Abstract
In mid-1999, we noted multiple isolations at the Veterans Affairs Medical Center (VAMC) Houston Tex. of an unusual nonpigmented Mycobacterium species. Since, on the basis of 16S rRNA gene sequence analysis, the strains were identical to the Mycobacterium szulgai type strain and since M. szulgai has been reported only rarely as a commensal or environmental isolate, we were concerned about laboratory contamination, nosocomial spread, or even the possibility that this could be a novel organism associated with disease. Our investigation found that from 1999 to 2000, 37 strains of M. szulgai were isolated from patients at the VAMC (the base rate for the previous 10 years had been <1 isolation per year). We compared the phenotypic properties and genetic relatedness of these 37 strains (31 of which were nonpigmented) as well as eight stock strains and the M. szulgai type strain. All strains were similar in cellular fatty acid patterns, growth rates, and biochemical characteristics. However, we found three genogroups by gene sequence analysis. Genogroup I comprised the M. szulgai type strain, all the tested nonpigmented strains (27 of the 31 strains were tested), two pigmented strains isolated in 1999 and 2000, and five pigmented stock strains. Genogroup II comprised five pigmented strains: three were isolated from 1999 to 2000 and two were stock strains. The single strain (isolated in 1996) in genogroup III was pigmented and was the only strain associated with disease. Whereas the randomly amplified polymorphic DNA (RAPD) patterns of all nonpigmented strains were identical, indicating that they came from a common source (the pseudoepidemic strain), the RAPD patterns of the other strains were varied. In our investigation for a possible source, we found that there were no common reagents, specimen-processing or patient locations, or procedures linking the 31 pseudoepidemic strains. However, a nonpigmented M. szulgai strain with a gene sequence and RAPD pattern identical to those of the pseudoepidemic strain was recovered from a water storage tank serving the hospital. We concluded that the strains most likely originated from hospital water, which transiently inoculated our patients. Although no disease was associated with this cluster of isolates, the event was costly because identification was problematic and we could not easily discount the isolations, since most of the patients were immunocompromised and were candidates for opportunistic infection.
Mycobacterium szulgai is a rarely isolated mycobacterial species that has been associated with pulmonary disease in immunocompromised patients (1, 2, 4, 7, 8). M. szulgai is said to be unique in that it is a scotochromogen (produces pigment in both light and darkness) at 37°C but a photochromogen (forms pigment only after exposure to light) at 25°C (10, 11). Its key characteristics are its slow growth and its positive test results for nitrate reduction and urease (10, 11).
Early in 1999, we noticed that a number of isolates from clinical specimens at the Veterans Affairs Medical Center (VAMC) in Houston, Texas, although identified as M. szulgai by both cellular fatty acid (CFA) analysis and 16S rRNA gene sequencing, were not typical in phenotypic characteristics because they were nonpigmented. Since these nonpigmented strains were all isolated from paucibacillary specimens (e.g., from BACTEC 12B broth medium only), we were concerned that the laboratory was introducing a contaminant during processing or that a hospital procedure was introducing the organism (9, 13, 14). We also considered the possibility that this was a novel organism associated with disease. Our genotypic, phenotypic, and epidemiologic investigations of these clinical strains as well as stock strains and one type strain are the subject of this report.
MATERIALS AND METHODS
Phenotypic and genotypic microbiological characterization of strains.
A total of 47 strains were examined in this study (Tables 1 and 2): 38 strains were isolated during this outbreak, including 1 from a hospital water tank; 8 were stock strains, including 5 from the Houston Department of Health; and 1 was the M. szulgai type strain (ATCC 35799). The BACTEC 460 apparatus, Löwenstein-Jensen solid medium, and 12B bottles (Becton Dickinson Diagnostic Instrument Systems, Cockeysville, Md.) were used for the initial isolations.
TABLE 1.
Characteristics of M. szulgai study isolates and associated patients
| Patient no. | VAMC accession no. | Specimen source | Hospital unit(s) | Date of positive culture (mo/day/yr) | Risk factor(s)a | RAPD-PCR gel patternb | 16S rRNA geno group cluster | Pigment |
|---|---|---|---|---|---|---|---|---|
| 1 | 00TB0087 | Sputum | 3A | 01/16/00 | MSOF, CA | C | NDc | Buff |
| 2 | 00TB0146 | Sputum | 4A | 01/27/00 | CA | C | I | Buff |
| 3 | 00TB0180 | Sputum | 4A, 3C | 02/02/00 | CHF, PE | C | I | Buff |
| 4 | 00TB0296 | Sputum | 4A | 02/19/00 | CA | C | I | Buff |
| 5 | 00TB0479 | Urine | 4A | 03/26/00 | CML | C | I | Buff |
| 6 | 00TB0524 | Urine | 3A | 04/04/00 | DM | C | I | Buff |
| 7 | 00TB0525 | Sputum | 3A | 04/04/00 | RA | C | I | Buff |
| 8 | 00TB0556 | Sputum | 3A | 04/08/00 | COPD | C | I | Buff |
| 9 | 00TB0999-2 | Sputum | 3B | 06/25/00 | MAC, CLL | C | I | Buff |
| 10 | 00TB1093 | Bronchial wash | 3B | 07/10/00 | SCCA | C | ND | Buff |
| 11 | 00TB1375 | Sputum | 3A | 08/25/00 | DM, COPD | C | ND | Buff |
| 12 | 00TB1423 | Sputum | 3B | 09/02/00 | None | C | I | Buff |
| 13 | 00TB1448-2 | Stool | 3B | 09/09/00 | HIV | C | I | Buff |
| 14 | 00TB1517 | Urine | 4A | 09/18/00 | MSOF | C | I | Buff |
| 15 | 00TB1538 | Bronchial wash | 3C | 09/20/00 | CA | C | ND | Buff |
| 16 | 00TB1571 | Sputum | 4A, 5K | 09/24/00 | CA | C | I | Buff |
| 17 | 00TB1594 | Bronchial wash | 3B, 3D | 09/27/00 | SCI | C | I | Buff |
| 18 | 00TB1744 | Sputum | 1A | 10/25/00 | SCI | C | I | Buff |
| 19 | 00TB1796 | Sputum | 3A | 11/06/00 | None | C | I | Buff |
| 20 | 98TB2561 | Sputum | 3A | 12/25/98 | COPD | C | I | Buff |
| 21 | 99TB0239 | Sputum | 4D | 02/05/99 | Treated TB | C | I | Buff |
| 22 | 99TB0446 | Sputum | 4A | 03/04/99 | COPD | C | I | Buff |
| 23 | 99TB0950 | Sputum | 3B | 05/28/99 | ESRD | C | I | Buff |
| 24 | 99TB1239-1 | Sputum | 4D | 07/20/99 | CVA | C | I | Buff |
| 25 | 99TB1327 | Sputum | 3B | 08/03/99 | CA | C | I | Buff |
| 26 | 99TB1390 | Sputum | 4A | 08/12/99 | HIV | C | I | Buff |
| 27 | 99TB1544 | Sputum | 3A | 09/19/99 | CA | C | I | Buff |
| 28 | 99TB1592 | Sputum | 3A | 09/29/99 | HIV | C | I | Buff |
| 29 | 99TB1866 | Sputum | 4A | 11/12/99 | CA | C | ND | Buff |
| 30 | 99TB1918 | Sputum | 4A | 11/19/99 | HIV | C | I | Buff |
| 31 | 99TB2036-2 | Sputum | 3A | 12/09/99 | HIV | C | I | Buff |
| 32 | 00TB0011 | Sputum | 4B | 01/05/00 | Myelofibrosis | A-3 | I | Yellow |
| 36 | 00TB1638 | Sputum | Outpatient | 10/04/00 | COPD | B | I | Yellow |
| 37 | 99TB1239-2 | Sputum | 4D | 07/20/99 | CVA | A-2 | II | Yellow |
| 35 | 00TB1577 | Sputum | 3B | 09/25/00 | TB | A-2 | II | Yellow |
| 34 | 00TB1448-1 | Stool | 3B | 09/09/00 | HIV | ND | II | Yellow |
| 33 | 00TB0544 | Sputum | Outpatient | 04/06/00 | HIV | ND | ND | Yellow |
| 38 | None | Hospital water (main tank) | 12/12/00 | C | I | Buff |
Abbreviations: MSOF, multisystem organ failure; CA, carcinoma; CHF, congestive heart failure; PE, pleural effusion; CML, chronic myelogenous leukemia; DM, diabetes mellitus; RA, rheumatoid arthritis; COPD, chronic obstructive pulmonary disease; MAC, Mycobacterium avium complex; CLL, chronic lymphocytic leukemia; SCCA, squamous cell carcinoma; HIV, human immunodeficiency virus; SCI, spinal cord injury; TB, tuberculosis; ESRD, end-stage renal disease; CVA, cerebrovascular accident.
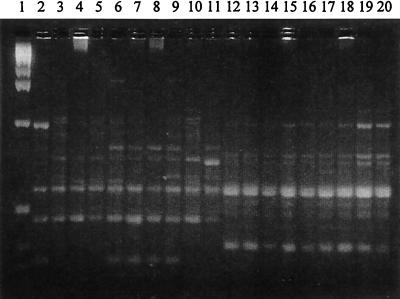
RAPD-PCR gel patterns refer to the results shown in Fig. 2 as follows: pigmented M. szulgai strain patterns A-2 (lanes 6 to 9), A-3 (lane 10), and B (lane 11); pseudoepidemic strain pattern C (lanes 12 to 20).
ND, not done.
TABLE 2.
Characteristics of stock strains of M. szulgai
| Stock strain | Specimen sourcea | Hospital unit | Date of positive culture (mo/day/yr) | Risk factorb | RAPD-PCR gel patternc | 16S rRNA geno group cluster | Pigment |
|---|---|---|---|---|---|---|---|
| 97TB2917 | Sputum | 4B | 12/23/97 | DM | ND | I | Yellow |
| CH2 | CHDH | A-1 | I | Yellow | |||
| CH3 | CHDH | A-1 | I | Yellow | |||
| CH4 | CHDH | A-1 | I | Yellow | |||
| Type strain | ATCC 35799 | A-4 | I | Yellow | |||
| CH1 | CHDH | ND | II | Yellow | |||
| CH5 | CHDH | A-2 | II | Yellow | |||
| 96TB1204 | Sputum | 4A | 5/9/96 | None | Unique | III | Yellow |
| 97TB1869 | Sputum | 4B | 7/30/97 | Lung cancer | ND | ND | Yellow |
CHDH, city of Houston Department of Health.
DM, diabetes mellitus.
RAPD-PCR gel patterns refer to the results shown in Fig. 2, as follows: pigmented M. szulgai strain patterns A-1 (lanes 3 to 5), A-2 (lanes 6 to 9), and A-4, (lane 2).
ND, not done.
After initial isolation, the strains were frozen at −70°C. For the growth characteristic study and biochemical tests, all isolates were subcultured on 7H11 medium, incubated at 37 and 25°C for 4 weeks, and checked for microbial growth daily for 7 days and every 3 days thereafter. After 2 weeks of incubation, urease and nitrate reduction tests were performed by standard methods (10). All pigmented strains were tested for photoactivation of pigment production at 25°C. All nonpigmented strains were exposed to light for 2 h and observed for pigmentation.
16S rRNA gene sequence analysis was performed. The nucleotide sequence of about 500 bp of the 16S rRNA gene was determined (MicroSeq 16S rRNA gene kit 500; PE Applied Biosystems, Foster City, Calif.). This sequence was compared to those of the 52 mycobacterial type strains in the MicroSeq database by MicroSeq analysis software (PE Applied Biosystems) and to the sequences in the GenBank database. Neighbor-joining analysis was performed (MicroSeq analysis software).
The CFA were analyzed by the Hewlett-Packard HP 5890 II microbial identification system (MIDI, Inc., Newark, Del.). We performed randomly amplified polymorphic DNA (RAPD) analysis. In brief, the isolates were suspended in 0.9% sterile saline to a turbidity that matched that of a 3.0 McFarland standard and were stored at −20°C. Before they were used, the isolates were thawed and boiled for 10 min. Several primers were tested for the arbitrary primed PCR, and a 10-mer (5′-TGGTCGCGGC) was chosen for the analysis based on its higher yield in number of bands. Fifty-microliter reaction mixtures containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 3 mM MgCl2, 60 pmol of primer, 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.), and 1 μl of template were run in a thermocycler (MJ Research, Watertown, Mass.) with the following program: 94°C for 5 min; 40 cycles at 94°C for 30 s, 37 °C for 1 min, and 72°C for 1 min; and 72°C for 10 min.
Epidemiological investigation.
To determine the prevalence of isolates, all positive mycobacterial cultures obtained at the VAMC between 1995 and 2000 were reviewed. To determine if a particular person, laboratory procedure, or reagent was associated with the outbreak, the names of all personnel involved, the procedures employed, and the specimen type, receiving date, processing batch, and lot number for each reagent used were reviewed. The following reagents used in processing were cultured: phosphate buffer (BBL, Sparks, Md.), water (Baxter Healthcare Corporation, Deerfield, Ill.), N-acetyl-l-cysteine-NaOH (used for specimen digestion; Alpha-Tec Systems, Vancouver, Wash.), and the antimicrobial supplement PANTA (Becton Dickinson Diagnostic Instrument Systems). The BACTEC 460 apparatus, including the sampling needles, was inspected.
The hospital water system was also investigated. The hospital receives water from two tanks, both of which are filled with city water. The first tank is the main tank for the hospital water supply, while the second tank is a reservoir. Fifteen thousand gallons of water from the second tank flows into the main tank daily. Four liters of water from each tank was collected. The water was concentrated by one of two methods. Either it was centrifuged for 30 min at 4,000 × g or it was filtered through a 0.45-μm-pore-size membrane. The pellet or the filter wash was obtained and decontaminated with 1.5% NaOH; the specimen was then inoculated on 7H11 plates (BBL) and BACTEC 12B vials (Becton Dickinson). Tap water and ice water from four patient wards were cultured similarly.
Patient records were reviewed for evidence of clinical diseases and therapeutic intervention. The nursing units involved, patient room numbers, admittance and discharge dates, and patient risk factors were recorded and reviewed.
RESULTS
Phenotypic and genotypic microbiological characterization.
Forty-seven strains (Tables 1 and 2) were studied for their phenotypic characteristics. Thirty-eight strains were isolated at the VAMC during this outbreak, including one from the hospital water tank, eight were stock strains, including five from the Houston Department of Health, and one was the M. szulgai type strain. All nonpigmented strains and seven of nine pigmented strains were isolated from BACTEC 12B broth only.
All strains showed certain similar characteristics and conformed to the description of M. szulgai (4, 10). All strains were slow growing. Both the pigmented and nonpigmented strains isolated from patient specimens required 3 to 4 weeks of culture to become positive. Ten days after subculture, pinpoint colonies (0.1 mm diameter) were seen. At 2, 3, and 4 weeks, the colony sizes were 0.2, 0.5 to 1, and 1 to 2 mm, respectively. Urease and nitrate reduction test results were positive. The CFA patterns of all tested strains (21 nonpigmented and 4 pigmented) were similar, with C16:0 (42%) and C18:1ω9c (30%) predominating, but other fatty acids (3 to 6%) (C14:0, C16:1ω7c, and C16:1ω5c) were also detected. All isolates were identified as M. szulgai by the CFA analysis software. All colonies were moist; however, for any one strain, some colonies were irregular while others were smooth and entire.
However, some strains were yellow at 37°C and were photochromogens at 25°C whereas others were nonpigmented (buff) at both 37 and 25°C (Table 1). The buff strains had no pigmentation after 2 h (not even when left for 2 days with exposure to light).
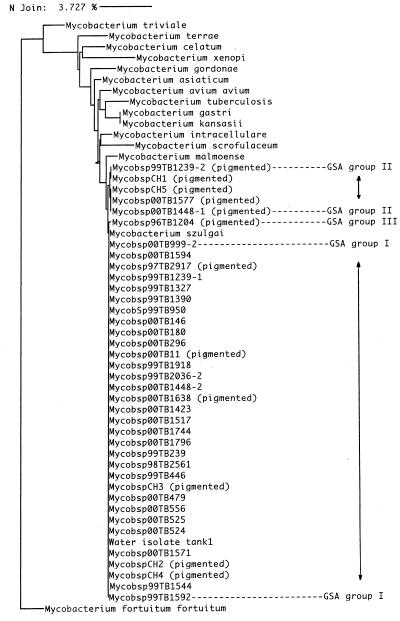
A total of 40 isolates were sequenced, including 32 VAMC outbreak isolates, seven stock strains, and the type strain. There were three genotypic clusters, as shown in the dendrogram (Fig. 1) and as listed in Tables 1 and 2. Cluster I included all of the 27 nonpigmented (pseudoepidemic) strains analyzed, including the isolate from the hospital water tank. Cluster II included two pigmented strains isolated from 1999 to 2000, four stock strains, and the M. szulgai type strain. In cluster II, there were five pigmented strains, three of which were isolated from 1999 to 2000 and two of which were stock strains. Cluster II differed from cluster I by only 0.19%; the difference was a T-to-C base pair mutation at position 462. The single strain (isolated in 1996) in genogroup cluster III was pigmented and was the only strain associated with disease.
FIG. 1.
Dendrogram of sequence data for 27 nonpigmented M. szulgai, 12 pigmented M. szulgai, and some mycobacterial type strains in the MicroSeq database, with Mycobacterium fortuitum subsp. fortuitum as an out-group. GSA, gene sequence analysis. The dendrogram was generated by the neighbor joining (N Join) method, with the horizontal line at the top representing a 3.727% genetic difference.
All the nonpigmented (pseudoepidemic) strains tested, including the isolate from the water tank, had identical RAPD patterns (pattern C), which differed from those of the type strain and the other tested strains (Fig. 2). The RAPD patterns of the pigmented strains were varied although the strains that had the same 16S rRNA genetic clusters were more alike in their RAPD patterns.
FIG. 2.
RAPD-PCR patterns of nine nonpigmented M. szulgai strains, nine pigmented M. szulgai strains, and one ATCC 35799 strain. Lane 1, 1-kb DNA ladder (Life Technologies, Inc.); lane 2, strain ATCC 35799; lanes 3 to 5, 10, and 11, pigmented M. szulgai strains in 16S rRNA gene sequence analysis group I; lanes 6, 8, and 9, pigmented M. szulgai strains in 16S rRNA gene sequence analysis group II; lane 7, pigmented M. szulgai strain in 16S rRNA gene sequence analysis group III; lanes 12 to 20, nonpigmented M. szulgai strains.
Epidemiological investigation.
From 1999 to 2000, three medical technologists alternately processed specimens at the VAMC. No technologist was specifically associated with the occurrence of the unusual nonpigmented (pseudoepidemic) strains. Throughout the study period, (1995 to 2000), the procedures used were standard and had not changed (10). No organism was cultured from any reagents, and no specific lot number of any reagent correlated with the pseudoepidemic isolates. The BACTEC instrument had been functioning properly, and BACTEC needles were changed at every use.
Of the 31 nonpigmented (pseudoepidemic) patient isolates, 24 were obtained from sputum, three were from bronchial wash, three were from urine, and one was from stool (Table 1). There were 30 different collection and processing dates. Only two of the specimens were consecutive and were processed on the same day; however, one was a urine sample and the other was a sputum sample. We concluded that contamination due to laboratory error was unlikely.
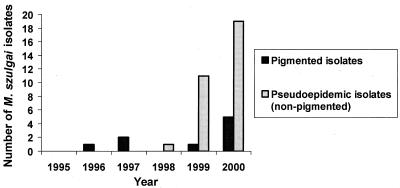
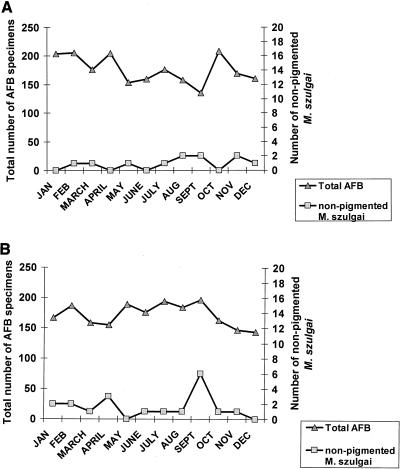
From 1995 through 2000, approximately 2,697 specimens per year were processed for mycobacterial culture in the VAMC microbiology laboratory. Pigmented M. szulgai strains were isolated from 0 to 5 patients each year from 1995 to 2000. The unusual nonpigmented (pseudoepidemic) strains were isolated from 1 patient in December 1998, 11 patients in 1999, and 19 patients in 2000 (Fig. 3). Figure 4 shows that the number of pseudoepidemic M. szulgai isolates increased, while the total number of acid-fast bacillus (AFB) cultures processed during 1999 (Fig. 4A) and 2000 (Fig. 4B) did not. There was a statistically significant increase of M. szulgai isolates collected in September 2000 as compared with the numbers collected in other months (P < 0.01) and a lesser increase increase in August 1999, possibly indicating better growth of the organism in warm weather.
FIG. 3.
Numbers of M. szulgai isolates collected at the Houston VAMC from 1995 to 2000.
FIG. 4.
Seasonal changes in the numbers of nonpigmented M. szulgai isolates collected and of total AFB cultures processed in 1999 (A) and 2000 (B).
Some of the characteristics of patients from whom M. szulgai was recovered are shown in Table 1. Almost all of the patients had an underlying disease. However, M. szulgai was not thought to be causing disease in any of these patients. Although pseudoepidemic strains were isolated from patients from a variety of hospital rooms and nursing units, there were more isolates of M. szulgai found in patients from one of the nursing units (unit 4A) in 2000 than in patients from the other nursing units (P < 0.001).
We examined the tap water and ice water from this ward and several other wards as well as water from the hospital water storage tanks. No M. szulgai strain was found in any cultures from the nursing units. However, the pseudoepidemic strain was recovered from the second hospital water storage tank.
DISCUSSION
Many species of nontuberculous mycobacteria, such as Mycobacterium gordonae, M. xenopi, M. kansasii, and M. abscessus, have been reported to cause pseudoinfection (5, 9, 13, 14). The isolates causing contamination have been traced to a variety of sources, such as PANTA PLUS (Becton-Dickinson Instrument Systems), carryover from one positive specimen to the next by the BACTEC apparatus, the hospital water and ice supplies, bronchoscopes, and a variety of laboratory handling errors (9, 13, 14). To our knowledge, there has been no previous report of M. szulgai pseudoinfection.
Although M. szulgai is an uncommon cause of human diseases, it has been most commonly associated with pulmonary disease (1, 4, 7, 15). Other diseases, such as osteomyelitis, also have been reported in connection with this organism (6). In 1996, there was one case of lung infection caused by M. szulgai at the VAMC. The patient had a clinical presentation similar to tuberculosis, had six positive AFB smears and seven positive cultures from sputum, and was successfully treated with a four-drug tuberculosis treatment regimen (isoniazid, ethambutol, rifampin, and pyrazinamide). In contrast, all patients harboring the nonpigmented pseudoepidemic M. szulgai strain had negative AFB smears and only a single positive specimen, isolated from a single medium (BACTEC vials). Most of the patients were immunocompromised, and there was no primary disease associated with the pseudoepidemic strain of M. szulgai. Similarly, there was no disease associated with the pigmented strains of M. szulgai isolated during the 1997 to 2000 study period.
Neither CFA nor 16S rRNA gene sequencing analysis could uniquely differentiate pseudoepidemic strains from other strains. However, in this study, the RAPD-PCR patterns of the nonpigmented M. szulgai isolates were identical and were distinct from those of the pigmented strains, suggesting that a single unique clone was responsible. Furthermore, the RAPD pattern indicated that the pigmented strains were varied; they were found in three different genotypic clusters with at least four different RAPD patterns and thus did not represent a single source. The RAPD pattern was particularly valuable in distinguishing the six pigmented strains that were in the same 16S rRNA genogroup (cluster I) as the pseudoepidemic strains. Although 16S rRNA gene sequence analysis is excellent for bacterial identification (3, 12), it was not definitive in recognizing these pseudoepidemic strains. However, both the buff phenotype and a common RAPD-PCR pattern were uniquely associated with the pseudoepidemic strains.
Several characteristics made these pseudoepidemic strains difficult to investigate and evaluate. First, the strains of M. szulgai were nonpigmented, and pigment production has been considered a necessary characteristic before an identification of M. szulgai can be made. Because of this unusual phenotype, the strains could have been misidentified as Mycobacterium terrae or Mycobacterium triviale by conventional methods. Second, the strains were isolated over a period exceeding 2 years and were not associated with laboratory procedures, methodological changes, or reagents. Third, the pseudoepidemic strains originally could not be connected to an environmental source. They appeared abruptly in December 1998 but were unconnected to any known geographic location or water supply. The initial search for an environmental source within the hospital was negative, a finding that could have been due to the low concentration of organisms. In addition, the water tests were not performed at the peak period in August and September, when more organisms might be expected. At the time of the investigation, we found that the hospital water tanks were 2 years past their scheduled maintenance time for descaling and cleaning. Subsequently, we have made sure that the tanks get regular preventive maintenance cleaning, since M. szulgai could be just an indicator organism, signaling the presence of other pathogens of a more serious nature, such as Legionella spp. There have been no further isolations of M. szulgai at the VAMC.
We found an unusually high rate of M. szulgai isolations at the VAMC in recent years. The identical RAPD-PCR patterns and phenotypes of the 31 pseudoepidemic strains indicated that they were from a single source. Isolation of the identical strain from one of the hospital water supply tanks suggested that the pseudoepidemic of M. szulgai at the VAMC between 1998 and 2000 most likely originated from hospital water, which transiently inoculated or colonized our patients. The precursor source has not been determined, but nonpigmented M. szulgai strains occasionally have been isolated elsewhere in Texas (K. C. Jost, Texas State Health Laboratory, personal communication). We do not have a clear explanation as to why the strains first appeared in December 1998 or why there were increased isolations in September 2000 and from patients in a single nursing unit (unit 4A). However, our extensive investigation has proved valuable in that it focused attention on the probability of our stored water supply being a source of waterborne contaminants. In addition, the description of this unique strain increases the known characteristics associated with this species and makes correct identification in other laboratories more likely. Although no disease was associated with this cluster of isolates, the event was costly because, since most of the patients were immunocompromised and candidates for a mycobacterial disease, we could not easily discount the isolations. The identification of M. szulgai in a respiratory specimen as a nonpathogen and the analysis of the pseudoepidemic strains subsequently prevented unnecessary diagnostic workups and treatments.
Acknowledgments
We thank Margaret Price at St. Luke’s Hospital (Houston, Tex.) for her thoughtful review of the manuscript and assistance with statistical data analysis and Kenneth C. Jost at Texas State Health Laboratory (Austin) and Gregg Dufour at City Health Laboratory (Houston) for their contribution of strains.
Kristina Hulten was supported by a postdoctoral grant from the Swedish Society for Medical Research.
REFERENCES
- 1.Benator, D. A., V. Kan, and F. M. Gordin. 1997. Mycobacterium szulgai infection of the lung: case report and review of an unusual pathogen. Am. J. Med. Sci. 313:346-351. [DOI] [PubMed] [Google Scholar]
- 2.Butler, W. R., and J. T. Crawford. 1999. Nontuberculous mycobacteria reported to the public health laboratory information system by state public health laboratories United States, 1993-1996. Centers for Disease Control and Prevention, Atlanta, Ga.
- 3.El Amin, N. M., H. S. Hanson, B. Pettersson, B. Petrini, and L. V. Von Stedingk. 2000. Identification of non-tuberculous mycobacteria: 16S rRNA gene sequence analysis vs. conventional methods. Scand. J. Infect. Dis. 32:47-50. [DOI] [PubMed] [Google Scholar]
- 4.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkinham, J. O., III. C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurr, H., and T. Sorg. 1998. Mycobacterium szulgai osteomyelitis. J. Infect. 37:191-192. [DOI] [PubMed] [Google Scholar]
- 7.Kourbeti, I. S., and M. J. Maslow. 2000. Nontuberculous mycobacterial infections of the lung. Curr. Infect. Dis. Rep. 2:193-200. [DOI] [PubMed] [Google Scholar]
- 8.Maloney, J. M., C. R. Gregg, D. S. Stephens, F. A. Manian, and D. Rimland. 1987. Infections caused by Mycobacterium szulgai in humans. Rev. Infect. Dis. 9:1120-1126. [DOI] [PubMed] [Google Scholar]
- 9.Maloney, S., S. Welbel, B. Daves, K. Adams, S. Becker, L. Bland, M. Arduino, R. Wallace, Y. Zhang, and G. Buck. 1994. Mycobacterium abscessus pseudoinfection traced to an automated endoscope washer: utility of epidemiologic and laboratory investigation. J. Infect. Dis. 169:1166-1169. [DOI] [PubMed] [Google Scholar]
- 10.Master, R. N. 1992. Mycobacteriology, p. 3.0.1-3.16.4. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 11.Metchock, B. G., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437.In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 12.Rogall, T., J. Wolters, T. Flohr, and E. C. Böttger. 1990. Toward a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 13.Tokars, J. I., M. M. McNeil, O. C. Tablan, K. Chapin-Robertson, J. E. Patterson, S. C. Edberg, and W. R. Jarvis. 1990. Mycobacterium gordonae pseudoinfection associated with a contaminated antimicrobial solution. J. Clin. Microbiol. 28:2765-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace, R. J., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 15.Wayne, L. G., and H. A. Sramek. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 5:1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]