME algorithm (50). The trajectories were saved every picosecond and were used for subsequent analysis. For ease of discussion, the three simulations are referred to as K1600, K1400, and K1200 (where 16 refers to DPPC, 14 to DMPC, and 12 to DLPC) in the rest of the article.
TABLE 1.
KALP peptides simulated
| Peptide | N-terminus | Hydrophobic stretch | C-terminus | Length of hydrophobic stretch (Å) |
|---|---|---|---|---|
| KALP15 | Ac-GKK | (LA)4L | KKA-NH2 | 13.5 |
| KALP19 | Ac-GKK | (LA)6L | KKA-NH2 | 19.5 |
| KALP23 | Ac-GKK | (LA)8L | KKA-NH2 | 25.5 |
| KALP27 | Ac-GKK | (LA)10L | KKA-NH2 | 31.5 |
| KALP31 | Ac-GKK | (LA)12L | KKA-NH2 | 37.5 |
The peptide simulations were performed at peptide/lipid (P/L) ratios of 1:25 and 1:128. The amino-acid sequences of the five peptides used in this study are shown in Table 1. The five different peptides, inserted in three different lipid bilayers at two P/L ratios, yielded 30 independent simulations, providing a way to investigate systematically hydrophobic mismatch. The peptides were created as ideal helices using the software Swiss PDB viewer ((51); http://www.expasy.org/spdbv). The C-termini were acetylated and the N-termini amidated for all the peptides. The peptides were solvated in a bath of water and a 1-ns simulation was performed with restraints on the backbone atoms. This ensured that the side chains were relaxed and were not in a biased, ideal configuration. These peptides were then inserted into the well-equilibrated DPPC, DMPC, and DLPC bilayers using the methodology developed by Faraldo Gomez et al. (52). In the 15 simulations with a P/L ratio of 1:128, a single peptide was inserted in a vertical, trans-membrane orientation into a lipid bilayer containing 128 lipids. In the 15 simulations with a P/L ratio of 1:25, 28 lipids were removed from the bilayers and four peptides were inserted, again in a trans-membrane orientation. At the higher P/L ratio, care was taken to ensure that the peptides were fairly uniformly distributed throughout the bilayer and not aggregated. After the insertion stage, equilibration simulations were performed on all 30 peptide/lipid systems for 5 ns, with position restraints on the peptide backbone atoms. The simulation conditions were identical to those of the pure lipid simulations described above. After the equilibration stage, all systems were simulated in production runs with no restraints on the system for at least 50 ns. Some of the simulations were extended to longer times, up to 200 ns, to achieve better equilibration of the properties of interest. The duration of the individual simulations are provided in Tables 3 and 4. The total simulation time for all the peptide-free and peptide/lipid simulations was ∼2.7 μs. All the computations were performed on a Linux cluster with 24 AMD 2600+ processors, with visualization of results using the software Visual Molecular Dynamics (53).
TABLE 3.
Simulations at low P/L ratio
| Label | Lipid | Peptide | Lipid hydrophobic width (Å) | Peptide hydrophobic length (Å) | Hydrophobic mismatch (Å) | Time (ns) |
|---|---|---|---|---|---|---|
| L1215−07 | DLPC | KALP15 | 20.9 | 13.8 | −7.1 | 50 |
| L1219−01 | DLPC | KALP19 | 20.5 | 19.4 | −1.1 | 50 |
| L1223+06 | DLPC | KALP23 | 20.3 | 25.8 | 5.5 | 50 |
| L1227+10 | DLPC | KALP27 | 21.0 | 31.0 | 10.0 | 80 |
| L1231+16 | DLPC | KALP31 | 21.0 | 37.0 | 16.0 | 100 |
| L1415−11 | DMPC | KALP15 | 25.0 | 13.7 | −11.3 | 50 |
| L1419−05 | DMPC | KALP19 | 24.3 | 19.7 | −4.6 | 50 |
| L1423+01 | DMPC | KALP23 | 24.3 | 25.7 | 1.4 | 50 |
| L1427+07 | DMPC | KALP27 | 24.7 | 31.1 | 6.7 | 100 |
| L1431+13 | DMPC | KALP31 | 25.1 | 37.8 | 12.7 | 80 |
| L1615−16 | DPPC | KALP15 | 29.3 | 13.7 | −15.6 | 50 |
| L1619−09 | DPPC | KALP19 | 28.8 | 19.8 | −9 | 50 |
| L1623−03 | DPPC | KALP23 | 28.2 | 25.4 | −2.8 | 50 |
| L1627+05 | DPPC | KALP27 | 27 | 31.6 | 4.6 | 50 |
| L1631+10 | DPPC | KALP31 | 28.2 | 37.8 | 9.6 | 80 |
TABLE 4.
Simulations at high P/L ratio
| Label | Lipid | Peptide | Lipid hydrophobic width (Å) | Peptide hydrophobic length (Å) | Hydrophobic mismatch (Å) | Time (ns) |
|---|---|---|---|---|---|---|
| H1215−08 | DLPC | KALP15 | 21.5 | 13.5 | −8 | 50 |
| H1219−03 | DLPC | KALP19 | 22.3 | 19.7 | −2.6 | 50 |
| H1223+04 | DLPC | KALP23 | 22.3 | 25.8 | 3.5 | 50 |
| H1227+09 | DLPC | KALP27 | 22.2 | 31.1 | 8.9 | 70 |
| H1231+16 | DLPC | KALP31 | 20.7 | 36.9 | 16.2 | 80 |
| H1415−11 | DMPC | KALP15 | 24.3 | 13.7 | −10.6 | 50 |
| H1419−05 | DMPC | KALP19 | 25.0 | 19.7 | −5.3 | 50 |
| H1423+00 | DMPC | KALP23 | 25.7 | 25.8 | 0.1 | 50 |
| H1427+05 | DMPC | KALP27 | 26.0 | 31.2 | 5.2 | 50 |
| H1431+11 | DMPC | KALP31 | 25.7 | 37.0 | 11.3 | 80 |
| H1615−13 | DPPC | KALP15 | 26.6 | 13.7 | −12.9 | 200 |
| H1619−09 | DPPC | KALP19 | 28.3 | 19.7 | −8.6 | 200 |
| H1623−03 | DPPC | KALP23 | 28.4 | 25.3 | −3.1 | 200 |
| H1627+02 | DPPC | KALP27 | 30.1 | 31.7 | 1.6 | 200 |
| H1631+07 | DPPC | KALP31 | 30.8 | 37.5 | 6.7 | 100 |
RESULTS
Hydrophobic mismatch
Hydrophobic mismatch is defined as the difference between the hydrophobic length of the peptide and the hydrophobic width of the lipid bilayer. The hydrophobic width of the lipid bilayer is defined as the distance between the acyl-chain C-2 carbon atoms of the two opposing bilayer leaflets (3). For the KALP peptides considered in this study, the ideal hydrophobic length is simply the total number of central leucine and alanine residues multiplied by 1.5 Å (the rise per residue for an ideal helix). We measured the hydrophobic length of the peptides in all our simulations and the data is shown in Tables 3 and 4. The deviation of the measured hydrophobic length of the peptide from the ideal value is rather small (∼0.5 Å or less). The addition of the peptide to the lipids, irrespective of the mismatch condition, affects the global bilayer properties such as bilayer thickness, area per lipid, etc. (especially in a small bilayer patch with ∼100 lipids). The lipids closer to the peptide are likely to be more perturbed than those far away from the peptide and the bilayer thickness is also likely to be different for different peptide concentrations. Thus, we define the hydrophobic mismatch as the difference between the measured hydrophobic length of the peptide and the measured hydrophobic width of the average distant lipids in the bilayer. This definition ensures that large-scale bilayer adjustments, due to the introduction of the peptide, are taken into account. We use a distance-based criterion to define the unperturbed lipids. In the simulations at low concentrations of peptides, we divide the lipids into three categories: neighboring, intermediate and distant or unperturbed. A lipid that has any of its atoms <5 Å from any of the backbone atoms of the peptide is classified as “neighboring”. Any other lipid in which all atoms are <15 Å from any of the peptide backbone atoms is classified as “intermediate”, and all the other lipids are classified as “distant” or “unperturbed”. Note that even though we call the distant lipids unperturbed lipids, they are slightly perturbed when compared to the ideal peptide-free case. This is primarily due to the small system sizes used in this study. At high P/L ratio, due to the relative proximity of the peptides, we adopt a two-tiered classification: neighboring (<5 Å) and unperturbed (all other lipids). Using this classification, at low P/L ratios, we obtain ∼12–15 neighboring lipids, ∼55 distant lipids, and ∼55 intermediate lipids with an almost equal distribution in both leaflets. At high P/L ratios, we obtain ∼50 neighboring and ∼50 distant lipids. Using these definitions, the hydrophobic mismatches for all 30 peptide/lipid simulations were calculated and listed in Tables 3 and 4.
The neighboring lipids roughly represent the first lipid shell around each peptide. As discussed later, the neighboring lipids account almost exclusively for all the peptide-lipid hydrogen bonds throughout the course of the simulations. Thus, the terms “neighboring lipids” and “hydrogen-bonded lipids” (referring to the hydrogen bonds formed between the lipids and the peptides) shall be used interchangeably. For ease of discussion, we label each simulation as AXXYY ± ZZ, where A can be L or H, denoting a low (1:128) or high (1:25) P/L ratio. XX refers to the number of lipid tail carbon atoms and can take values of 12 (DLPC), 14 (DMPC), or 16 (DPPC). YY refers to the peptide length and can take values of 15, 19, 23, 27, or 31, corresponding to the total number of residues in the peptide, ± refers to a positive or negative mismatch, and ZZ denotes the extent of mismatch (rounded off to the nearest Å). Thus, L1231+16 refers to the DLPC/KALP31 system at 1:128 P/L ratio and has a positive mismatch of 16 Å. To simplify discussions even further, we define simulations with mismatches >+3 Å as positive mismatches, those with mismatches <−3 Å as negative mismatches, and those in between as near-matching conditions. Now that hydrophobic mismatch has been defined, we explore the ways that the bilayer and peptide alleviate the mismatch.
Altered peptide secondary structure
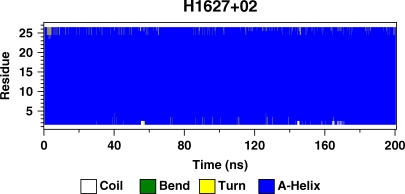
One possible mechanism to overcome the hydrophobic mismatch is for the peptide to change its secondary structure from an α-helix to a π-helix in the case of a positive mismatch or to a 310 helix in the case of a negative mismatch. Fig. 2 shows the secondary structure of a representative peptide as a function of time. Blue represents an α-helical structure. Except for some minor fraying at the terminal residues, the peptide show remarkable stability. We do not observe any kinks in the α-helix either. All the peptides from all the simulations show similar helical stability. Thus, from our simulations, it is safe to say that the peptide secondary structure is strictly preserved as an α-helix in essentially all cases of mismatch.
FIGURE 2.
Secondary structure profile of a peptide from the simulation H1627+02. Blue represents an α-helical structure.
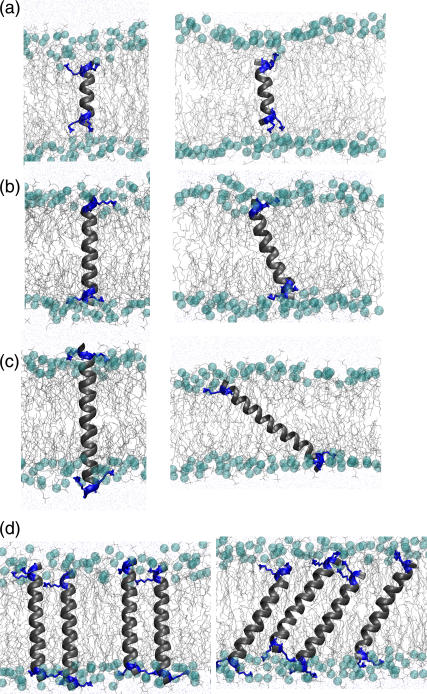
Peptide tilt
Fig. 3 shows the snapshots from the beginning and end of four simulations, corresponding to cases of (Fig. 3 a) extreme negative mismatch at low P/L ratio (L1616−16), (Fig. 3 b) near-matching at low P/L ratio (L1423+01), (Fig. 3 c) extreme positive mismatch at low P/L ratio (L1231+16), and (Fig. 3 d) near-matching at high P/L ratio (H1627+02). The four snapshots on the left side of the figure depict the starting conditions, after 5 ns of equilibration. The right side of the figure shows the configurations after 50 ns of production simulations for Fig. 3, a–c, and after 200 ns for Fig. 3 d. It is clear that the peptides adopt various tilt angles, depending on the mismatch. We define the tilt angle as the angle formed by the helical axis with the membrane normal. A zero degree tilt angle corresponds to a perpendicular orientation. Since the peptide helical structures were extremely well conserved (Fig. 2), the helical axis of the peptide was simply defined by two points, one near the top and the other near the bottom of the helix. The top point was taken to be the center of mass of the backbone atoms of the top lysine residues and the flanking glycine residue, while the bottom point was taken to be the center of mass of the backbone atoms of the bottom lysine residues and the flanking alanine residue. This gives a fairly robust measure of the tilt angle of the helix relative to the bilayer normal.
FIGURE 3.
Snapshots from the start (left) and end (right) of four simulations under different conditions of hydrophobic mismatch. (a) Extreme negative: L1615−16. (b) Near matching at low P/L: L1423+01. (c) Extreme positive: L1231+16. (d) Near matching at high P/L: H1627+02.
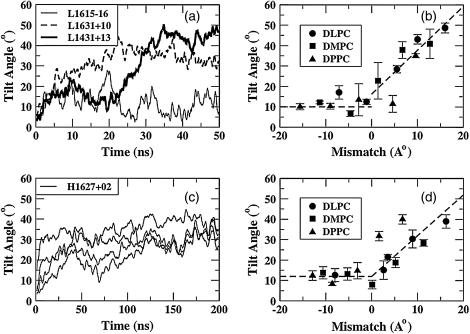
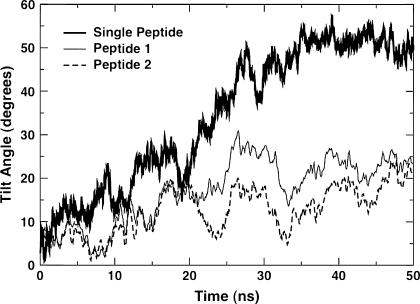
In Fig. 4 a, we show the tilt angle as a function of time for three representative cases (L1615−16, L1631+10, and L1431+13). For clarity, the lines have been smoothed by taking running averages over 200 ps. In all cases, the peptides start with an orientation close to vertical, corresponding to a tilt angle of 0°. After an initial equilibration period, it is clear that the tilt angle increases as a function of mismatch. Visual inspection shows that most simulations show well-equilibrated behavior over the latter half of the trajectories. Thus, time averages of the tilt angle were taken over the latter half of the trajectories. In Fig. 4 b, we plot these time-averaged tilt angles for all 15 simulations performed at a P/L ratio of 1:128. The standard errors were calculated using a block-averaging procedure (54). Most of the simulations with negative mismatch exhibit a small standard error, whereas those with near matching or positive mismatch show relatively larger error due to inadequate equilibration over the run time. Nevertheless, we see that for all negative mismatch conditions, the peptides exhibit a small but consistent tilt angle of ∼10°, whereas for positive mismatch, the peptides seem to exhibit a nearly monotonous increase in tilt angle as the mismatch increases. The dashed lines in Fig. 4 b are shown as a guide to the eye and do not represent a fit.
FIGURE 4.
(a) Peptide tilt angles as functions of time at low P/L ratio for three different peptide-lipid systems. (b) Average tilt angle as a function of mismatch at low P/L for these systems. (c) Peptide tilt angles for all four peptides in the bilayer as a function of time at high P/L ratio for H1627+02, with a slight positive mismatch. (d) Average tilt angle as a function of mismatch at high P/L ratio for these systems.
In Fig. 4 c, we show the tilt angles as a function of time for a representative simulation at high P/L ratio (1:25). Note that in all the simulations at high P/L ratio, there are four peptides and 100 lipids in the system. The tilt angles of all four peptides as a function of time show well-equilibrated tilt behavior after ∼50 ns. In Fig. 4 d, we plot the tilt angles, averaged over all four peptides in the bilayer, for all the simulations at the higher P/L ratio. The time-averaged standard errors in tilt angle for each peptide, calculated over the latter half of all the simulations, were extremely small for all the cases of negative mismatch and near-matching conditions. This error was calculated by the same block-averaging procedure employed for the simulations with low P/L ratio. Again, as in the case of low P/L ratios, for negative mismatch the peptides show a small but consistent tilt angle of ∼10°. For positive mismatch, the peptides show a tilt that increases with increasing mismatch, but data is more scattered than in the case of low P/L ratios, presumably due to the longer equilibration times required for the high P/L simulations. Again, the dashed lines are drawn as a guide to the eye and do not represent a fit. Thus, it is clear from Fig. 4, a–d, that for negative mismatch, the peptides exhibit a small but consistent tilt angle, and for positive mismatch, the tilt angle increases with the extent of mismatch. Experimental studies (55) show that under near-matching conditions, the tilt angles are rather small, as in our simulations. However, under extremely negative mismatch conditions, experiments show that nonlamellar phases are formed. Moreover, none of the experimental results on KALP or WALP peptides show the large tilt angles under extreme positive mismatch that are seen in our simulations. We shall address this point in detail later.
Local lipid adjustment
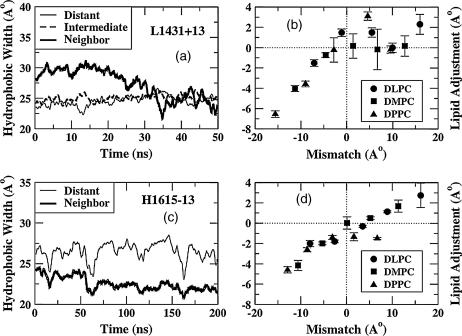
Local lipid rearrangement is another mechanism by which the system can alleviate the imposed hydrophobic mismatch. We define local lipid adjustment as the difference between the measured hydrophobic width of the perturbed lipids (lipids that neighbor the peptides) and the measured hydrophobic width of the unperturbed lipids (lipids that are distant from the peptides). The hydrophobic width is simply the distance between the average positions of the first hydrophobic carbon atoms of the lipids (adjacent to the carbonyl group), in the two leaflets of the bilayer. In Fig. 5 a, we show the hydrophobic widths of neighboring, intermediate, and distant lipids for a case with positive mismatch (L1431+13). The abscissa shows the duration of the production run. It should be remembered that all production runs were preceded by an equilibration simulation lasting 5 ns, during which the peptide backbones were constrained and the rest of the system was allowed to fluctuate. During this equilibration run, the neighboring lipids undergo a drastic rearrangement. This can be seen in Fig. 5 a, where the hydrophobic width of the neighboring lipids is larger by ∼5 Å than that of the intermediate or distant lipids before the production run begins. As the production run progresses, and the peptide is allowed to sample all of its degrees of freedom, we observe that the hydrophobic width of the neighboring lipids decreases, presumably due to the tilting of the peptide, and becoming almost as small as that of the intermediate and distant lipids after ∼30 ns.
FIGURE 5.
(a) Hydrophobic widths versus time at low P/L ratio and positive mismatch. (b) Local lipid adjustment as a function of mismatch at low P/L ratio. (c) Hydrophobic widths versus time at high P/L ratio and negative mismatch. (d) Local lipid adjustment as a function of mismatch at high P/L ratio.
In Fig. 5 c, the hydrophobic widths of neighboring and distant lipids for a case of extreme negative mismatch at high P/L ratios are shown (H1615–13). Again, during the 5-ns equilibration period (not shown here), the neighboring lipids undergo drastic rearrangements. Thus, at the beginning of the production run, the hydrophobic width of the neighboring lipids is lower than those of the intermediate or distant lipids. However, unlike the case of the positive mismatch, the difference in hydrophobic widths between the neighboring and distant lipids remains nearly constant for the entire duration of the simulation. Thus, the lipid adjustment over the latter half of the simulations, when the system seems to have reached an equilibrium with respect to peptide tilting and bilayer bending (but not necessarily with respect to peptide aggregation or peptide expulsion), is ∼0 Å for L1431+13 (Fig. 5 a) and −6 Å for H1615−16 (Fig. 5 c). The lipid adjustment was calculated for all simulations at low P/L ratio, averaged over the latter half of the trajectory, and is plotted in Fig. 5 b. For negative mismatch, the lipid adjustment is negative and increasingly so as the mismatch becomes more negative. This implies that the first hydrophobic carbon atoms of the neighboring lipids are more deeply inserted in the bilayer than the average distant lipids under negative mismatch conditions. Under positive mismatch conditions, the trend is not as clear, as there is significant scatter of data points. Nevertheless, even under the most extreme positive mismatch condition, the lipid adjustment is only ∼2 Å. In Fig. 5 d, we plot the lipid adjustment as a function of mismatch for the 15 simulations at high P/L. Note that by definition, there are no intermediate lipids at high P/L. Each data point represents the average lipid adjustment over the latter half of the simulation. Again, we see a clear trend at negative mismatch conditions, with a monotonically greater negative lipid adjustment with a greater degree of negative mismatch. The magnitudes of lipid adjustment roughly match those for low P/L ratio under identical negative mismatch conditions. Under positive mismatch conditions, again, the trend is not as clear. Thus, at both low and high P/L ratios, under negative mismatch conditions, there is a negative lipid adjustment that is roughly proportional to the mismatch, whereas for positive mismatch, there is a slight positive lipid adjustment of <2 Å.
Lipid tail order
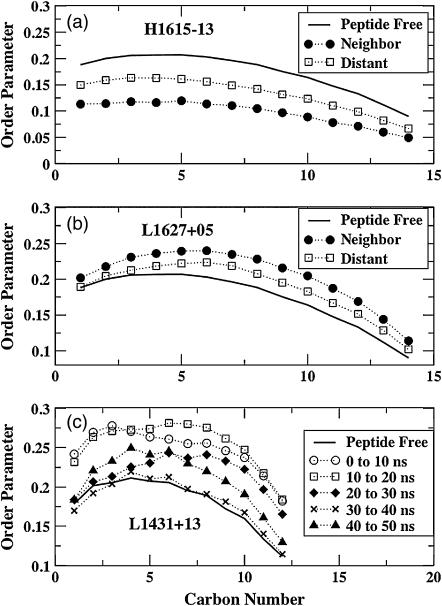
As the lipids undergo adjustments to alleviate the mismatch, the lipid tails become either more ordered or more disordered, depending on the sign of the hydrophobic mismatch. When the neighboring lipids undergo a positive lipid adjustment, we can expect the lipid tails to be more ordered and vice versa. Also, the effects on the intermediate and distant lipids should be less pronounced than on the neighboring lipids. In Fig. 6, we show the deuterium order parameters from some of the simulations. The deuterium order parameter SCD is defined as <3/2(cos2θ−1)>, where θ is the angle formed by the carbon-deuterium (C-D) vector with the bilayer normal. A smaller value for SCD indicates more disorder and a larger value more order. The carbon numbers increase from the headgroup toward the tail region. Fig. 6 a shows the deuterium order parameters for large negative mismatch (H1615−13), averaged over the latter half of the simulation. Clearly, the neighboring lipids (solid circles) are more disordered than the distant lipids. This is true for almost all simulations with negative mismatch, which is expected because of the negative lipid adjustment. (The order parameters for the peptide-free simulation (solid line) are also shown for reference.) In Fig. 6 b, the order parameters from a simulation with positive mismatch, again averaged over the latter half of the simulation, show the opposite trend; i.e., the neighboring lipids have larger order parameter than the distant lipids. However, the trend is not so clear in the case of all the positive mismatch conditions (not shown), which may not fully equilibrate over the simulated timescales (∼50–100 ns). The reason the order parameters of the lipid tails might equilibrate slowly, even though the carbon-carbon bond angles can undergo fast adjustments, is that the order parameter responds sensitively to other, more slowly changing characteristics of the lipid environment.
FIGURE 6.
Order parameters for the lipid tails for (a) negative mismatch and (b) positive mismatch. (c) Order parameter profiles at different times for simulation L1431+13.
To illustrate this, in Fig. 6 c, we show the deuterium order parameters for the neighboring lipids of simulation L1431+13, which has a positive mismatch, calculated over 10-ns windows. From Fig. 4 a, we can see that while the tilt angle for this peptide is still evolving between 25 and 40 ns, it appears to be reasonably equilibrated over the last 10 ns of the simulations. From Fig. 5 a, the lipid adjustment for this peptide is still evolving up to ∼35 ns but shows reasonably equilibrated behavior over the last ∼15 ns of the simulation. In Fig. 6 c, we see that the tails are highly ordered over the first 10 ns (circles), consistent with the initially low tilt angle (Fig. 4 a) and highly positive lipid adjustment (Fig. 5 a). Between 10 and 20 ns, the peptide tilt angle anomalously decreases (rather than increasing, as expected, to alleviate the mismatch) and the lipid adjustment still remains highly positive. This is reflected in the tail order (squares), where some of the carbons show increased order compared to that in the 0–10 ns window. Between 20 and 30 ns, the peptide tilt increases drastically and the lipid adjustment reaches a value of ∼0 (i.e., the hydrophobic width of the neighboring lipids decreases to that of the distant ones; see Fig. 5 a). The order parameters decrease correspondingly (diamonds on Fig. 6 c), especially for the carbons that are closer to the headgroup. Between 30 and 40 ns, the tilt angle remains approximately constant, whereas the lipid adjustment undergoes some changes, even showing some slightly negative values (i.e., the bold line in Fig. 5 a dips below the other lines). This is again reflected in the extreme disorder (crosses) in Fig. 6 c. Over the last 10 ns, when the tilt seems to be equilibrated and the lipid adjustment is ∼0, the order parameters (triangles) indicate a greater order. Thus, the deuterium order parameters seem to respond rather quickly to the slower evolution of the peptide tilt and the lipid layer thickness surrounding the peptide, and this was true for all the simulations (data not shown).
Partitioning of the lysine residues
Positive hydrophobic mismatch arises because the hydrophobic residues of the peptide are exposed to the more polar headgroup region of the lipids and even the bulk water phase in the case of extreme positive mismatches. Thus, the system adjusts by partitioning the hydrophobic residues of the peptide into the energetically favorable lipid tail region. However, in the case of negative mismatch, all the hydrophobic residues of the peptide are already favorably placed in the hydrophobic core of the bilayer. The mismatch arises, in this case, from the unfavorable partitioning of the lysine residues into the lipid tail region. The hydrophobicity scale of Wimley and White (56,57) estimates that the free energy penalty associated with the partitioning of a lysine residue in the hydrophobic core is ∼2.8 kcal/mol. Thus, the system undergoes the necessary adjustments to expose the lysine residues, especially the side chain, to a more polar environment. This can be achieved by either expelling the peptide into the water phase, or by a combination of local bilayer bending (negative lipid adjustment) and snorkeling the lysine residues up into the polar phase (32).
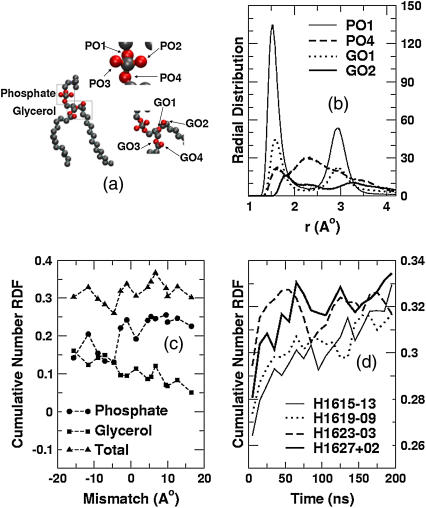
Experiments have shown that tryptophan residues have a highly energetically favorable interaction site near the carbonyl groups of the lipid tails (4,32). The length and flexibility of the lysine side chain should provide a large conformational flexibility for the ammonium group, enabling this residue also to seek the most energetically favorable position in the lipid/water interface. We have analyzed the radial distribution function of the hydrogen atoms of the lysine side-chain ammonium groups (referred to henceforth as “lysine hydrogens”) with the lipid oxygen atoms. Hydrogen-bonding analysis shows that under all conditions of mismatch, strong hydrogen bonding occurs between the lysine headgroups and the oxygen atoms of the lipids. Here we define hydrogen bonding to occur if the donor-acceptor distance is <0.25 nm and the donor-hydrogen-acceptor angle is >145°, which is a conventional definition used in simulation studies (58). Fig. 7 a shows a snapshot of a single lipid. The lipid oxygen atoms are shown in red. There are four phosphate oxygen atoms in the phosphate group of the lipid, and four glycerol oxygens. The phosphate oxygens are labeled as PO1, PO2, PO3, and PO4, while the glycerol oxygens are labeled as GO1 and GO2 (sn-1 chain) and GO3 and GO4 (sn-2 chain).
FIGURE 7.
(a) Schematic representation of the lipid molecule, highlighting the oxygens that participate in hydrogen bonding. (b) Representative radial distribution functions of oxygens around the lysine hydrogens for simulation H1627+02. (c) Cumulative oxygen number distributions as a function of mismatch at low P/L ratios. (d) Total number of oxygens in the first coordination shell (0–2.3 Å) of the lysine hydrogens as functions of time for the long simulations. (The thin solid lines represent H1615−13, the dotted lines H1619−09, the dashed lines H1623−03, and the thick solid lines H1627+02.)
In Fig. 7 b, we show the radial distribution functions for some of the lipid oxygen atoms with respect to the lysine hydrogens from a representative near-matching simulation (H1627+02). Only the four most dominant distributions are shown. From the dominant first peak at ∼1.5 Å, it is clear that the strongest interaction with the lysine hydrogen is that formed by the PO1 oxygen of the lipid (refer to Fig. 7). The three other major interactions (PO4, GO1, and GO2 with lysine hydrogens) are also shown. The interactions of PO3, PO4, GO2, and GO3 with the lysine hydrogens are not significant and are not shown. Each radial distribution function was calculated over the last 50 ns of the 200-ns simulation. In Fig. 7 b, in addition to the first peak at ∼1.5 Å, there is also a first minimum at ∼2.3 Å, which represents the outer edge of the first coordination shell of the lysine residue. All simulations show qualitatively similar radial distribution functions for PO1, with a strong first peak at ∼1.5 Å and a first minimum at ∼2.3 Å. To understand the interactions of the lysine residues with the lipid headgroup regions under different conditions of mismatch, we analyzed the cumulative radial distribution functions of the lysine-lipid interactions, which are volume-weighted integrals from zero out to a distance r. The region of interest encompasses the first coordination shell of the lysine hydrogen atoms, which extends out to the first minimum at ∼2.3 Å. In Fig. 7 c, we plot the cumulative number distributions from all the simulations at low P/L ratios. The cumulative distribution functions are averaged over the latter half of the simulations. For each mismatch, we show the cumulative distribution averaged over all interactions between the lysine hydrogens and all the lipid oxygens (triangles), between the lysine hydrogens and all the phosphate oxygens (circles), and between the lysine hydrogens and all the glycerol oxygens (squares). Each point represents the number of oxygens within the first coordination shell of the lysine hydrogens (∼2.3 Å).
For negative mismatch, the backbone atoms of all four lysine residues in the peptide are in the hydrophobic tail region. Thus, the glycerol oxygens, which are closer to the acyl carbons, are more easily accessible to the lysine side chains than are the phosphate oxygens, which are ∼4 Å farther removed. In Fig. 7 c, we see that at large negative mismatches, the lysine hydrogens interact to an equal extent with the glycerol and phosphate oxygens, indicating a preference for interaction with the phosphate oxygens, since these are harder to reach. Under near-matching conditions when both the glycerol and phosphate oxygens are equally accessible, the lysine hydrogens prefer to interact with phosphate oxygens, as indicated by the greater cumulative number of interactions with phosphates. Under most positive mismatch conditions, the lysine hydrogens of an untilted peptide will be closer to the phosphate oxygens than to the glycerol oxygens. Peptide tilting, however, enables the lysine hydrogens to sample the vicinity of both phosphate and glycerol oxygens. Still, we observe in Fig. 7 c that the lysines have a strong preference for the phosphate oxygen atoms under positive mismatch, indicating, again, that this is an energetically favored interaction. Although the relative interactions of the lysine hydrogens with the phosphate and glycerol oxygens varies as a function of mismatch, the total number of interactions remains approximately the same under all conditions of mismatch.
The interaction of the lysine side chains with lipids is a more localized phenomenon than is overall peptide tilt, and is expected to equilibrate over a different characteristic relaxation timescale. In Fig. 7 d, we plot the number of lipid oxygens within the first coordination shell of the lysine oxygens (which is the cumulative number up to the coordination shell minimum) as a function of time, for all four 200-ns simulations. We average over 10-ns windows. All four systems tend toward an apparent steady-state, equilibrium, number of oxygens within the first coordination shell, which is ∼0.33, at times between 150 and 200 ns. The simulations with near-matching conditions reach the presumed equilibrium value more quickly than the simulations with negative mismatch. Hence, the system perturbations associated with lysine-oxygen interactions have rather long relaxation times (∼100 ns or more), especially for the negative mismatch conditions. We also find that at longer timescales, the number of phosphate oxygens in the first coordination shell increases and the number of glycerol oxygens in the first coordination shell decreases for all cases of mismatch (data not shown).
Lipid reorientation dynamics
The addition of the peptide to a lipid bilayer influences the bulk properties of the system, such as the overall hydrophobic width and bilayer thickness, both of which increase under all cases of mismatch. To understand the influence of the peptide on the dynamic properties of the surrounding lipids, we calculated the rotational correlation functions of the P-N and C-C vectors of the lipids. The P-N vector is the vector connecting the phosphorus and nitrogen atoms of the headgroup. This tracks the relaxation characteristics of the lipid headgroups. The C-C vector is the vector connecting the first hydrophobic carbon atoms (closest to the headgroups) of the two tails of the lipid molecule. The time dependence of this vector broadly reflects the reorientation time of the lipid about the bilayer normal. The rotational autocorrelation functions were calculated using a first-order Legendre polynomial of the cosine of the angles of the appropriate vectors. To ensure adequate and equal sampling over the entire trajectory, the correlation functions were calculated only for time differences up to half the total trajectory length.
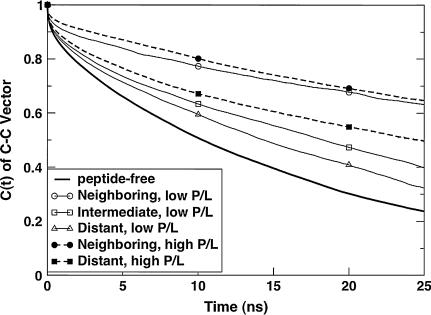
In Fig. 8, we show the C-C rotational correlation functions for two systems with identical lipids and peptides, but different P/L ratios. The results shown are for L1631+10 and H1631+07. The thick solid line shows the correlation function for the peptide-free case (K1600). The correlation functions for the neighboring lipids at low and high P/L ratio are nearly indistinguishable, although the system with higher P/L ratio seems to show slightly slower dynamics, as expected. The distant lipids at high P/L ratio shows clearly slower rotational relaxation than both the intermediate and distant lipids at low P/L ratio (L1631+10). Similar behavior was observed for almost all the peptide/lipid combinations between high and low P/L ratio. We also find that the P-N and C-C vectors show a peptide-length dependent relaxation behavior (data not shown), with faster relaxation for neighboring lipids of negative mismatch when compared to those of positive mismatch. We also find that the C-C vector relaxation timescale is larger than that of the P-N vector. Thus, even though the peptide tilt and local lipid adjustment seem to follow similar trends for the low and high P/L simulations (Figs. 4 and 5), the characteristic equilibration timescales are much larger for the high P/L cases due to the slower lipid reorientation dynamics.
FIGURE 8.
Comparison of the C-C rotational autocorrelation functions for the low and high P/L ratios of the same peptide-lipid pair.
DISCUSSION
Nonlocal lipid perturbations
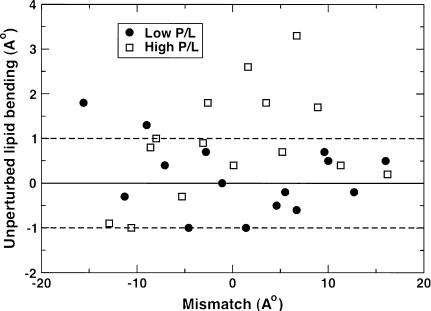
The addition of peptides to lipid bilayers perturbs the overall bilayer structure. In the immediate vicinity of the peptide, these perturbations depend on the nature of the mismatch, but globally they increase the bilayer width, and therefore the hydrophobic width of the lipids, irrespective of the mismatch. Theoretical calculations based on membrane elasticity theory (59) predict that a hydrophobic inclusion can thicken the bilayer for distances of up to 2 nm from the inclusion. We observe this increase in the bilayer thickness of the distant lipids in many of our simulations. However, in some of the simulations, we observe thinning of the distant lipid hydrophobic width relative to the peptide free bilayer. In Fig. 9, we show unperturbed lipid bending, which is the difference between the hydrophobic width of the distant lipids and the (ideal) hydrophobic width of the lipids of the peptide-free simulations, as a function of mismatch. Approximately nine of the 30 simulations show distant bilayer thinning. In most of the simulations, the bilayer thickening or thinning is roughly within 1 Å of the peptide-free lipid thickness. Some simulations, mostly at high P/L, show larger distant lipid thickness. This large variance is probably because of the limited simulation timescale (∼100 ns) and relatively small system size (∼64 lipids per leaflet). It should be mentioned here that although the distant lipids undergo changes in thickness of up to ∼3 Å in extreme cases on the addition of peptide, the overall bilayer thickness change is ∼1 Å or lower in most cases. This is consistent with the studies of Weiss et al. (30) who observe that there is little change in overall bilayer thickness for WALP peptides in different PC bilayers.
FIGURE 9.
Unperturbed lipid adjustment as a function of mismatch. Unperturbed lipid adjustment is defined as the difference between the hydrophobic width of the unperturbed lipids and the hydrophobic width of the lipids from the peptide-free simulation.
Our simulations over timescales of up to 200 ns yield results consistent with the established model of a rigid peptide in a fluid bilayer (57,60). Over this 200-ns timescale, the peptide/lipid system undergoes structural changes to minimize the mismatch conditions. For negative mismatches, the relaxation modes primarily involve the motion of local lipid molecules around the rather immobile peptide. For positive mismatches, the peptide undergoes large-scale tilting to minimize the mismatch. The lysine side chains of the peptide form strong interactions with the lipid headgroup region, anchoring the peptide to the neighboring lipids. Thus, the relaxation of the peptide tilt is strongly coupled to the diffusion of the lipid molecules with which it is associated. If the necessary tilt is to be achieved solely by the collective diffusive motion of the peptide and its neighboring lipids, it is likely to be an extremely slow process, in the order of microseconds for extremely large positive mismatch. However, in the simulations, an additional fast relaxation mode is available, namely a sliding motion of the two lipid leaflets with respect to each other. Even though the center of mass of the entire system is reset periodically to prevent drifting of the bilayer within the simulation cell, the two leaflets can develop a relative velocity. The relative motion of the leaflets is likely to be enhanced for relatively small simulation cells, such as those used in this study. Thus, we observe that even the extremely long KALP31 peptides tilt rapidly in the ∼50–100-ns simulations. Such rapid tilt relaxation would not be possible if the relaxation modes were purely based on molecular diffusion. This is also evident from the simulations at high P/L ratio, where the tilt relaxation is relatively slow compared to the simulations at low P/L. This is because of the much slower relative motion of the two lipid leaflets at high P/L ratios, due to the large number of lysine anchors.
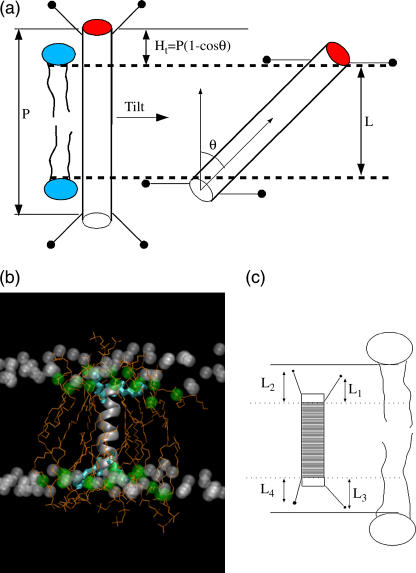
Earlier, we showed that the peptides have a small inherent tilt for negative mismatch, and large tilts, roughly proportional to the extent of hydrophobic mismatch, for positive mismatch. In Fig. 10 a, we show schematically the peptide tilt for positive mismatch. If P is the hydrophobic length of the peptide and L is the hydrophobic width of the lipid bilayer, then a peptide tilt of θ should reduce the hydrophobic mismatch by Ht = P(1−cosθ). Here Ht is the hydrophobic mismatch alleviated by tilt. We also observe a lipid adjustment (of Hl) for both positive and negative mismatch conditions.
FIGURE 10.
(a) Schematic representation of the peptide tilt for positive mismatch. (b) Snapshot of an extreme negative mismatch condition. The peptide is represented as a ribbon. The white spheres are the first hydrophobic carbons of the distant lipids and the green spheres are the first hydrophobic carbons of the neighboring lipids, which are shown as orange lines. The lysine residues are shown as cyan bonds. (c) Schematic definition of snorkeling length (L1 + L2 + L3 + L4)/2.
Due to the presence of the flanking lysine residues that can snorkel, the effective hydrophobic length of the peptides is greater for negative mismatch than for positive mismatch. Also, there is significant lipid adjustment, leading to a curvature around the peptide in the bilayer. In Fig. 10 b, we show a snapshot from L1615−16, a case of negative mismatch. The peptide is shown as a ribbon. The lysine residues are shown as cyan bonds. The orange lines represent the neighboring lipids and the green spheres the first hydrophobic carbons of the neighboring lipids. The white spheres represent the first hydrophobic carbons from all the intermediate and distant lipids. For sake of clarity, only the carbons from one of the two tails is shown for each lipid. The local lipid adjustment is clear from this figure, with the average neighboring lipid compressed in length relative to the average unperturbed lipid. We can also see that the lysine residues, shown as cyan bonds, snorkel toward the more polar interfacial region.
Thus, the negative mismatch seems to be alleviated by a combination of local bilayer bending and snorkeling of the lysine side chains. The four lysine residues in each peptide can have different extents of snorkeling depending on the local bilayer environment. To estimate the contribution of snorkeling to mismatch alleviation, we defined an average snorkeling length (Hs) as (L1 + L2 + L3 + L4)/2. Here L1, L2, L3, and L4 are the projections onto the bilayer normal, of the distances between the nitrogen atoms of the lysine side chains and the C-α atoms of the nearest inner lysine residues of the peptide; see Fig. 10 c. This definition provides a good estimate of snorkeling length under most cases of negative mismatch. For extreme negative mismatch, when the lysines are in their conformationally strained fully snorkeled position, the snorkeling length from both leaflets is ∼8.5 Å. For smaller negative mismatch, the snorkeling length is ∼4 Å. Note that this definition of snorkeling length is not very well defined under near-matching conditions since a relatively large snorkeling length will be estimated even when the peptide does not need to snorkel.
Total mismatch alleviation
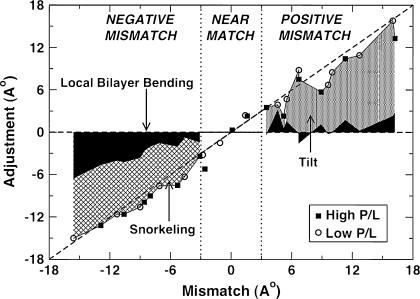
To quantify the contributions of Ht, Hl, and Hs toward mismatch alleviation, we plot, in Fig. 11, the total adjustment of the system as a function of mismatch. The circles represent the low P/L ratio and the squares the high P/L ratio. For positive mismatch (>3 Å), the contributions of peptide tilt, Ht (dotted area), and local lipid bending, Hl (black shaded area), are plotted and for negative mismatch (<−3 Å), the contributions of snorkeling, Hs (hatched area), and local lipid bending, Hl (black shaded area), are plotted. All results were averaged over the latter halves of the respective simulations. We see that the data fall reasonably close to the line representing perfect mismatch alleviation, with a slight systematic underestimation at high positive mismatch and an overestimation, especially for low negative mismatch. Although it is clear that the contribution due to tilt dominates under positive mismatch conditions, both the snorkeling and bilayer bending contributions are significant for all negative mismatch conditions.
FIGURE 11.
The system response to the imposed mismatch. Dark shaded area represents the contributions from the lipid adjustment, the dotted region represents the contributions from the tilt adjustment and the hatched area the contribution from snorkeling. Tilt and lipid bending contributions are shown for positive mismatch conditions, snorkeling and lipid bending contributions are shown for negative mismatch, and all three contributions are shown for near matching conditions.
At relative low positive mismatch, the mismatch alleviation is almost fully alleviated within the timescale of the simulations. However, for larger mismatches, the system adjustment is consistently slightly less than the imposed mismatch, possibly due to inadequate sampling time. For some of the positive mismatch simulations, we averaged the tilt angle and lipid adjustment over just the last 20 ns, when the peptide tilts were presumably closer to the final equilibrium value, and these values yield a better fit to the line for perfect adjustment (data not shown). For smaller negative mismatches, the snorkeling contribution is more important than bilayer bending, indicating that the first response of the lysine residue to a modest negative hydrophobic mismatch is to snorkel, rather than induce curvature in the lipid bilayer. As the negative mismatch increases, both the snorkeling length and the lipid adjustment increase. For extreme negative mismatch, the lysines become fully stretched outward toward the interface, the snorkeling length reaches a plateau, and further increase in negative mismatch leads to a more rapid increase in the lipid adjustment than to snorkeling. Nevertheless, under all conditions of negative mismatch, the system readjusts to alleviate the mismatch nearly completely. At near-matching conditions (between −3 Å and +3 Å), the total mismatch alleviation is plotted as the sum of the lipid adjustment, snorkeling, and the tilt adjustment. Although it must be conceded that some of the individual components, especially the snorkeling length, are ill-defined at near-matching conditions where their contributions to alleviating the mismatch are the most important, nevertheless, the total adjustment seems to match the imposed mismatch well, although this is possibly due in part to compensating errors from the various alleviation mechanisms.
It should be emphasized here that although some schematic representations and models treat the bilayer as a rigid slab, in reality the bilayer is an extremely fluid and fluctuating phase whose interfaces are not very sharp. Thus, relative large margins of error (of a few Ångstroms) should be allowed in the definitions of hydrophobic width, hydrophobic mismatch, and lipid bending contribution to alleviation. Also, we have neglected the effect of the lysine side chains, which can either increase or decrease the effective hydrophobic length of the peptide under positive mismatch conditions. Considering these limitations, Fig. 11 captures the response to positive mismatch reasonably well. Presumably, longer simulations and larger system sizes would yield a quantitatively more precise result. Nevertheless, It is gratifying that we can explain the system's response to hydrophobic mismatch through a combination of local bilayer bending and peptide tilting for positive mismatch and lysine snorkeling and local bilayer bending for negative mismatch.
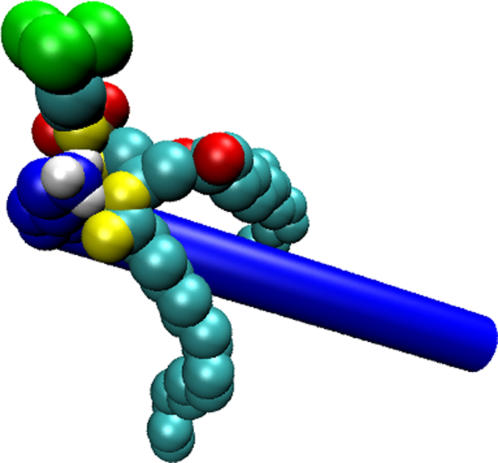
We had shown earlier that the lysine residues, under all conditions of mismatch, interact preferentially with the phosphate oxygens, rather than the glycerol oxygens. Analysis of the first coordination shell showed that just four oxygen atoms (out of eight) from the lipids accounted for most of the interactions with the lysine hydrogens. In Fig. 12, we show a snapshot of a lysine-lipid interaction. Only one lipid molecule is shown represented using van der Waals spheres. Two of the phosphate oxygens (PO1 and PO4; refer to Fig. 7 a) and two of the glycerol oxygens from the sn-1 chain (GO1 and GO2), shown in yellow, form a conformationally stable interaction site, into which the lysine side chain neatly fits. This region has four hydrogen-bond acceptor sites (the four aforementioned oxygen atoms) and thus provides a highly energetically favorable configuration for the lysine side-chain hydrogen atoms. Lysine residues under all conditions of mismatch seem to prefer this interaction site and the snapshot shown in Fig. 12 is probably the most energetically favorable configuration for the lysine residues in the lipid environment. Experimental studies of KALP and WALP peptides seem to indicate a position close to the carbonyl group (glycerol oxygens) for the tryptophan residues of WALP peptides and a position close to the phosphate group for the lysine residues of KALP peptides (55). Solid-state NMR studies of a synthetic K3 peptide, with many lysine residues, predict a similar interfacial position for the lysine residues, adjacent to the phosphate group (61). Thus, our simulations complement the experimental predictions by suggesting a preferred position.
FIGURE 12.
Snapshot of the lipid-lysine interaction. The lysine residue that interacts with this lipid is shown as a set of blue spheres, with the lysine hydrogens shown in white. A part of the peptide backbone is shown as a blue rod for reference. The lipid tail carbon atoms are depicted as cyan spheres. The choline headgroup is shown in green. The four oxygen atoms that are strongly interacting with the lysines are shown in yellow and the other four lipid oxygens are shown in red.
For near-matching conditions, as shown in Fig. 7 d, this energetically favorable state is reached rather quickly, due to the high conformational entropy of the lysine side chains and the surrounding lipids. Under negative mismatch conditions, when the lysine side chains are more constrained due to their snorkeling, the necessary equilibration timescale is longer. For extreme negative mismatch, when the lysine side chains are fully snorkeled and the neighboring lipids are highly bent, the equilibration times might be even longer. However, it is more likely that under extreme negative mismatch conditions, the lysines never get to fit into the interaction site, because the free energy change associated with the extreme lipid adjustment will, at some point, not be compensated by the favorable lysine-lipid interactions. At even larger mismatches, the free energy associated with local lipid bending is likely to be uncompensated by lysine snorkeling. Under these conditions, the peptide might be expelled from the bilayer.
Peptide expulsion
To test whether the peptides get expelled at larger mismatches, we ran a series of short simulations (∼1 ns) of two even shorter peptides, KALP13 and KALP11 in DPPC bilayers. We placed the peptide in the center of the bilayer, using the hole protocol described in Methods, and then ran an equilibration simulation of ∼500 ps to allow the lipids to equilibrate around the peptide, which was positionally restrained. We ran this equilibration with the lipids unrestrained in some cases and restrained in a plane perpendicular to the bilayer normal in some other cases. In the former cases, bilayer bending was allowed and in the latter cases, bilayer bending did not occur during equilibration. After 500 ps of equilibration, all the constraints were released and the system was simulated for 1 ns. We simulated five systems each of KALP11, KALP13, and KALP15 peptides in DPPC bilayers with restrained and unrestrained lipids during equilibration, making 30 short simulations in all. In all 10 systems of KALP11 (five with both lipids and peptides restrained during equilibration and five with only peptides restrained), the peptides moved out of the tail region, into the headgroup region of one of the leaflets within a few hundred picoseconds. This clearly shows that the lipid bending and snorkeling are not able to compensate for the extreme mismatch. For such a short peptide, the lysines will have to be in a fully snorkeled configuration and the neighboring lipids will have to bend by >10 Å to alleviate the mismatch. Evidently, the free energy penalty associated with such a conformational distortion is too large for the system to overcome. A surface-bound state, even though it places some of the hydrophobic residues in the interfacial region, is a more favorable one and thus the peptide is expelled from the tail region of the bilayer. Almost all the KALP13 peptides (nine of 10) were expelled from the bilayer also, except in one case where the lipids were unrestrained during equilibration. In that lone case, the peptide showed snorkeling behavior and local bilayer bending seemed to stabilize the peptide in the trans-membrane orientation. For the KALP15 in DPPC lipids, nine of the 10 simulations show that the peptide is stable in the bilayer but in one simulation, where the lipids were restrained during equilibration, the peptide moved out of the bilayer, toward the headgroup region of one of the bilayers in ∼600 ps. This simple test suggests that negative mismatches of >15 Å are probably unlikely to be compensated and will lead to peptide expulsion. Experiments have shown that for highly negative mismatches, nonlamellar phases are formed. E.g., KALP23 peptides form hexagonal phases in DEPE systems at high molar ratios of ∼1:10 (62). Such phase transitions cannot be observed in short molecular dynamics simulations of small systems sizes. However, the high curvature induced at negative mismatch can be considered the precursors to the phase transitions.
Elmore and Dougherty (10) have studied hydrophobic mismatching of the Tb-MscL protein in POPE and POPC lipids using molecular dynamics simulations. This protein has a trans-membrane portion that is a bundle of α-helices. The entire protein was embedded in a bilayer, and a mismatch imposed by shortening the lipid tail lengths. In ∼5 ns, the system partially compensated for the mismatch by local lipid adjustments and by introducing kinks in some of the helices. Clearly, in such short timescales, the system was not able to fully relax, owing to the long relaxation timescales associated with bundles of helices. In our simulations, since we deal with single peptides, we observe near total mismatch alleviation under almost all cases of mismatch. Petrache et al. (44) simulated WALP peptides under various conditions of negative mismatch and near match, and observed helix tilts <20°, for most of the simulations. A mismatch-dependent bilayer thickening was also observed for the boundary lipids. However, their simulations were rather short (∼1.5 ns) and thus the system could not fully relax. Nevertheless, reasonable matching with experimental results was found from their studies. The expected local bilayer thinning near negatively mismatched peptides is difficult to observe experimentally, but simulation studies have captured this effect (10,22). Our simulation results match those earlier results at least qualitatively. Local bilayer thinning has also been predicted in experimental studies of protegrin-1 in DLPC bilayers (63).
Solid-state NMR studies of model trans-membrane peptides (60,64) suggest additional relaxation modes for the peptides. The peptides, which may be tilted, or not, depending on the mismatch, are found to undergo fast rotational reorientation about the bilayer axis. Although fast by NMR timescales, with characteristic time constants of a few hundred nanoseconds to a few microseconds, these relaxations are still slow compared to the timescales that can be reached by current state-of-the-art molecular dynamics simulations. For near-matching conditions, when the relative tilt is small, such reorientations have smaller time constants. We tracked the reorientation dynamics of the peptides about the bilayer normal for select systems with near-matching or slight positive mismatches and do observe reorientation of the peptides over time periods of ∼1–10 ns. However, closer inspection reveals that these reorientations are just local readjustments, which can be classified as cage-rattling, with fixed lysine anchors. For longer peptides, with larger tilt angles, the rotational reorientation time will be related to the diffusion rates of the lipid molecules and thus will have much longer reorientation timescales. We observe that at higher concentration of peptides, the tilts of all four peptides in the simulation are highly correlated, i.e., not only do the peptides tilt to the same extent, but also in the same general direction, presumably due to the relative movement of the two lipid leaflets. Thus, relative deviation of the individual peptides from an average tilt angle is relatively small, with the peptides mostly parallel to each other with extremely small crossing angles. At longer timescales, one would expect a distribution of tilt angles, and larger peptide crossing angles. Because of the interactions of the lysines, which repel each other, larger crossing angles permit closer peptide approach than do nearly parallel orientations, especially at higher peptide concentrations, leading to possible oligomerization. Nevertheless, we do not observe such peptide tilt and aggregation in the timescale of the simulations.
Recently, hydrophobic mismatch has been studied by dissipative particle dynamics (45) and coarse-grained MD (46). These coarse-grained models for the peptides and the lipids lack the atomistic details present in our study. The peptides, in particular, are modeled as semirigid hydrophobic cylinders with polar ends, lacking atomistic details. Nevertheless, comparisons between those studies and our studies are worthwhile. The DPD simulations of Venturoli et al. (45) considered three different peptide models, a thin tube (corresponding to a single helix, relevant to our studies), an intermediate tube, and a thick tube (corresponding to a large membrane protein). They observed that the thin tubes tilt to a degree proportional to their mismatch. The thicker tubes of similar lengths undergo a much smaller tilt. They also found that the lipid tails can order or disorder, especially in the vicinity of the peptide, depending on the extent of mismatch. Their results are qualitatively similar to those found in our study. Nielsen et al. (46) considered two different protein models, a thinner tube or a thicker tube. The radius of the smaller tube was much larger than that of a single helix. Thus, direct comparison between their results and ours is not possible. Nevertheless, they observed larger tilt for the thinner tube and a correspondingly smaller tilt for the larger tube. They also observed mismatch-dependent lipid ordering, qualitatively similar to our results and those of Venturoli et al. (45).
NMR experiments performed on KALP23 peptides in DLPC and DMPC bilayers at a P/L ratio of 1:100 show tilt angle values of 11.2° and 7.6°, respectively (65). Our simulations predict corresponding values of 28.5° and 22.8° at low P/L ratio and 21.5° and 8° at high P/L ratio. Thus, for the DLPC system, our simulations predict much larger tilt values than are seen in experiments. Replica exchange molecular dynamics simulations of atomistic WALP peptides in implicit lipid bilayers modeled as simple low-dielectric continua (43) also predict much larger tilt angles at high mismatch than do experiments on those peptides (15). In fact, most experiments on KALP and WALP peptides show relatively small peptide tilt even for large positive mismatch (15,65). However, NMR experiments on other trans-membrane peptides, such as the pore region of the M2 channel (66) and the trans-membrane helix of Vpu (67), show that tilt angles do systematically increase as a function of mismatch. In the NMR study by Park and Opella (67), performed on a trans-membrane helix of Vpu in three different bilayers of varying hydrophobic width, the peptide tilt angle increased systematically, from 27° to 51°, with increasing hydrophobic mismatch. In fact, these tilt angles correspond to exact mismatch alleviation; i.e., the mismatch is entirely overcome by peptide tilt alone. These authors attribute the differences in tilt angles between their experiments and those performed with KALP and WALP peptides (where tilt angles required for total mismatch alleviation is not observed), to the fact that the peptide they studied was part of a naturally occurring protein, while the KALP and WALP peptides are synthetic, with high symmetry. Thus, while our simulation prediction that tilt alone can largely compensate for positive hydrophobic mismatch agrees very well with experimental results on segments of naturally occurring proteins, they do not agree well with corresponding experiments on KALP and WALP peptides.
Peptide aggregation
A possible reason for this discrepancy could be that the KALP peptides can aggregate as oligomers, at least transiently, under experimental conditions. This could lead to the smaller tilt angles seen in experiments. This point is also made by Im and Brooks (43), with reference to WALP peptides. Although the KALP peptides are less likely to aggregate when compared to the WALP peptides, due to the repulsion between the lysine residues, a relative tilt between the peptides could still lead to transient aggregation. In our simulations at high P/L ratios, we start with the peptides fairly uniformly distributed in the lipid bilayers, under the assumption that the peptides do not oligomerize. Even in the longest (200 ns) simulation, we do not observe peptide oligomerization, which presumably requires even longer timescales. To test whether oligomerization will lead to smaller tilt angles, we performed a simple test simulation by placing two KALP31 peptides adjacent to each other, in a vertical and parallel orientation in a DMPC bilayer, with a distance of 10 Å between the peptides. The evolution of tilt angles of the two peptides (thin solid lines and thick dashed lines) as a function of time is shown in Fig. 13. The tilt angle for the unoligomerized single KALP31 peptide in DMPC bilayer (thick solid line) is also shown for comparison. Clearly, the oligomerized peptides show much lower tilt values than the single peptide, at least in the timescales shown. The lipids immediately surrounding the peptide dimer are more ordered than those far away (data not shown). Thus, presumably, transient oligomerization could lead to smaller tilt values with additional compensation provided by the neighboring lipids increasing their tail order (positive lipid adjustment). Currently, we are performing a systematic study of different peptide oligomers (dimers, trimers, tetramers) at different interpeptide distances and different relative orientations (parallel, antiparallel) to fully understand the effect of peptide oligomerization on the mismatch alleviation mechanisms (unpublished data).
FIGURE 13.
Tilt angles as functions of time for a single KALP31 peptide in a DMPC bilayer (thick solid line, on top) and two KALP31 peptides in an aggregated state in a DMPC bilayer (thin solid line and dashed line at the bottom).
One of the most important results obtained from our study is that the lysine residues have a specific interaction site, formed by the phosphate headgroup and the glycerol oxygens of the sn-1 chain. In our simulations, depending on the mismatch, the lysines have different initial positions, but converge to an apparent preferred position in the bilayer, clearly showing this specific interaction. Most experimental results thus far suggest only that the lysine residues have a preferential position in the polar region close to the phosphate group. Thus, although our results provide a clearer picture of the interactions, they should still be viewed with caution. We use a well-established GROMOS-based force field that has been optimally parameterized for lipid systems and has been used by many to accurately reproduce numerous experimental observations. Nevertheless, validation of our results with other commonly used lipid force fields, like the CHARMM parameter set, will be useful for estimation of the accuracy and possible refinement of the current force-field parameters.
Finally, we revisit our definition of hydrophobic mismatch as the difference between the measured hydrophobic length of the peptide and the measured hydrophobic width of the average unperturbed lipids in the bilayer. This definition corrects for the global bilayer perturbations that are not related to the mismatch alleviation mechanisms and enables us to explain the mismatch response as a combination of tilt adjustment, local bilayer perturbations, and snorkeling of the lysine side chains. However, as seen from Fig. 9, even at ∼100 ns, the simulations are probably not well equilibrated, leading to some statistical uncertainties of ∼1 Å or more. Alternatively, we could have simply defined hydrophobic mismatch as the difference between the measured hydrophobic length of the peptide and the measured average hydrophobic width of all the lipids in the bilayer. Yet another possible definition is the difference between the measured hydrophobic length of the peptide and the (ideal) hydrophobic width of the peptide-free bilayer. We carried out calculations using these definitions, and found that they would have changed the imposed mismatch by only ∼1–2 Å, and therefore would not have changed the overall qualitative conclusions of our study.
CONCLUSIONS
We have performed a systematic study of trans-membrane, lysine-terminated, KALP peptides using molecular dynamics simulations. Three different lipids (DPPC, DMPC, DLPC) and five different peptides (KALP15, KALP19, KALP23, KALP27, KALP31) were simulated at two peptide/lipid ratios (1:128 and 1:25), providing 30 different simulations. The cumulative simulation time from this study was ∼2.7 μs, making this one of the largest studies of atomistic peptide/lipid simulations to date. The simulations covered a range of mismatch extending from extreme negative to extreme positive. Compensation for an imposed positive mismatch is by a combination of peptide tilting and local bilayer bending, with the former giving the larger contribution. For extreme positive mismatches, the tilt did not completely equilibrate in the timescale of some of the simulations. The compensatory mechanism was similar at both low and high peptide concentrations, with typically slower equilibration observed at high P/L ratio.
For negative mismatch, compensation is by a combination of local bilayer bending and lysine snorkeling, with the bending being stronger for negative mismatch than for positive mismatch. The systems under negative mismatch are well equilibrated within the timescale of the simulations. Although some of the extreme positive and negative mismatch conditions are not feasible experimentally, they nevertheless provide useful information about the transitional pathways to peptide expulsion or formation of nonlamellar phases. An important result from our simulations is the observation of a preferred binding site for the lysine side-chain residues. The lysine side chains preferentially interact with a binding site formed by two of the oxygen atoms from the phosphate group and two from the carbonyl oxygen of the sn-1 chain of the lipid tail. For negative mismatch, when the lysine residues snorkel and are conformationally restrained, the preference for the interaction site is not dominant, because it would involve excessive bilayer bending. However, under near-matching conditions, when the lysines have more conformational freedom, they clearly prefer to interact with this interaction site, anchoring the peptide in a trans-membrane orientation.
The peptides do not undergo significant structural changes, remaining largely α-helical, reinforcing the idea of a hard peptide in a soft lipid environment. The lipids, on the other hand, undergo drastic structural changes when peptides are added. The lipids that immediately border the peptides, forming the first shell, are the most perturbed by the peptide, showing a slower decay of the rotational correlation functions of the P-N (headgroup) and C-C (tails) vectors. There is both a distance- and concentration-dependence of the lipid dynamics. At larger P/L ratios, all the lipid molecules exhibit slower relaxation times, leading to slower overall lipid dynamics. Although relaxation modes such as peptide tilting and local lipid perturbation are accessible in the ∼50–200-ns timescales, slower motions such as peptide reorientation about the bilayer normal, possible oligomerization, and nonlamellar phase formation are clearly not possible over this timescale. Nevertheless, this comprehensive study provides a rather complete picture of the compensatory molecular events that occur in response to an imposed hydrophobic mismatch over timescales up to 200 ns. Thorough understanding of such model systems will eventually lead to insights into physiologically relevant phenomenon that occur in large membrane proteins that are directly related to hydrophobic mismatch.
TABLE 2.
Lipid properties
| Lipid | Number of carbons in tail | Ideal hydrophobic width (Å) (de Planque and Killian (3)) | Hydrophobic thickness (Å) (from simulations) |
|---|---|---|---|
| DLPC | 12 | 19.5 | 20.5 |
| DMPC | 14 | 23.0 | 25.3 |
| DPPC | 16 | 26.5 | 27.5 |
References
- 1.Jensen, M. O., and O. G. Mouritsen. 2004. Lipids do influence protein function—the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 1666:205–226. [DOI] [PubMed] [Google Scholar]
- 2.Mouritsen, O. G., and M. Bloom. 1993. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 22:145–171. [DOI] [PubMed] [Google Scholar]
- 3.de Planque, M. R. R., and J. A. Killian. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20:271–284. [DOI] [PubMed] [Google Scholar]
- 4.Killian, J. A. 1998. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1376:401–416. [DOI] [PubMed] [Google Scholar]
- 5.Martinac, B., J. Adler, and C. Kung. 1990. Mechanosensitive ion channels of Escherichia coli activated by amphipaths. Nature. 348:261–263. [DOI] [PubMed] [Google Scholar]
- 6.Perozo, E., A. Kloda, D. M. Cortes, and B. Martinac. 2002. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9:696–703. [DOI] [PubMed] [Google Scholar]
- 7.Perozo, E., D. M. Cortes, P. Sompornpisut, A. Kloda, and B. Martinac. 2002. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418:942–948. [DOI] [PubMed] [Google Scholar]
- 8.Hong, H. D., and L. K. Tamm. 2004. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. USA. 101:4065–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamill, O. P., and B. Martinac. 2001. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 81:685–740. [DOI] [PubMed] [Google Scholar]
- 10.Elmore, D. E., and D. A. Dougherty. 2003. Investigating lipid composition effects on the mechanosensitive channel of large conductance (MscL) using molecular dynamics simulations. Biophys. J. 85:1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sukharev, S., S. R. Durell, and H. R. Guy. 2001. Structural models of the MscL gating mechanism. Biophys. J. 81:917–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betanzos, M., C. S. Chiang, H. R. Guy, and S. Sukharev. 2002. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat. Struct. Biol. 9:704–710. [DOI] [PubMed] [Google Scholar]
- 13.Suchyna, T. M., S. E. Tape, R. E. Koeppe, O. S. Andersen, F. Sachs, and P. A. Gottlieb. 2004. Bilayer-dependent inhibition of mechanosensitive channels by neuroactive peptide enantiomers. Nature. 430:235–240. [DOI] [PubMed] [Google Scholar]
- 14.Morein, S., J. A. Killian, and M. M. Sperotto. 2002. Characterization of the thermotropic behavior and lateral organization of lipid-peptide mixtures by a combined experimental and theoretical approach: effects of hydrophobic mismatch and role of flanking residues. Biophys. J. 82:1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strandberg, E., S. Ozdirekcan, D. T. S. Rijkers, P. C. A. van der Wel, R. E. Koeppe, R. M. J. Liskamp, and J. A. Killian. 2004. Tilt angles of transmembrane model peptides in oriented and nonoriented lipid bilayers as determined by H-2 solid-state NMR. Biophys. J. 86:3709–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, F., R. N. A. H. Lewis, R. S. Hodges, and R. N. McElhaney. 2001. A differential scanning calorimetric and P-31 NMR spectroscopic study of the effect of transmembrane α-helical peptides on the lamellar-reversed hexagonal phase transition of phosphatidylethanolamine model membranes. Biochemistry. 40:760–768. [DOI] [PubMed] [Google Scholar]
- 17.Liu, F., R. N. A. H. Lewis, R. S. Hodges, and R. N. McElhaney. 2002. Effect of variations in the structure of a polyleucine-based α-helical transmembrane peptide on its interaction with phosphatidylcholine bilayers. Biochemistry. 41:9197–9207. [DOI] [PubMed] [Google Scholar]
- 18.Liu, F., R. N. A. H. Lewis, R. S. Hodges, and R. N. McElhaney. 2004. Effect of variations in the structure of a polyleucine-based α-helical transmembrane peptide on its interaction with phosphatidylglycerol bilayers. Biochemistry. 43:3679–3687. [DOI] [PubMed] [Google Scholar]
- 19.van Duyl, B. Y., D. T. S. Rijkers, B. de Kruijff, and J. A. Killian. 2002. Influence of hydrophobic mismatch and palmitoylation on the association of transmembrane α-helical peptides with detergent-resistant membranes. FEBS Lett. 523:79–84. [DOI] [PubMed] [Google Scholar]
- 20.Caputo, G. A., and E. London. 2003. Cumulative effects of amino acid substitutions and hydrophobic mismatch upon the transmembrane stability and conformation of hydrophobic α-helices. Biochemistry. 42:3275–3285. [DOI] [PubMed] [Google Scholar]
- 21.Subczynski, W. K., M. Pasenkiewicz-Gierula, R. N. McElhaney, J. S. Hyde, and A. Kusumi. 2003. Molecular dynamics of 1-palmitoyl-2-oleoylphosphatidylcholine membranes containing transmembrane α-helical peptides with alternating leucine and alanine residues. Biochemistry. 42:3939–3948. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, R. N. A. H., Y. P. Zhang, R. S. Hodges, W. K. Subczynski, A. Kusumi, C. R. Flach, R. Mendelsohn, and R. N. McElhaney. 2001. A polyalanine-based peptide cannot form a stable transmembrane α-helix in fully hydrated phospholipid bilayers. Biochemistry. 40:12103–12111. [DOI] [PubMed] [Google Scholar]
- 23.de Planque, M. R. R., B. B. Bonev, J. A. A. Demmers, D. V. Greathouse, R. E. Koeppe, F. Separovic, A. Watts, and J. A. Killian. 2003. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 42:5341–5348. [DOI] [PubMed] [Google Scholar]
- 24.Ridder, A., W. van de Hoef, J. Stam, A. Kuhn, B. de Kruijff, and J. A. Killian. 2002. Importance of hydrophobic matching for spontaneous insertion of a single-spanning membrane protein. Biochemistry. 41:4946–4952. [DOI] [PubMed] [Google Scholar]
- 25.Strandberg, E., S. Morein, D. T. S. Rijkers, R. M. J. Liskamp, P. C. A. van der Wel, and J. A. Killian. 2002. Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 41:7190–7198. [DOI] [PubMed] [Google Scholar]
- 26.Bystrom, T., G. Grobner, and G. Lindblom. 2003. Orientation of a polyleucine-based peptide in phosphatidy1choline bilayers of different thickness. Solid-state NMR and CD spectroscopy. Colloids Surf. A Physicochem. Eng. Asp. 228:37–42. [Google Scholar]
- 27.Lew, S., G. A. Caputo, and E. London. 2003. The effect of interactions involving ionizable residues flanking membrane-inserted hydrophobic helices upon helix-helix interactions. Biochemistry. 42:10833–10842. [DOI] [PubMed] [Google Scholar]
- 28.de Planque, M. R. R., E. Goormaghtigh, D. V. Greathouse, R. E. Koeppe, J. A. W. Kruijtzer, R. M. J. Liskamp, B. de Kruijff, and J. A. Killian. 2001. Sensitivity of single membrane-spanning α-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 40:5000–5010. [DOI] [PubMed] [Google Scholar]
- 29.van der Wel, P. C. A., E. Strandberg, J. A. Killian, and R. E. Koeppe. 2002. Geometry and intrinsic tilt of a tryptophan-anchored transmembrane α-helix determined by H-2 NMR. Biophys. J. 83:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss, T. M., P. C. A. van der Wel, J. A. Killian, R. E. Koeppe, and H. W. Huang. 2003. Hydrophobic mismatch between helices and lipid bilayers. Biophys. J. 84:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killian, J. A. 2003. Synthetic peptides as models for intrinsic membrane proteins. FEBS Lett. 555:134–138. [DOI] [PubMed] [Google Scholar]
- 32.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434. [DOI] [PubMed] [Google Scholar]
- 33.Karplus, M. 2003. Molecular dynamics of biological macromolecules: a brief history and perspective. Biopolymers. 68:350–358. [DOI] [PubMed] [Google Scholar]
- 34.Werten, P. J. L., H. W. Remigy, B. L. de Groot, D. Fotiadis, A. Philippsen, H. Stahlberg, H. Grubmuller, and A. Engel. 2002. Progress in the analysis of membrane protein structure and function. FEBS Lett. 529:65–72. [DOI] [PubMed] [Google Scholar]
- 35.Anezo, C., A. H. de Vries, H. D. Holtje, D. P. Tieleman, and S. J. Marrink. 2003. Methodological issues in lipid bilayer simulations. J. Phys. Chem. B. 107:9424–9433. [Google Scholar]
- 36.Tieleman, D. P., S. J. Marrink, and H. J. C. Berendsen. 1997. A computer perspective of membranes: molecular dynamics studies of lipid bilayer systems. Biochim. Biophys. Acta. 1331:235–270. [DOI] [PubMed] [Google Scholar]
- 37.Tieleman, D. P., H. J. C. Berendsen, and M. S. P. Sansom. 1999. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys. J. 76:1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tieleman, D. P., M. S. P. Sansom, and H. J. C. Berendsen. 1999. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys. J. 76:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tieleman, D. P., P. C. Biggin, G. R. Smith, and M. S. P. Sansom. 2001. Simulation approaches to ion channel structure-function relationships. Q. Rev. Biophys. 34:473–561. [DOI] [PubMed] [Google Scholar]
- 40.Tieleman, D. P., and M. S. P. Sansom. 2001. Molecular dynamics simulations of antimicrobial peptides: from membrane binding to trans-membrane channels. Int. J. Quant. Chem. 83:166–179. [Google Scholar]
- 41.Law, R. J., L. R. Forrest, K. M. Ranatunga, P. La Rocca, D. P. Tieleman, and M. S. P. Sansom. 2000. Structure and dynamics of the pore-lining helix of the nicotinic receptor: MD simulations in water, lipid bilayers, and transbilayer bundles. Protein Struct. Funct. Gen. 39:47–55. [DOI] [PubMed] [Google Scholar]
- 42.Lensink, M. F., B. Christiaens, J. Vandekerckhove, A. Prochiantz, and M. Rosseneu. 2005. Penetratin-membrane association: W48/R52/W56 shield the peptide from the aqueous phase. Biophys. J. 88:939–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im, W., and C. L. Brooks. 2005. Interfacial folding and membrane insertion of designed peptides studied by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 102:6771–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrache, H. I., D. M. Zuckerman, J. N. Sachs, J. A. Killian, R. E. Koeppe, and T. B. Woolf. 2002. Hydrophobic matching mechanism investigated by molecular dynamics simulations. Langmuir. 18:1340–1351. [Google Scholar]
- 45.Venturoli, M., B. Smit, and M. M. Sperotto. 2005. Simulation studies of protein-induced bilayer deformations, and lipid-induced protein tilting, on a mesoscopic model for lipid bilayers with embedded proteins. Biophys. J. 88:1778–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen, S. O., B. Ensing, V. Ortiz, P. B. Moore, and M. L. Klein. 2005. Lipid bilayer perturbations around a transmembrane nanotube: a coarse-grain molecular dynamics study. Biophys. J. 88:3822–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 48.Berger, O., O. Edholm, and F. Jahnig. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690.
- 50.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 51.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PDBViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 52.Faraldo-Gomez, J. D., G. R. Smith, and M. S. P. Sansom. 2002. Setting up and optimization of membrane protein simulations. Eur. Biophys. J. 31:217–227. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. Model. 14:33–41. [DOI] [PubMed] [Google Scholar]
- 54.Hess, B. 2002. Determining the shear viscosity of model liquids from molecular dynamics simulations. J. Chem. Phys. 116:209–217. [Google Scholar]
- 55.de Planque, M. R. R., J. A. W. Kruijtzer, R. M. J. Liskamp, D. Marsh, D. V. Greathouse, R. E. Koeppe, B. de Kruijff, and J. A. Killian. 1999. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane α-helical peptides. J. Biol. Chem. 274:20839–20846. [DOI] [PubMed] [Google Scholar]
- 56.White, S. H., and W. C. Wimley. 1998. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1376:339–352. [DOI] [PubMed] [Google Scholar]
- 57.White, S. H., and W. C. Wimley. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28:319–365. [DOI] [PubMed] [Google Scholar]
- 58.Mukhopadhyay, P., L. Monticelli, and D. P. Tieleman. 2004. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys. J. 86:1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.May, S. 2002. Membrane perturbations induced by integral proteins: role of conformational restrictions of the lipid chains. Langmuir. 18:6356–6364. [Google Scholar]
- 60.Sharpe, S., K. R. Barber, C. W. M. Grant, D. Goodyear, and M. R. Morrow. 2002. Organization of model helical peptides in lipid bilayers: insight into the behavior of single-span protein transmembrane domains. Biophys. J. 83:345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toke, O., R. D. O'Connor, T. K. Weldeghiorghis, W. L. Maloy, R. W. Glaser, A. S. Ulrich, and J. Schaefer. 2004. Structure of (KIAGKIA)(3) aggregates in phospholipid bilayers by solid-state NMR. Biophys. J. 87:675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strandberg, E., and J. A. Killian. 2003. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 544:69–73. [DOI] [PubMed] [Google Scholar]
- 63.Buffy, J. J., A. J. Waring, R. I. Lehrer, and M. Hong. 2003. Immobilization and aggregation of the antimicrobial peptide protegrin-1 in lipid bilayers investigated by solid-state NMR. Biochemistry. 42:13725–13734. [DOI] [PubMed] [Google Scholar]
- 64.Goodyear, D. J., S. Sharpe, C. W. M. Grant, and M. R. Morrow. 2005. Molecular dynamics simulation of transmembrane polypeptide orientational fluctuations. Biophys. J. 88:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozdirekcan, S., D. T. S. Rijkers, R. M. J. Liskamp, and J. A. Killian. 2005. Influence of flanking residues on tilt and rotation angles of transmembrane peptides in lipid bilayers. A solid-state H-2 NMR study. Biochemistry. 44:1004–1012. [DOI] [PubMed] [Google Scholar]
- 66.Duong-Ly, K. C., V. Nanda, W. F. Degrad, and K. P. Howard. 2005. The conformation of the pore region of the M2 proton channel depends on lipid bilayer environment. Protein Sci. 14:856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park, S. H., and S. J. Opella. 2005. Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 350:310–318. [DOI] [PubMed] [Google Scholar]