Abstract
Plasma human immunodeficiency virus RNA in 491 clinical specimens was measured by LCx. There was a strong correlation with the results provided by other methods (r2 values of 0.93 with Cobas-Monitor version 1.5 and of 0.95 with Quantiplex version 3.0). However, values were uniformly higher with LCx than with Quantiplex when non-B subtypes were tested.
The measurement of human immunodeficiency virus (HIV) RNA in plasma allows accurate prediction of the clinical outcome in a given patient as well as monitoring of the effect of antiretroviral treatment (9). There are several commercially available assays for the quantitation of HIV RNA. The reverse transcription (RT)-PCR (Cobas-Monitor; Roche) and the branched-DNA assay (Quantiplex; Bayer) are among the most frequently used (2, 11). None of them, however, is suitable for the accurate detection and quantitation of HIV type 1 (HIV-1) variants distinct from subtype B (5). Since the prevalence of infections with non-B subtypes is growing in Europe (3, 10) and the United States (1), there is increased concern about the possibility of inaccurate viral load determinations. It is hoped that viral load tests will detect and quantitate accurately all genetically divergent HIV-1 strains.
In this study, we have evaluated the performance of a new viral load test, the Abbott LCx quantitative assay, for the measurement of HIV-1 in plasma from subjects carrying different HIV-1 variants. The inter- and intrarun reproducibilities of the assay, its specificity and linearity, and its ability to detect and accurately quantitate specimens from subjects infected with non-B subtype HIV-1, including group O viruses, were evaluated. Finally, a comparison with the results obtained by use of Cobas-Monitor version 1.5 (11) and Quantiplex version 3.0 (2) was performed. Both assays have a detection limit of 50 copies/ml.
Plasma samples were randomly collected from HIV-1-seropositive patients attending two clinical centers in Madrid, Spain. Ten specimens taken from HIV-1-infected patients and two samples taken from the BBI HIV-1 RNA quantification panel (Boston Biomedica Inc., West Bridgewater, Mass.) were tested in triplicate on four separate days. The estimated HIV RNA concentration spanned the dynamic range of LCx, as determined by using the 0.2-ml procedure (detection limit of 178 HIV RNA copies/ml). The intraassay, interassay, and total assay variabilities were assessed. The same lot of LCx reagent and the same controls were used throughout the complete reproducibility study. The same technician performed all the runs.
Human EDTA-treated plasma samples from 41 healthy donors found to be negative for antibodies to HIV-1 or HIV-2 by an enzyme immunoassay (Abbott) were obtained from a blood bank. All samples were tested by LCx by using the 1.0-ml procedure (detection limit of 50 HIV RNA copies/ml). Initially reactive samples were tested twice by LCx and another viral load test.
Samples obtained from an HIV-1-infected patient carrying high levels of plasma HIV RNA and culture supernatants from cells infected with the BRU and BLA strains of HIV-1 were serially diluted fivefold in seronegative plasma and in culture medium, respectively. Then, they were tested in parallel by using LCx and Quantiplex.
A total of 362 plasma samples were selected for the comparative analysis of LCx and the other viral load assays (all of them were tested with Quantiplex and 101 were tested with Cobas-Monitor). The 1.0-ml protocol of LCx was used for the comparative analyses. Moreover, 64 samples from 24 HIV-1-infected patients carrying group M non-B subtypes (15 A, 10 C, 2 F, 35 G, and 2 H) and 24 samples from 5 HIV-1 group O-infected patients were examined. Viruses from all individuals included in this study had been previously genetically characterized (3, 4, 7, 8).
Viral burden in samples from HIV-1 group O-infected patients was assessed by using a p24 antigen test which has a sensitivity of 2 to 5 pg/ml (Murex, Dartford, United Kingdom) and MUPROVAMA, a noncommercial RT-PCR assay based on the simultaneous amplification of overlapping fragments of a single HIV RNA target sequence in a single reaction (6).
Assay linearities for LCx and Quantiplex were assessed graphically by parallel-line analysis related to dilution factor. Averages were statistically compared by using the paired t test. Linear regression and correlation analyses were used to determine assay relationships. Qualitative analyses comparing the number of samples with fewer or more than 50 HIV RNA copies/ml in different methods (LCx, Quantiplex, and Cobas-Monitor) were carried out by using a concordance method based on the κ index.
The inter-and intrarun reproducibilities were assessed by using specimens that were tested once in four different runs. Total variation ranged from 1.37% ± 0.09% to 13.06% ± 0.39% (Table 1). A greater variation was noted in the range of 2.62 to 2.95 log copies/ml, near the lower limit of quantitation of the LCx assay (2.25 log copies/ml).
TABLE 1.
Precision of LCx using the 0.2-ml sample extraction procedure
| Specimen (n = 12) | Mean copies/ml
|
SD (% CVa)
|
|||
|---|---|---|---|---|---|
| Log | No. | Intrarun | Interrun | Total | |
| 2171 | 2.62 | 561 | 0.36 (13.8) | 0.17 (6.5) | 0.34 (13.0) |
| 1970 | 2.95 | 1,243 | 0.46 (15.7) | 0.07 (2.4) | 0.39 (13.1) |
| Panel 3 | 3.17 | 1,772 | 0.24 (7.5) | 0.19 (5.9) | 0.30 (9.5) |
| 2199 | 3.25 | 2,262 | 0.25 (7.7) | 0.19 (5.7) | 0.28 (8.5) |
| 2201 | 3.94 | 9,456 | 0.09 (2.2) | 0.15 (3.9) | 0.18 (4.5) |
| 2186 | 4.16 | 15,577 | 0.12 (2.9) | 0.11 (2.7) | 0.17 (4.0) |
| Panel 4 | 4.59 | 39,792 | 0.06 (1.4) | 0.02 (0.5) | 0.07 (1.5) |
| 1971 | 4.77 | 61,769 | 0.07 (1.6) | 0.09 (2.0) | 0.12 (2.6) |
| 2203 | 5.62 | 470,242 | 0.18 (3.1) | 0.15 (2.6) | 0.23 (4.1) |
| 1974 | 5.70 | 536,872 | 0.17 (2.9) | 0.03 (0.5) | 0.14 (2.4) |
| 2205 | 6.62 | 4,280,114 | <0.01 (0.7) | 0.08 (1.2) | 0.09 (1.4) |
CV, coefficient of variation.
The overall specificity of the LCx assay was 97.5%. One of 41 HIV-seronegative blood donor samples was found repeatedly positive, although it yielded a small number of HIV RNA copies. Initial testing of that particular sample gave a result of 121 copies/ml, and the retest gave a result of 69 copies/ml. The presence of HIV antibodies was ruled out by enzyme immunoassay and Western blot testing as well as in follow-up samples collected after 6 months.
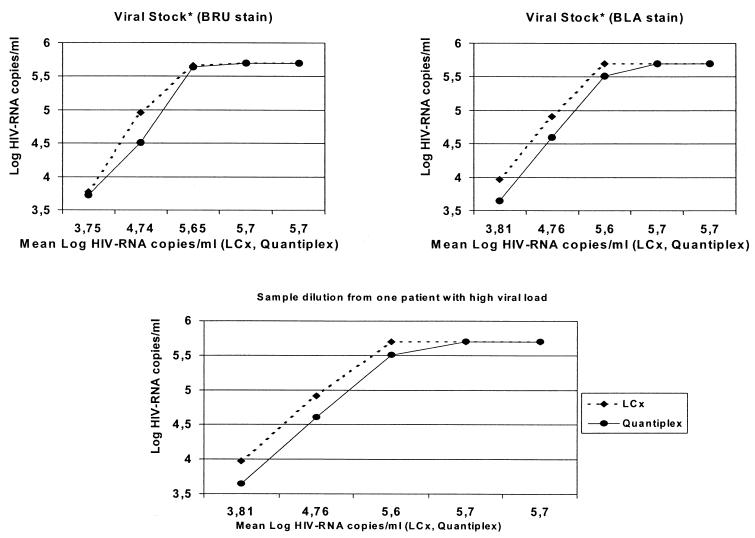
Serially diluted culture supernatants were quantitated in parallel by using LCx and Quantiplex. Excellent parallel-line linearity was observed across all dilutions assayed (Fig. 1). A strong correlation between both tests was noted (R = 0.98). However, HIV RNA values determined by LCx were uniformly higher (0.21 log unit, on average) than those determined by Quantiplex, although the difference was not statistically significant.
FIG. 1.
Linear comparison between LCx and Quantiplex.
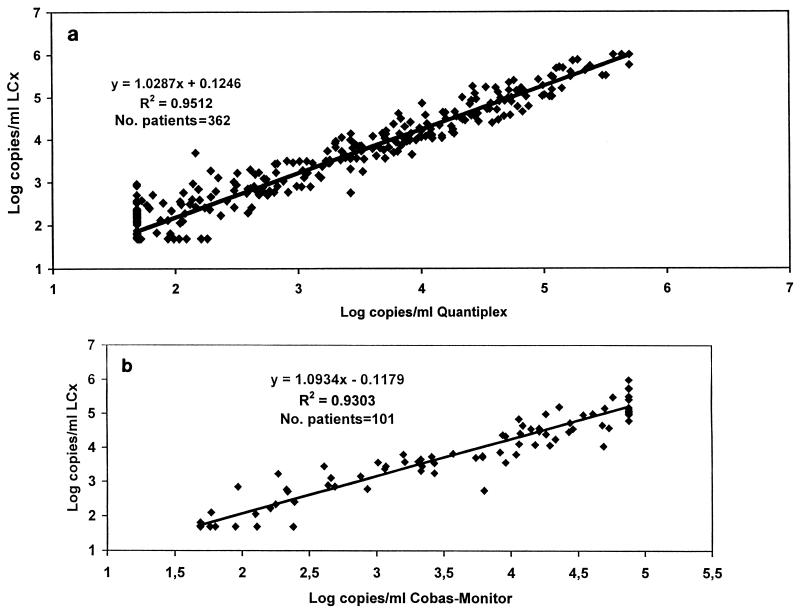
The 362 clinical samples selected for the comparison study were from individuals infected with HIV-1 subtype B. Most of these individuals had been exposed to antiretroviral drugs. The results obtained with all three assays were strongly correlated. Overall, 64.8% of the samples could be quantitated by both LCx and Quantiplex. Figure 2a shows a plot of the respective results. The fitted regression line is described by the equation y = 1.0287 x + 0.1246 (r2 = 0.95). The estimated intercept was 0.1246, with a standard error of 0.04. The slope was 1.0287, with a standard error of 0.012. This value differs slightly from the ideal value of 0.0 at the 0.05 level of confidence.
FIG. 2.
Correlation between LCx and Quantiplex (a) and Cobas-Monitor (b).
The comparative performance of the LCx and Cobas-Monitor assays was examined with 101 specimens. Overall, 73.3% of the specimens could be quantitated by both assays. LCx and Cobas-Monitor differed, on average, by 0.19 log copy/ml (0.12 to 0.27 log copy/ml; 95% confidence interval) and were not significantly different, as determined by a paired t test (P = 0.275). Figure 2b shows the scatter plot for LCx versus Cobas-Monitor. The linear regression equation for log HIV RNA copies per milliliter was y = 1.093 x − 0.118 (r2 = 0.93; R = 0.965), which represents the best fit in our study.
A qualitative comparison based on the lower limit (<50 HIV RNA copies/ml) showed a concordance of 88% between LCx and Quantiplex, with a κ index of 72%. Thirty-four samples found positive by LCx were found negative by Quantiplex, and 9 samples found positive by Quantiplex were found negative by LCx. In both situations, positive values did not reach statistically significant differences. When we compared LCx with Cobas-Monitor, we obtained a concordance of 93%, with a κ index of 79%. Only five samples found positive by Cobas-Monitor were found negative by LCx, and only two samples found positive by LCx were found negative by Cobas-Monitor.
A total of 88 specimens belonging to non-B subtypes were tested by using the LCx 0.2-ml protocol. The results obtained for 64 specimens carrying group M non-B subtypes were compared with those obtained by using Quantiplex. The results obtained for the remaining 24 specimens, obtained from group O-infected persons, were compared with those generated by using an in-house RT-PCR, MUPROVAMA (6), as well as by using a quantitative p24 antigen test.
Of the 64 non-B subtype specimens, 41 (67.2%) were quantitated by LCx, whereas Quantiplex quantitated 29 (45.3%) (P < 0.05). Of note, 19 samples below the limit of detection of LCx (178 copies/ml) were drawn from subjects on antiretroviral therapy, which might have truly suppressed viral replication in those subjects. In addition, viral loads in LCx tended to be higher than those in Quantiplex. Differences above 0.5 log unit were seen in 36% of the total specimens, 40% of the subtype C specimens, and about 26% of the subtype A and G specimens (Table 2).
TABLE 2.
Results obtained for quantifying HIV-1 non-B subtypes using LCx and Quantiplex
| HIV-1 subtype | No. of samples
|
% Discordance (>0.5 log unit) | |||
|---|---|---|---|---|---|
| Total | From patients receiving treatment | Detected by:
|
|||
| LCx | BDNA | ||||
| A | 15 | 4 | 12 | 8 | 27 |
| C | 10 | 9 | 4 | 3 | 40 |
| F | 2 | 1 | 1 | 2 | 100 |
| G | 35 | 20 | 23 | 15 | 26 |
| H | 2 | 1 | 1 | 1 | 50 |
| Total | 64 | 35 (55%) | 41 (64%) | 29 (45%) | 36 |
Of the HIV-1 group O specimens, 23 of 24 could be quantitated by LCx. Viral loads ranged from 178 copies/ml to nearly 100,000 copies/ml. Using MUPROVAMA and a quantitative p24 antigen assay, similar information was obtained with respect to viral burden in each sample (Table 3).
TABLE 3.
Positive results for testing HIV-1 group O specimens using LCx, a p24 antigen assay, and a noncommercial RT-PCR assay
| Patient | No. (rangea) of samples
|
||||
|---|---|---|---|---|---|
| Total | From patients receiving treatment | Detected by:
|
Noncommercial RT-PCR assay | ||
| LCx | p24 antigen assay | ||||
| 1 | 3 | 0 | 3 (3.6-3.8) | 3 (15-37) | 3 |
| 2 | 12 | 10 | 12 (3.1-4.9) | 12 (10-265) | 12 |
| 3 | 6 | 5 | 6 (3.2-4.5) | 5 (0-110) | 6 |
| 4 | 2 | 1 | 1 (<2.2-4.2) | 0 (0) | 1 |
| 5 | 1 | 1 | 1 (3.6) | 0 (0) | 1 |
| Total | 24 | 14 | 23 (<2.2-4.9) | 21 (0-265) | 23 |
Reported as log units for LCx and as picograms per microliter for the p24 antigen assay.
In conclusion, the performance of LCx was found to be satisfactory and comparable to that of the latest versions of current viral load assays for testing HIV-1 subtype B specimens. However, LCx showed a greater efficiency for measuring plasma viremia in subjects carrying other HIV-1 variants, including group O viruses.
Acknowledgments
This work was funded in part by grants from Asociación Investigación y Educación en SIDA (AIES), Comunidad Autónoma de Madrid (CAM), and Instituto de Salud Carlos III.
We thank John Hackett for reviewing the manuscript.
REFERENCES
- 1.Brodine, S. K., J. R. Mascola, P. J. Weiss, S. I. Ito, K. R. Porter, A. W. Artenstein, F. C. Garland, F. E. McCutchan, and D. S. Burke. 1995. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet 346:1198-1999. [DOI] [PubMed] [Google Scholar]
- 2.Collins, M. L., B. Irvine, D. Tyner, E. Fine, c. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. Ho. 1997. A branched DNA signal amplification assay for quantitation of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holguín, A., B. Rodés, U. Dietrich, and V. Soriano. 1999. HIV type 1 subtypes circulating in Spain. J. Med. Virol. 59:189-193. [PubMed] [Google Scholar]
- 4.Holguin, A., B. Rodés, and V. Soriano. 2000. Protease gene analysis of HIV type 1 non-B subtypes in Spain. AIDS Res. Hum. Retrovir. 16:1395-1403. [DOI] [PubMed] [Google Scholar]
- 5.Jenny-Avital, E., and S. Beatrice. 2001. Erroneously low or undetectable plasma HIV-1 RNA load, determined by PCR, in West African and American patients with non-B subtype HIV-1 infection. Clin. Infect. Dis. 32:1227-1230. [DOI] [PubMed] [Google Scholar]
- 6.Lu, W., L. Cao, L. Ty, M. Arlie, and J. M. Andrieu. 1999. Equivalent amplification of intrinsically variable nucleic acid sequences by multiple-primer-induced overlapping amplification assay: applications for universal detection and quantitation. Nat. Med. 5:1081-1085. [DOI] [PubMed] [Google Scholar]
- 7.Mas, A., M. E. Quiñones-Mateu, V. Soriano, and E. Domingo. 1996. Env gene characterization of the first HIV type 1 group O Spanish isolate. AIDS Res. Hum. Retrovir. 12:1647-1649. [DOI] [PubMed] [Google Scholar]
- 8.Mas, A., M. E. Quiñones-Mateu, E. Domingo, and V. Soriano. 1999. Phylogeny of HIV type 1 group O isolates based on env gene sequences. AIDS Res. Hum. Retrovir. 15:769-773. [DOI] [PubMed] [Google Scholar]
- 9.Mellors, J., A. Muñoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo. 1997. Plasma viral load and CD4+ lymphocytes as prognostic marker of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 10.Parry, J. V., G. Murphy, K. Barlow, K. Lewis, P. Rogers, F. Belda, A. Nicoll, C. McGarrigle, S. Cliffe, P. Mortimer, and J. Clewley. 2001. National surveillance of HIV-1 subtypes for England and Wales. J. Acquir. Immune Defic. Syndr. 26:381-388. [DOI] [PubMed] [Google Scholar]
- 11.Sun, R., J. Ku, H. Jayakar, J. C. Kuo, D. Brambilla, S. Herman, M. Rosenstraus, and J. Spadoro. 1998. Ultrasensitive reverse transcription PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 36:2964-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]