Abstract
The laccase enzyme and melanin synthesis have been implicated as contributors to virulence in Cryptococcus neoformans. Since isolations of Cryptococcus species other than C. neoformans from clinical specimens have been increasing, we examined the laccase activities of C. albidus, C. laurentii, C. curvatus, and C. humicola. Incubation of cells with epinephrine produced adrenochrome color in C. albidus, C. laurentii, and C. curvatus but not in C. humicola. Activity was always less than in C. neoformans. Laccase was detected in the soluble fractions of disrupted C. albidus, C. laurentii, and C. curvatus cells. Activity staining of partially purified enzyme after nondenaturing polyacrylamide gel electrophoresis revealed that laccases from C. albidus, C. laurentii, and C. curvatus migrated more slowly than that from C. neoformans. One strain of C. curvatus exhibited two melanin bands. Thus, several clinically emerging Cryptococcus species express laccase and can synthesize melanin.
Cryptococcosis is a life-threatening infection that complicates a variety of immunocompromising conditions (4). Although 34 species of the genus Cryptococcus are recognized (6), C. neoformans is generally considered to be the pathogenic species, while C. albidus, C. laurentii, and C. curvatus have been regarded as saprophytic species. However, species other than C. neoformans have, in fact, been isolated from normally sterile clinical specimens. We note isolations of C. albidus in septicemia and meningitis (15, 22, 23), C. laurentii in fungemia (14, 19) and meningitis (15), C. curvatus in myeloradiculitis (5), and C. humicola in meningitis (28), all in patients with AIDS or cancer. Thus, cryptococcosis due to species other than C. neoformans may be considered an emerging infection.
The virulence of C. neoformans has been ascribed to capsule formation, melanization due to laccase (phenoloxidase), proteinase secretion, and phospholipases (3). Although many pathogenic fungi synthesize precursors of melanin, melanization in C. neoformans occurs when exogenous or environmental catechols or aminophenols are oxidatively polymerized by the laccase enzyme and deposited as melanin in the cell wall (1, 26). Melanization has often been used for the identification of isolates, since growth of brown colonies on birdseed agar made from Guizotia abyssinica, or thistle seeds rich in caffeic acid, is characteristic of C. neoformans (29). However, we noted that several strains of species other than C. neoformans produced light-brown colonies on this medium or on defined medium containing synthetic substrates. In this study, we examined laccase enzymes and melanization in cryptococcal species other than C. neoformans.
MATERIALS AND METHODS
Strains used.
The strains used in this study are listed in Table 1 as accessed in the culture collections Centraalbureau voor Schimmelcultures (CBS) and Japan Collection of Microorganisms (JCM). The strains were obtained from each institute directly and maintained on YM agar (Difco) slants. Some of the strains have recently been reclassified (30-34); therefore, current nomenclature is also given.
TABLE 1.
Strains used
| Previous species | Strain | Source | Current species | Reference |
|---|---|---|---|---|
| C. neoformans | B-3501 | |||
| C. albidus | CBS142 | Atmosphere | C. albidus | |
| CBS945 | Tubercular lung | C. albidus | ||
| CBS965 | Sputum | C. diffluens | 32 | |
| C. laurentii | CBS2174 | Tumor | C. laurentii | 30 |
| CBS8645 | Cerebrospinal fluid; AIDS with meningitis | C. nodaensis | 30 | |
| CBS8648 | Lung | C. laurentii | 30 | |
| CBS2409 | Surface of shrimp | Cryptococcus sp. strain | 30 | |
| C. curvatus | CBS570 | Sputum of tubercular patient | C. curvatus | 31 |
| JCM1532 (CBS570) | C. curvatus | 31 | ||
| JCM1605 | Frozen food | C. curvatus | 31 | |
| C. humicola | CBS571 | Soil | C. humicola | 31 |
| JCM1530 | Strawberry | C. fragicola sp. nov. | 34 | |
| CBS1896 | Bronchial secretion | Trichosporon debeurmannianum sp. nov. | 33 | |
| CBS5920 | Radioactive solution at pH 2 | C. longus sp. nov. | 34 | |
| CBS5123 | Skin | C. daszewskae sp. nov. | 34 |
Laccase assay using permeabilized cells.
The laccase activities of several Cryptococcus species were determined in suspensions of permeabilized cells by a modification of the method of Rhodes et al. (27). Each strain was inoculated into 5 ml of 2% glucose-2% yeast extract and incubated with agitation at 25°C for 24 h. The culture (0.1 ml) was inoculated into 50 ml of asparagine salts medium (per liter: glucose, 3 g; asparagine, 1 g; MgSO4 heptahydrate, 0.5 g; KH2PO4, 3 g; thiamine, 1 mg) and again incubated at 25°C for 24 h with agitation. The cells were harvested by centrifugation at 1,000 × g for 10 min and washed once with asparagine salts medium without glucose. The cells were resuspended in 50 ml of the medium without glucose and incubated at 25°C for 12 h with agitation to induce laccase. The cells were harvested by centrifugation and washed once with 50 mM sodium phosphate buffer, pH 7.0. One milliliter of cell suspension adjusted to 108 cells/ml and 0.1 ml of toluene-ethanol (1:4) were mixed for 90 s. The permeabilized cell suspension (0.1 ml) was mixed with 10 mM l-epinephrine (0.1 ml) and 50 mM sodium phosphate buffer (0.8 ml) and incubated at 30°C. After 0.5, 1, 2, 3, and 4 h, the reaction mixture was centrifuged at 1,000 × g for 5 min, and the optical absorbance of the supernatant at 480 nm was determined.
Preparation of laccase.
Each strain was inoculated into 10 ml of 2% glucose and 2% yeast extract and incubated with agitation at 25°C for 24 h. The culture (0.3 ml) was inoculated into 12 bottles of 145 ml of asparagine salts medium and incubated for 24 h with agitation. The cells were harvested by centrifugation at 15,000 × g for 20 min at 4°C and washed once with asparagine salts medium without glucose. The cells were resuspended in medium without glucose and incubated at 25°C for 12 h. After centrifugation, the cells were washed once with 1 mM sodium phosphate buffer, pH 7.0. The cells were disrupted mechanically as follows (8). The cells were placed in a 50-ml ice-jacketed bead beater (Biospec Products) with 0.5-mm-diameter glass beads added almost up to the top, and phosphate buffer was added to displace all the air. Intervals of agitation (1 min) were interspersed with 1-min pauses for cooling. Five 1-min treatments generally provided good breakage. The suspension of disrupted cells was ultracentrifuged at 100,000 × g for 60 min. This crude extract was partially purified in a Phenyl Sepharose CL-4B column (9) and lyophilized.
Activity staining was performed after nondenaturing gel electrophoresis to compare laccases from several Cryptococcus species as described previously. Each partially purified laccase was dissolved in a small amount of water and applied to a 10% polyacrylamide gel without sodium dodecyl sulfate. The gel was run at a constant current of 18 mA. After electrophoresis, the gel was immersed in freshly prepared 1 mM l-dihydroxyphenylalanine dissolved in citrate phosphate buffer (pH 6.0) and incubated overnight at room temperature with gentle agitation. Laccase was visualized by melanization at the position to which the enzyme migrated.
RESULTS
Comparative laccase activities in various Cryptococcus species.
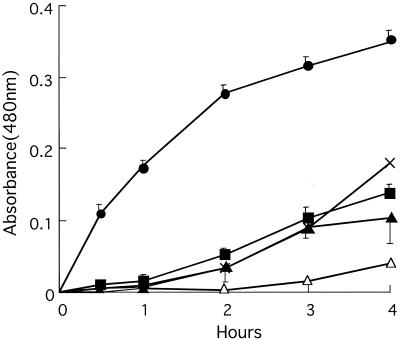
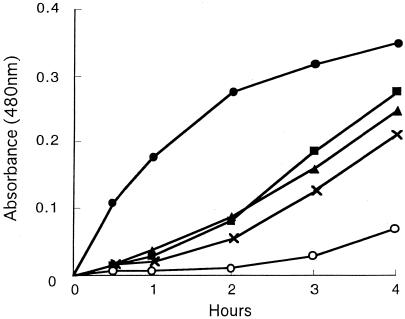
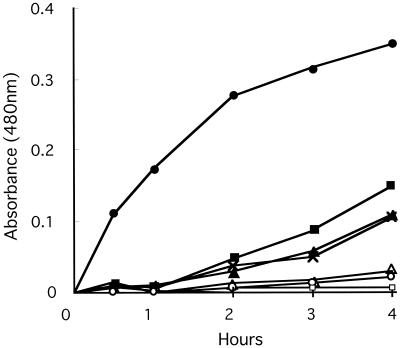
Laccase activities in strains of C. albidus, C. laurentii, C. curvatus, and C. humicola were compared to that of C. neoformans B-3501 by observing the time course of color development in the presence of epinephrine after 0.5, 1, 2, 3, and 4 h. Initially, we performed many trials because the shapes of the plots for C. neoformans and for C. albidus were different: concave downward for C. neoformans and concave upward for C. albidus. We performed nine experiments on C. neoformans, seven on C. albidus CBS142, and five on C. albidus CBS945, each experiment in triplicate (Fig. 1). All other strains were studied once in triplicate. The precision of the first three plots is indicated. It seems clear that all preparations from C. albidus (Fig. 1), three of four from C. laurentii (Fig. 2), and all strains of C. curvatus (Fig. 3) oxidized the epinephrine substrate, while preparations from C. humicola (Fig. 3) contained little, if any, laccase activity. A strain of Candida albicans served as a negative control (Fig. 1). We quantitated laccase activities by calculating linear slopes, after 1 h in the case of C. neoformans and after 4 h in the other cases. These data are given in Table 2.
FIG. 1.
Laccase activities of strains of C. albidus. •, C. neoformans B3501; ×, C. albidus CBS965; ▪, C. albidus CBS142; ▴, C. albidus CBS945; ▵, Candida albicans CBS562. The error bars indicate standard deviations.
FIG. 2.
Laccase activities of strains of C. laurentii. •, C. neoformans B3501; ▪, C. laurentii CBS2174; ▴, C. laurentii CBS8645; ×, C. laurentii CBS8648; ○, C. laurentii CBS2409.
FIG. 3.
Laccase activities of strains of C. curvatus and C. humicola. •, C. neoformans B3501; ▪, C. curvatus JCM1605; ▴, C. curvatus CBS570; ×, C. curvatus JCM1532; ▵, C. humicola JCM1530; ○, C. humicola CBS571; □, C. humicola CBS1896, CBS5920, and CBS5123.
TABLE 2.
Laccase activities in C. neoformans and species other than C. neoformans
| Species | Strain | Slopea |
|---|---|---|
| C. neoformans | B-3501 | 0.175 |
| C. albidus | CBS142 | 0.036 |
| CBS945 | 0.029 | |
| CBS965 | 0.043 | |
| C. laurentii | CBS2174 | 0.070 |
| CBS8645 | 0.061 | |
| CBS8648 | 0.052 | |
| CBS2409 | 0.015 | |
| C. curvatus | JCM1605 | 0.037 |
| CBS570 | 0.026 | |
| JCM1532 | 0.025 |
Slopes were calculated after 1 h in C. neoformans and 4 h in other Cryptococcus species.
Cell-free laccases from C. neoformans and species other than C. neoformans.
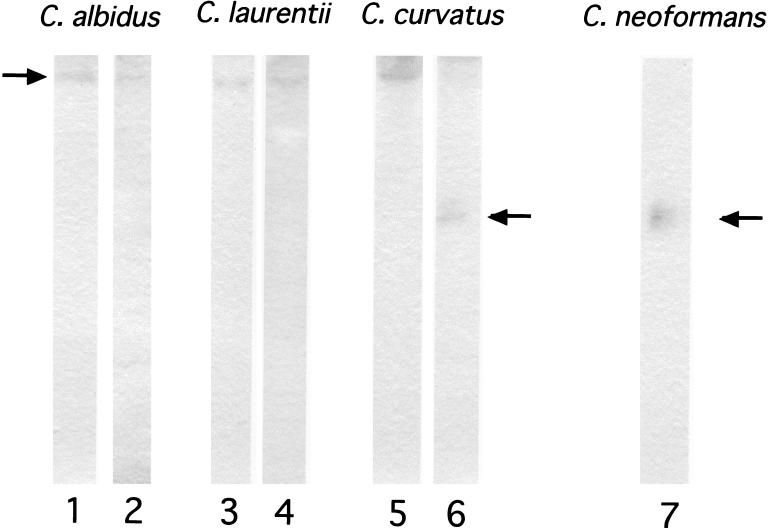
Laccase activities were detected in the crude soluble fractions of broken-cell extracts made from C. albidus, C. laurentii, and C. curvatus, although again, the activities were lower than in comparable extracts from C. neoformans (not shown). To test whether the laccases of species other than C. neoformans were similar to the C. neoformans protein, nondenaturing gel electrophoresis and activity staining were performed. As shown in Fig. 4, melanin bands were seen. Laccases from C. albidus, C. laurentii, and C. curvatus migrated more slowly than laccase from C. neoformans. We previously showed that laccases from C. neoformans var. gattii are slower than those from C. neoformans var. neoformans (8). However, laccases from the Cryptococcus species other than C. neoformans studied here were even slower, although with mobilities similar among themselves. Interestingly, in C. curvatus JCM1532, two laccases with different mobilities were detected: one with mobility similar to those of C. albidus and C. laurentii and one with mobility similar to that of C. neoformans (Fig. 4, lane 6).
FIG. 4.
Activity-stained electropherograms of laccases produced by C. neoformans and Cryptococcus species other than C. neoformans. Lanes: 1, C. albidus CBS142; 2, C. albidus CBS945; 3, C. laurentii CBS8645; 4, C. laurentii CBS2174; 5, C. curvatus CBS570; 6, C. curvatus JCM1532; 7, C. neoformans B3501.
DISCUSSION
How do the virulence factors of Cryptococcus species other than C. neoformans compare to those of C. neoformans? The major virulence factors identified in C. neoformans are capsule formation and melanin synthesis (3). The polysaccharide capsule aids pathogenicity through antiphagocytic activity (16, 18) and effects on the complement system (17), by preventing interaction with platelets (24), and by suppressing immune responses (3). Similarities in the chemical structures of polysaccharides from C. neoformans and several other Cryptococcus species have been suggested by chemical and serological studies. Thus, the serological reactivities of C. albidus, C. curvatus, and C. humicola strains were the same as those of C. neoformans serotypes A, AD, and D, respectively, indicating similarities in the chemical structures of the capsular polysaccharides (11). In fact, the chemical structures of polysaccharides from C. albidus CBS142 and C. neoformans serotype A were highly similar and differed only in the molar ratios of component sugars and in the existence of unreplaced mannose residues in C. neoformans serotype A (10).
Melanin synthesis is a second virulence factor in C. neoformans (20). It is thought that melanin, made by a laccase from exogenous catecholamines, provides C. neoformans with some protection against physiological stress. For instance, melanin has been shown to act as a redox buffer, resulting in resistance to external oxidants and to intracellular killing by anions in phagocytosis (2, 12, 13). According to an alternative hypothesis, the laccase itself, rather than its product, may be the factor that protects the fungus from strong oxidants (21). In this study, we surveyed the laccases of clinically important Cryptococcus species other than C. neoformans. We observed (slight) browning of cultures on bird seed agar in some strains, suggesting melanization. We demonstrated oxidation of laccase substrate by suspensions of C. albidus, C. laurentii, and C. curvatus, although the activity of C. neoformans clearly was much greater. Because it was not possible to set a clear lower limit for this assay, we confirmed the production of laccase by other methods. First, we confirmed that laccase activity was present in the soluble fractions after cell disruption. Next, we showed that macromolecules could be electrophoresed in polyacrylamide gel and stained for laccase activity in the gel.
None of the laccase assays were perfectly linear with respect to time. The laccase assay in C. neoformans gave a curve which was concave downward. It is possible that the reaction ran out of substrate, although the concentration provided was 14 times the Km (26) and was limited by the solubility of epinephrine. An alternative possibility is that the enzyme was inactivated by its extremely reactive intermediate products. We compensated for this curvature by calculating the activity in C. neoformans from the slope at 1 h. The time curves obtained for the species other than C. neoformans were repeatedly concave upward, which may be the result of slow production of chromogenic intermediates.
In the case of one strain of C. curvatus, we observed two bands of melanin on activity staining. It is possible that these represent isoenzymes, but the different mobilities may also represent an aggregation-disaggregation phenomenon.
Finally, we consider the taxonomic consistency of the results (Table 1). With regard to C. albidus, all representative strains showed laccase activity. However, one of the three strains (CBS965) has been reclassified as C. diffluens on the basis of DNA sequencing of ribosomal and internal transcribed spacer regions (7, 32). Of the four representative strains of C. laurentii, strain CBS2409 showed little or no laccase activity; however, on the basis of recent DNA sequence data, the identification of that strain was incorrect (30). In C. curvatus, recently reclassified from the genus Candida into Cryptococcus (6), we found laccase activity in all three representative strains. Thus, all representative strains of C. albidus, C. laurentii, and C. curvatus which survived recent DNA-based reclassification are considered to express laccase. We attempted to obtain a fragment from the laccase gene of C. curvatus by PCR, using primers designed from a conserved region of the laccase genes from C. neoformans var. neoformans and C. neoformans var. gattii. Although we obtained the expected products from five strains of C. neoformans var. neoformans and C. neoformans var. gattii (35, 36), we obtained none from any of the strains of C. curvatus studied. This negative result is consistent with the qualitative differences seen on electrophoresis of laccases from species other than C. neoformans and suggests that the species other than C. neoformans produce laccases with molecular structures different from that of C. neoformans laccase.
Recently, Petter et al. (25) surveyed heterobasidiomycetous yeast genera for production of melanin-like pigment on medium containing l-dihydroxyphenylalanine or norepinephrine and for the CNLAC1 gene, which encodes the laccase of C. neoformans. Several species showed faint or delayed pigmentation. Interestingly, the cryptococcal species C. podzolicus alone showed positive production of pigment; it contained a DNA sequence homologous to the CNLAC1 gene. That report, with its focus on DNA homology, might be considered complementary to the present report, whose methodology was primarily enzymologic.
In conclusion, several clinically emerging Cryptococcus species have the potential to express laccase and to synthesize melanin, although the rates and amounts are less than in C. neoformans.
Acknowledgments
This study was partially supported by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Bell, A. A., and M. H. Wheeler. 1986. Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24:411-451. [Google Scholar]
- 2.Casadevall, A., A. L. Rosas, and J. D. Nosanchuk. 2000. Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3:354-358. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall, A., and J. R. Perfect. 1998. Virulence factors, p. 145-176. In A. Casadevall and J. R. Perfect (ed.), Cryptococcus neoformans. ASM Press, Washington, D.C.
- 4.Casadevall, A., and J. R. Perfect. 1998. Human cryptococcosis, p. 407-756. In A. Casadevall and J. R. Perfect (ed.), Cryptococcus neoformans. ASM Press, Washington, D.C.
- 5.Dromer, F., A. Moulignier, B. Dupont, E. Guého, M. Baudrimont, L. Improvisi, F. Provost, and G. Gonzalez-Canali. 1995. Myeloradiculitis due to Cryptococcus curvatus in AIDS. AIDS 9:395-396. [PubMed] [Google Scholar]
- 6.Fell, J. W., and A. Statzell-Tallman. 1998. Cryptococcus Vuillemin, p. 742-767. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts: a taxonomic study. Elsevier, Amsterdam, The Netherlands.
- 7.Fonseca, A., G. Scorzetti, and J. W. Fell. 2000. Diversity in the yeast Cryptococcus albidus and related species as revealed by ribosomal DNA sequence analysis. Can. J. Microbiol. 46:7-27. [PubMed] [Google Scholar]
- 8.Ikeda, R., and E. S. Jacobson. 1992. Heterogeneity of phenol oxidases in Cryptococcus neoformans. Infect. Immun. 60:3552-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda, R., T. Shinoda, T. Morita, and E. S. Jacobson. 1993. Characterization of a phenol oxidase from Cryptococcus neoformans var. neoformans. Microbiol. Immunol. 37:759-764. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, R., H. Matsuyama, A. Nishikawa, T. Shinoda, and Y. Fukazawa. 1991. Comparison of serological and chemical characteristics of capsular polysaccharides of Cryptococcus neoformans var. neoformans serotype A and Cryptococcus albidus var. albidus. Microbiol. Immunol. 35:125-138. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda, R., T. Sugita, and T. Shinoda. 2000. Serological relationships of Cryptococcus spp.: distribution of antigenic factors in Cryptococcus and intraspecies diversity. J. Clin. Microbiol. 38:4021-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson, E. S. 2000. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13:708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson, E. S., and H. S. Emery. 1991. Catecholamine uptake, melanization, and oxygen toxicity in Cryptococcus neoformans. J. Bacteriol. 173:401-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, L. B., S. F. Bradley, and C. A. Kauffman. 1998. Fungaemia due to Cryptococcus laurentii and a review of non-neoformans cryptococcaemia. Mycoses 41:277-280. [DOI] [PubMed] [Google Scholar]
- 15.Kordossis, T., A. Avlami, A. Velegraki, I. Stefanou, G. Georgakopoulos, C. Papalambrou, and N. J. Legakis. 1998. First report of Cryptococcus laurentii meningitis and a fatal case of Cryptococcus albidus cryptococcaemia in AIDS patients. Med. Mycol. 36:335-339. [PubMed] [Google Scholar]
- 16.Kozel, T. R. 1993. Opsonization and phagocytosis of Cryptococcus neoformans. Arch. Med. Res. 24:211-218. [PubMed] [Google Scholar]
- 17.Kozel, T. R. 1993. Activation of the complement system by the capsule of Cryptococcus neoformans. Curr. Top. Med. Mycol. 5:1-26. [PubMed] [Google Scholar]
- 18.Kozel, T. R., G. S. Pfrommer, A. S. Guerlain, B. A. Highison, and G. J. Highison. 1988. Role of the capsule in phagocytosis of Cryptococcus neoformans. Rev. Infect. Dis. 10(Suppl. 2):S436-S439. [DOI] [PubMed] [Google Scholar]
- 19.Kunova, A., and V. Krcmery. 1999. Fungaemia due to thermophilic cryptococci: 3 cases of Cryptococcus laurentii bloodstream infections in cancer patients receiving antifungals. Scand. J. Infect. Dis. 31:328.. [DOI] [PubMed] [Google Scholar]
- 20.Kwon-Chung, K. J., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, L., R. P. Tewari, and P. R. Williamson. 1999. Laccase protects C. neoformans from antifungal activity of alveolar macrophages. Infect. Immun. 67:6034-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loison, J., J. P. Bouchara, E. Guého, L. de Gentile, B. Cimon, J. M. Chennebault, and D. Chabasse. 1996. First report of Cryptococcus albidus septicaemia in an HIV patient. J. Infect. 33:139-140. [DOI] [PubMed] [Google Scholar]
- 23.Melo, J. C., S. Srinivasan, M. L. Scott, and M. J. Raff. 1980. Cryptococcus albidus meningitis. J. Infect. 2:79-82. [DOI] [PubMed] [Google Scholar]
- 24.Nail, S., R. Robert, F. Dromer, A. Marot-Leblond, and J. M. Senet. 2001. Susceptibilities of Cryptococcus neoformans strains to platelet binding in vivo and to the fungicidal activity of thrombin-induced platelet microbicidal proteins in vitro. Infect. Immun. 69:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petter, R., B. S. Kang, T. Boekhout, B. J. Davis, and K. J. Kwon-Chung. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 147:2029-2036. [DOI] [PubMed] [Google Scholar]
- 26.Polacheck, I., V. J. Hearing, and K. J. Kwon-Chung. 1982. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. Infect. Immun. 150:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes, J. C., I. Polacheck, and K. J. Kwon-Chung. 1982. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect. Immun. 36:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogowska-Szadkowska, D., A. Wiercinska-Drapalo, A. Borzuchowska, and D. Prokopowicz. 1997. Candida humicola infection of the central nervous system in an HIV-infected patient: a case report. Przegl. Epidemiol. 51:465-469. [PubMed] [Google Scholar]
- 29.Staib, F., M. Seibold, E. Antweiler, B. Frohlich, S. Weber, and A. Blisse. 1987. The brown colour effect (BCE) of Cryptococcus neoformans in the diagnosis, control and epidemiology of C. neoformans infections in AIDS patients. Zentbl. Bakteriol. Mikrobiol. Hyg. 266:167-177. [DOI] [PubMed] [Google Scholar]
- 30.Sugita, T., M. Takashima, R. Ikeda, T. Nakase, and T. Shinoda. 2000. Intraspecies diversity of Cryptococcus laurentii as revealed by sequences of internal transcribed spacer regions and 28S rRNA gene and taxonomic position of C. laurentii clinical isolates. J. Clin. Microbiol. 38:1468-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita, T., M. Takashima, R. Ikeda, T. Nakase, and T. Shinoda. 2000. Phylogenetic and taxonomic heterogeneity of Cryptococcus humicola by analysis of the sequences of the internal transcribed spacer regions and 18S rDNA, and the phylogenetic relationships of C. humicola, C. curvatus, and the genus Trichosporon. Microbiol. Immunol. 44:455-461. [DOI] [PubMed] [Google Scholar]
- 32.Sugita, T., M. Takashima, R. Ikeda, T. Nakase, and T. Shinoda. 2001. Intraspecies diversity of Cryptococcus albidus isolated from human as revealed by sequences of internal transcribed spacer regions. Microbiol. Immunol. 45:291-297. [DOI] [PubMed] [Google Scholar]
- 33.Sugita, T., M. Takashima, T. Nakase, T. Ichikawa, R. Ikeda, and T. Shinoda. 2001. Two new yeasts, Trichosporon debeurmannianum sp. nov. and Trichosporon dermatis sp. nov., transferred from the Cryptococcus humicola complex. Int. J. Syst E vol. Microbiol. 51:1221-1228. [DOI] [PubMed] [Google Scholar]
- 34.Takashima, M., T. Sugita, T. Shinoda, and T. Nakase. 2001. Reclassification of the Cryptococcus humicola complex. Int. J. Syst. E vol. Microbiol. 51:2199-2210. [DOI] [PubMed] [Google Scholar]
- 35.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]