Abstract
Transmembrane profiles of molecular oxygen in lipid bilayers are not only significant for membrane physiology and pathology, but also are essential to the determination of membrane protein structure by site-directed spin labeling. Oxygen profiles obtained with spin-labeled lipid chains have a Boltzmann sigmoidal dependence on the depth into each lipid leaflet, which represents a two-compartment distribution between outer and inner regions of the membrane, with a transfer free energy that depends linearly on distance from the dividing planes. Transmembrane profiles for intramembrane polarity, and for water penetration into the membrane, have an identical form, but are of the reverse sign. Comparison with recently published oxygen profiles from a site-specifically spin-labeled α-helical transmembrane peptide validates the use of spin-labeled lipids for all these profiles and provides the necessary bridge to generate the full bilayer from a single lipid leaflet.
Recently, Nielsen et al. (1) presented data on the transmembrane profile of oxygen-induced enhancements in spin-lattice relaxation rate (1/T1e) from spin labels attached at site-specific residues in a rigid α-helical WALP peptide. Such relaxation enhancements are linearly related to the diffusion-solubility product, DT[O2], of oxygen in the membrane (2,3), and are used frequently in site-directed spin labeling to determine the topography and conformation of membrane-associated proteins (4,5). Comparing with earlier data (6,7), Nielsen et al. (1) called into question the usual strategy of using spin-labeled lipids to establish calibrations for the immersion depth of spin labels in membranes.
Previously, we showed that it is necessary to take into account the anisotropic rotational diffusion of the spin-labeled lipid chains when evaluating T1-relaxation enhancements from progressive saturation measurements using continuous wave (CW) electron paramagnetic resonance (EPR) (8). With this method, it was then possible to determine a high-resolution profile for penetration of oxygen into the membrane by using phospholipids (n-PCSL) that were systematically spin labeled throughout the sn-2 acyl chain from C-position n = 4 to n = 14 (9). Additionally, enhancements in Lorentzian line broadening by paramagnetic oxygen were determined using convolution methods, hence demonstrating the equality of T1- and T2-relaxation enhancements that is expected for a Heisenberg-exchange mechanism (3,10).
From the above study, we found that the diffusion-solubility profile for oxygen exhibited a sigmoidal Boltzmann form of the type established earlier for the transmembrane polarity profile (11). It had been proposed already that the Boltzmann sigmoid,
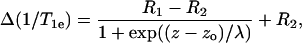 |
(1) |
was appropriate to describe the envelope of oxygen-induced T1-enhancements from the site-specifically spin-labeled KcsA potassium channel (11), based on extensive measurements by Perozo et al. (5). Equation 1 describes a two-phase distribution between membrane regions with respective transverse coordinates z > zo and z < zo, where the free energy of transfer for oxygen depends linearly on the distance from the dividing plane at z = zo. The oxygen-induced enhancements in the two regions are R1 and R2, respectively, and λ is the decay length that characterizes the width of the transition region.
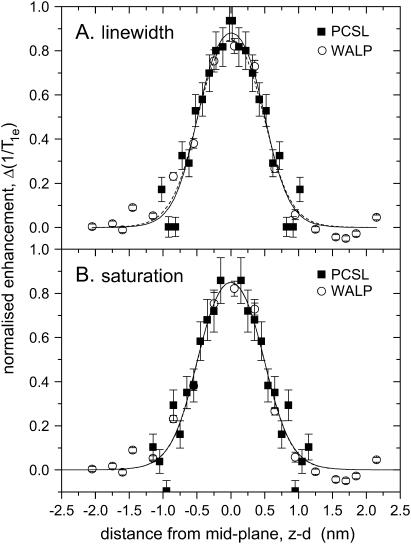
Our purpose in this letter is to show that the oxygen relaxation enhancement profile that we determined using spin-labeled lipids in fluid bilayer membranes (9) is fully compatible with that established recently by Nielsen et al. (1) using a WALP-based ruler. The measurements with the spin-labeled WALP peptide were performed in fluid membranes of dioleoyl phosphatidylcholine, whereas those with the spin-labeled phospholipids were in fluid dimyristoyl phosphatidylcholine bilayers. Nevertheless, the thicknesses of the two membranes are closely similar in the fluid phase (12). Fig. 1 compares the transmembrane profiles of oxygen-induced relaxation enhancement from the two series of site-specific spin labels. The spin-labeled peptide is indicated by open symbols and the spin-labeled lipid by solid symbols. A common abscissa is established by using the increments: Δz = 0.15 nm/residue for an α-helix (13), and Δz = 0.1 nm/CH2 for a fluid lipid chain (14). It is readily seen that, with suitable normalization of the relaxation enhancements, the two sets of data are essentially superimposable.
FIGURE 1 .
Transmembrane profile of oxygen-induced T1-relaxation enhancement of spin-labeled lipids, n-PCSL (▪), and spin-labeled peptide, WALP (○), in fluid phospholipid bilayer membranes; data from Dzikovski et al. (9) and Nielsen et al. (1), respectively. (A) Lipid relaxation enhancements deduced from convolution linewidths: ΔΔHL = Δ(1/T1e)/γe, compared with peptide relaxation enhancements deduced from saturation recovery EPR. (B) Lipid relaxation enhancements deduced from CW-EPR saturation parameters: 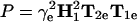 , compared with peptide relaxation enhancements deduced from saturation recovery EPR. Solid and dashed lines are nonlinear least-squares fits of Eq. 2 to the data for spin-labeled lipids and peptides, respectively. Relaxation enhancements are normalized such that R1 − R2 = 1 and R2 = 0, for the fits of Eq. 2; transmembrane distances are referred to the membrane midplane as origin.
, compared with peptide relaxation enhancements deduced from saturation recovery EPR. Solid and dashed lines are nonlinear least-squares fits of Eq. 2 to the data for spin-labeled lipids and peptides, respectively. Relaxation enhancements are normalized such that R1 − R2 = 1 and R2 = 0, for the fits of Eq. 2; transmembrane distances are referred to the membrane midplane as origin.
The sigmoidal Boltzmann profile given above by Eq. 1 describes only one lipid leaflet of the symmetrical bilayer membrane. That incorporating both leaflets back-to-back is given by
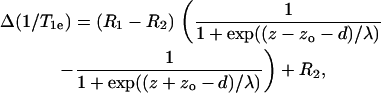 |
(2) |
where d is the thickness of one lipid leaflet of the membrane bilayer. Nonlinear least-squares fitting of Eq. 2 to the T1-enhancement data for the WALP23 peptides yields values of zo = 0.49(9) nm and λ = 0.20(5) nm, and the membrane midplane corresponds to residue position (d) 11.7(2). The corresponding fit is given by the dashed lines in Fig. 1. The goodness of fit (χ2 = 104 s−2) and the values of fitting parameters are consistent with those found by the original authors. For ease of comparison with the lipid data, the relaxation enhancements in Fig. 1 are normalized such that R1 − R2 = 1 and the baseline is defined by R2 = 0 for the fitted parameters.
Because the data for the spin-labeled phospholipids correspond to only one leaflet of the bilayer, it is necessary to specify the lipid chain length about which reflection symmetry will generate the profile for the complementary leaflet. This is done in Fig. 1 by aligning the lipid relaxation enhancements with those of the WALP peptides. A suitable criterion is to achieve similar values for the position, zo, of the dividing plane. For the linewidth enhancements (Fig. 1 A), the thickness of the leaflet (d) corresponds to chain length n = 14.2, and for the enhancements in saturation parameter (Fig. 1 B), d corresponds to n = 15.5. These values are largely consistent with the fact that the thicknesses found for the two bilayers by x-ray diffraction are rather similar (12), although uncertainties are involved in the extent of sn-1/sn-2 chain overlap and the contributions from the terminal methyl groups. Fitting Eq. 2 to the profiles for the lipid spin labels then yields values of zo = 0.50(6) nm and λ = 0.18(9) nm for the linewidth data, and zo = 0.50(12) nm and λ = 0.19(9) nm from the saturation parameters. Corresponding fits are given by the solid lines in Fig. 1, A and B, respectively. The fitting, particularly the comparable values of the decay length λ, confirms that both spin-labeled lipids and the transmembrane peptide reflect the same oxygen profile.
The relaxation enhancements in Fig. 1 are normalized and referred to a baseline defined by the limits of the fits with Eq. 2. The WALP peptide enhancements were obtained in 21% oxygen, whereas those of the spin-labeled lipids were in 100% oxygen. The unnormalized data for the lipids yields values of R1 − R2 = 10(3) × 106 s−1 and R2 = 9(1) × 106 s−1 from the linewidths, and R1 − R2 = 10(4) × 106 s−1 and R2 = 8.7(9) × 106 s−1 from the saturation parameters. For comparison, the values obtained from the WALP peptides are: R1 − R2 = 1.9(4) × 106 s−1 and R2 = 1.26(4) × 106 s−1, which are determined directly from saturation recovery EPR (1). The relaxation enhancement within the membrane, R1 − R2, displays the expected fivefold increase in oxygen relative to air, although the limiting baseline level, R2, is somewhat higher.
In conclusion, the transmembrane diffusion-solubility profiles of oxygen determined from spin-labeled lipids are in good, almost quantitative, agreement with those defined by the WALP peptide ruler. This is important, because spin-labeled lipids can be introduced more readily into membranes for in situ calibrations, than can an exogenous transmembrane peptide. Furthermore, spin-labeled lipids have been used extensively to determine high-resolution transmembrane polarity profiles (11,15), and the penetration profiles of water into membranes (16). These polarity-related profiles also have the form of Eq. 1 that is cross-validated here for oxygen, but are understandably of the reverse sign to that for hydrophobic molecules. Finally, the WALP peptides provide the necessary bridge to generate the complementary leaflet for all these extensive lipid-based studies.
References
- 1.Nielsen, R. D., K. Che, M. H. Gelb, and B. H. Robinson. 2005. A ruler for determining the position of proteins in membranes. J. Am. Chem. Soc. 127:6430–6442. [DOI] [PubMed] [Google Scholar]
- 2.Windrem, D. A., and W. Z. Plachy. 1980. The diffusion-solubility of oxygen in lipid bilayers. Biochim. Biophys. Acta. 600:655–665. [DOI] [PubMed] [Google Scholar]
- 3.Hyde, J. S., and W. K. Subczynski. 1989. Spin-label oximetry. In Spin Labeling. Theory and Applications. L. J. Berliner and J. Reuben, editors. Plenum Press, New York and London. 399–425.
- 4.Hubbell, W. L., and C. Altenbach. 1994. Site-directed spin-labeling of membrane proteins. In Membrane Protein Structure: Experimental Approaches. S. H. White, editor. Oxford University Press, New York. 224–48.
- 5.Perozo, E., D. Marien Cortes, and L. G. Cuello. 1998. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 5:459–469. [DOI] [PubMed] [Google Scholar]
- 6.Altenbach, C., T. Marti, H. G. Khorana, and W. L. Hubbell. 1990. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 248:1088–1092. [DOI] [PubMed] [Google Scholar]
- 7.Altenbach, C., D. A. Greenhalgh, H. G. Khorana, and W. L. Hubbell. 1994. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 91:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livshits, V. A., B. G. Dzikovski, and D. Marsh. 2003. Anisotropic motion effects in CW non-linear EPR spectra: relaxation enhancement of lipid spin labels. J. Magn. Reson. 162:429–442. [DOI] [PubMed] [Google Scholar]
- 9.Dzikovski, B. G., V. A. Livshits, and D. Marsh. 2003. Oxygen permeation profile in lipid membranes: nonlinear spin-label EPR. Biophys. J. 85:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livshits, V. A., B. G. Dzikovski, and D. Marsh. 2001. Mechanism of relaxation enhancement of spin labels in membranes by paramagnetic ion salts: dependence on 3d and 4f ions and on the anions. J. Magn. Reson. 148:221–237. [DOI] [PubMed] [Google Scholar]
- 11.Marsh, D. 2001. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 98:7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnott, S., and S. D. Dover. 1967. Refinement of bond angles of an α-helix. J. Mol. Biol. 30:209–212. [DOI] [PubMed] [Google Scholar]
- 14.Marsh, D. 1990. Handbook of Lipid Bilayers. CRC Press, Boca Raton, FL.
- 15.Kurad, D., G. Jeschke, and D. Marsh. 2003. Lipid membrane polarity profiles by high-field EPR. Biophys. J. 85:1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erilov, D. A., R. Bartucci, R. Guzzi, A. A. Shubin, A. G. Maryasov, D. Marsh, S. A. Dzuba, and L. Sportelli. 2005. Water concentration profiles in membranes measured by ESEEM of spin-labeled lipids. J. Phys. Chem. B. 109:12003–12013. [DOI] [PubMed] [Google Scholar]