Abstract
We have developed a rapid (1-h) real-time fluorescence-based PCR assay with the Smart Cycler thermal cycler (Cepheid, Sunnyvale, Calif.) for the detection of Shiga toxin-producing Escherichia coli (STEC), as well as other Shiga toxin-producing bacteria. Based on multiple-sequence alignments, we have designed two pairs of PCR primers that efficiently amplify all variants of the Shiga toxin genes stx1 and stx2, respectively. These primer pairs were combined for use in a multiplex assay. Two molecular beacons bearing different fluorophores were used as internal probes specific for each amplicon. Assays performed with purified genomic DNA from a variety of STEC strains (n = 23) from diverse geographic locations showed analytical sensitivities of about 10 genome copies per PCR. Non-STEC strains (n = 20) were also tested, and no amplification was observed. The PCR results correlated perfectly with the phenotypic characterization of toxin production in both STEC and non-STEC strains, thereby confirming the specificity of the assay. The assay was validated by testing 38 fecal samples obtained from 27 patients. Of these samples, 26 were PCR positive for stx1 and/or stx2. Compared with the culture results, both the sensitivity and the negative predictive value were 100%. The specificity was 92%, and the positive predictive value was 96%. Moreover, this assay detected STEC from a sample in which the STEC concentration was at the limit of detection of the conventional culture methods and from a sample in which STEC was not detected by the conventional culture methods. This real-time PCR assay is simple, rapid, sensitive, and specific and allows detection of all Shiga toxin-producing bacteria directly from fecal samples, irrespective of their serotypes.
Although Escherichia coli is a member of the normal flora of the gastrointestinal tract of human and animals, several pathogenic types of E. coli can lead to human diseases. E. coli O157:H7 and other Shiga toxin-producing E. coli (STEC) strains have emerged in recent years as important human pathogens associated with a spectrum of diseases ranging from diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome (HUS) (3, 12, 16). Due to the morbidity and mortality associated with outbreaks and sporadic cases of STEC diseases, these pathogens are now considered major public health problems of worldwide importance.
E. coli O157:H7 is the serotype most frequently isolated from patients, but STEC strains of others serotypes (e.g., serotypes O111, O103, and O26) have also been associated with both outbreaks and sporadic cases of disease (4). STEC strains share a variety of virulence factors, including two Shiga toxins, Stx1 and Stx2, and a pathogenicity island, termed the locus for enterocyte effacement, that encodes proteins responsible for the intimate adherence of STEC to epithelial cells (20). The production of Shiga toxins by STEC strains has a major role in pathogenesis, particularly in the pathogenesis of HUS (16, 17, 19). E. coli Stx1 is essentially identical to the Shiga toxin produced by Shigella dysenteriae, whereas Stx2 has only 56% amino acid identity to Stx1 (10).
Our ability to control diseases associated with STEC and to limit outbreaks depends upon the rapid detection of these pathogens (20). The method used in most clinical microbiology laboratories is based on sorbitol MacConkey agar culture (SMAC), coupled with specific detection of the O157 antigen. This approach neglects other STEC serotypes and also other Shiga toxin-producing bacteria (20). We report here on the development of a real-time PCR assay for the rapid detection of all Shiga toxin-producing bacteria based on the amplification of the genes encoding Stx1 and Stx2, the major virulence factors of these organisms. This assay relies on PCR amplification with molecular beacons (24) with the Smart Cycler, a rapid thermal cycler that allows real-time fluorescence detection.
MATERIALS AND METHODS
Bacterial strains.
Twenty-three STEC strains and four Shiga toxin-producing S. dysenteriae strains from various origins were used in this study (Table 1). These strains were obtained from the American Type Culture Collection (Manassas, Va.), the Centers for Disease Control and Prevention (Atlanta, Ga.), the Laboratoire de Santé Publique du Québec (Sainte-Anne de Bellevue, Quebec, Canada), the microbiology laboratory of the Centre Hospitalier de l’Université Laval (CHUL; Sainte-Foy, Quebec, Canada), and Helge Karch (Institut für Hygiene und Mikrobiologie der Universität Würzburg, Würzburg, Germany). The specificity of the PCR-based assay was verified by using 20 non-STEC strains, which comprised 15 different serotypes of E. coli including O157:H7 and 3 Shigella species (Shigella boydii, Shigella sonnei, and Shigella flexnerii). Strains for which the toxin production status was unknown were tested by an immunoassay that allowed the detection and identification of Shiga toxins Stx1 and Stx2 by reverse passive latex agglutination (VTEC-RPLA kit; Denka Seiken Co., Ltd., Tokyo, Japan).
TABLE 1.
Shiga toxin-producing strains used in this study
| Species (strain) | Serotypea | Origin | Shiga toxin(s) produced |
|---|---|---|---|
| E. coli (ATCC 43890) | O157:H7 | United States (Calif.) | Stx1 |
| E. coli (ATCC 43894) | O157:H7 | United States (Mich.) | Stx1, Stx2 |
| E. coli (ATCC 43895) | O157:H7 | United States | Stx1, Stx2 |
| E. coli (CDC 97-3254) | O157:H7 | United States (Minn.) | Stx1 |
| E. coli (CDC 97-3330) | O157:H7 | United States (Ga.) | Stx2 |
| E. coli (CDC 98-3055) | O157:H7 | United States (Tenn.) | Stx1, Stx2 |
| E. coli (LSPQ 2127) | O157:H7 | Canada (Quebec) | Stx1, Stx2 |
| E. coli (LSPQ 3760) | O157:H7 | Canada (Quebec) | Stx1, Stx2 |
| E. coli (LSPQ 3761) | O157:H7 | Canada (Quebec) | Stx2 |
| E. coli (LSPQ 3762) | O157:H7 | Canada (Quebec) | Stx1, Stx2 |
| E. coli (CDC 98-3169) | O157:NM | United States (Oreg.) | Stx2 |
| E. coli (CDC 98-3186) | O145:NM | United States (Mo.) | Stx1, Stx2 |
| E. coli (CDC 99-3077) | O111:NM | United States (Va.) | Stx1, Stx2 |
| E. coli (CDC 97-3162) | O111:NM | United States (Mont.) | Stx1, Stx2 |
| E. coli (CDC 89-3156) | OX3:H21 | United States (Mich.) | Stx2 |
| E. coli (HK 7828/95) | O103:H2 | Germany | Stx1, Stx2 |
| E. coli (HK 2905/96) | O103:H2 | Germany | Stx1 |
| E. coli (HK 6037/96) | O111:NM | Germany | Stx1, Stx2 |
| E. coli (HK 5380/96) | O111:NM | Germany | Stx1 |
| E. coli (HK 5720/96) | O26:NM | Germany | Stx2 |
| E. coli (HK 8574/96) | O26:NM | Germany | Stx1 |
| E. coli (HK 7465/96) | O145:NM | Germany | Stx1 |
| E. coli (HK 3485/99) | O145:NM | Germany | Stx2 |
| S. dysenteriae (ATCC 11835) | Type 1 | United States | Stx1 |
| S. dysenteriae (CDC C898) | Type 1 | Mexico | Stx1 |
| S. dysenteriae (CDC BU2X1) | Type 1 | Burundi | Stx1 |
| S. dysenteriae (CDC F4101) | Type 1 | Bangladesh | Stx1 |
NM, nonmotile.
Culture.
Detection of O157:H7 strains in fecal samples was done by SMAC. Sorbitol-negative colonies were then tested for the O157 antigen by a latex agglutination assay, the E. coli O157 test (Oxoid, Basingstoke, England).
DNA extraction and sample preparation.
Genomic DNA was purified from the strains listed in Table 1 by using the G NOME DNA kit (Bio 101 Inc., Vista, Calif.). Five fecal samples (collected without transport medium) negative for STEC by SMAC and PCR were obtained from the microbiology laboratory of CHUL. These samples were spiked separately with 10-fold dilutions of an STEC culture (strain ATCC 43894) in the exponential phase of growth to obtain concentrations of 108 to 103 CFU per g of feces in order to evaluate the analytical sensitivity of the assay. Thirty-eight samples with or without Enteric Pathogen Transport Medium (Quelab Laboratories, Montreal, Quebec, Canada) were also obtained from STEC-infected and noninfected patients. These liquid fecal samples were initially tested by SMAC and stored at 4°C for about 48 h before PCR testing. A total of 100 μl of each of the spiked samples and the naturally infected samples were prepared for PCR without enrichment by using a rapid (10-min) sample preparation protocol as described previously (2, 14). At the end of the sample preparation procedure, each amplification reaction was performed with an extract from the equivalent of 0.15 μl of liquid feces.
PCR primers and probes.
The stx1 (n = 10) and stx2 (n = 22) gene sequences available from public databases were analyzed with the GCG Wisconsin package (version 10.0; Accelrys, San Diego, Calif.). Multiple-sequence alignments revealed conserved and specific regions of each Shiga toxin gene, stx1 and stx2. These regions were chosen for use in the design of two PCR primer pairs and molecular beacon probes with the help of Oligo primer analysis software (version 5.0; National Biosciences, Plymouth, Minn.) and the DNA fold program (M. Zuker, http://bioinfo.math.rpi.edu/∼zukerm/). Oligonucleotide primers were synthesized with a model 391 DNA synthesizer (Perkin-Elmer Corp., Applied Biosystems Division, Mississauga, Ontario, Canada). Molecular beacons were synthesized by Operon Technologies (Alameda, Calif.). The PCR primers and fluorescent probes used in this study are listed in Table 2. The stem of each molecular beacon was formed by joining a six-nucleotide complementary sequence at each end of the probe sequence. The stem sequence was designed to ensure that the molecular beacons adopt a hairpin structure at the optimal annealing temperature of the PCR when no target amplicons are present to avoid nonspecific fluorescence emission.
TABLE 2.
Oligonucleotides used in this study
| Primer or probe name | Target gene | Oligonucleotide sequence (5′ → 3′)a | Amplicon size (bp) |
|---|---|---|---|
| Primers | |||
| 1Slt224 | stx1 | ATG TCA GAG GGA TAG ATC CA | 185 |
| 1Slt385 | stx1 | TAT AGC TAC TGT CAC CAG ACA AT | |
| 2Slt537 | stx2 | AGT TCT GCG TTT TGT CAC TGT C | 160 |
| 2Slt678b | stx2 | CGG AAG CAC ATT GCT GAT T | |
| Probes | |||
| 1SltB1FAM | stx1 | GCG AGG CGC TTT GCT GAT TTT TCA CAT GTT ACC CCT CGC | |
| 2SltB1TET | stx2 | GCG AGG CAC TGT CTG AAA CTG CTC CTG TCC TCG C |
The underlined sequences of each probe constitute the stem of each molecular beacon. FAM, 6-carboxyfluorescein; TET, tetrachloro-6-carboxyfluorescein.
PCR amplification.
Amplification was performed from purified genomic DNA or DNA extracted from fecal samples. PCR conditions and reagent concentrations were optimized to obtain the optimal parameters described hereafter. Multiplex amplification was carried out with 0.8 μM (each) primers 1Slt224 and 1Slt385, 0.5 μM (each) primers 2Slt537 and 2Slt678b, 0.3 μM each molecular beacon, 8 mM MgCl2, 12.25 μg of bovine serum albumin, a 0.2 mM concentration of each of the four deoxynucleoside triphosphates (Amersham Pharmacia Biotech, Little Chalfont, England), 50 mM Tris-HCl, 16 mM NH4SO4, 1× TaqMaster (Eppendorf, Hamburg, Germany), 2.5 U of KlenTaq DNA polymerase (AB Peptides, St. Louis, Mo.) combined with TaqStart (Clontech Laboratories Inc., Palo Alto, Calif.), and 1 μl of purified genomic DNA or 1.5 μl of DNA prepared from a fecal sample in a final volume of 25 μl. The PCR amplification (60 s at 95°C and then 45 cycles of three steps consisting of 10 s at 95°C, 15 s at 56°C, and 5 s at 72°C) was performed with a Smart Cycler thermal cycler (Cepheid, Sunnyvale, Calif.). This rapid PCR cycling required approximately 40 min. The presence of amplified products was confirmed when the fluorescent signal exceeded an automatic noise-based defined threshold. Specificity testing was performed with 1 ng of DNA purified from each of the 20 non-STEC strains per PCR. The sensitivity of the assay was determined by using twofold dilutions of purified genomic DNA from the 27 Shiga toxin-producing strains. Detection of the PCR products was performed in real time by measuring the fluorescent signal emitted by the molecular beacon when it hybridizes to its target at the end of each annealing step. An internal control was used to monitor PCR inhibition by the fecal samples. Each internal control amplification was carried out in a separate reaction with 100 copies of a linearized recombinant plasmid that we have previously constructed for that purpose (14). This internal control was amplified with 0.4 μM each PCR primer and 0.2 μM internal control-specific molecular beacon in the presence of a fecal sample.
RESULTS
Ubiquity, specificity, and analytical sensitivity of the assay.
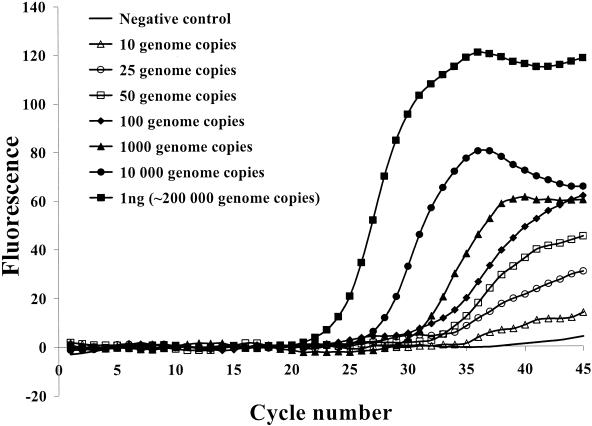
The ubiquity (i.e., ability to detect all Shiga toxin-producing bacteria) of the assay was demonstrated by testing STEC strains of different serotypes as well as Shiga toxin-producing S. dysenteriae strains of different origins (Table 1). For all 27 Shiga toxin-producing strains tested, the results obtained by the PCR assay correlated perfectly with the phenotypic characterization of toxin production determined by the Stx1- and Stx2-specific latex agglutination test (Table 1). None of the 20 non-STEC strains tested showed an amplification signal. The detection limit was found to be 10 to 50 genome copies per PCR for all 27 Shiga toxin-producing strains (Fig. 1).
FIG. 1.
Example of a sensitivity assay performed on the Smart Cycler thermal cycler with genomic DNA purified from an Stx1-positive E. coli strain. The fluorescence signal was from the 6-carboxyfluorescein dye of the stx1-specific probe.
Analytical sensitivity with spiked fecal samples.
The PCR assay was then used to detect STEC in fecal samples spiked with different concentrations of target bacteria. The rapid (10-min) sample preparation protocol was used. The detection limit was found to be about 105 CFU per g of feces. The constant intensity of the internal control signal indicated the absence of significant inhibition of the PCR amplifications.
Detection of STEC in feces from infected patients.
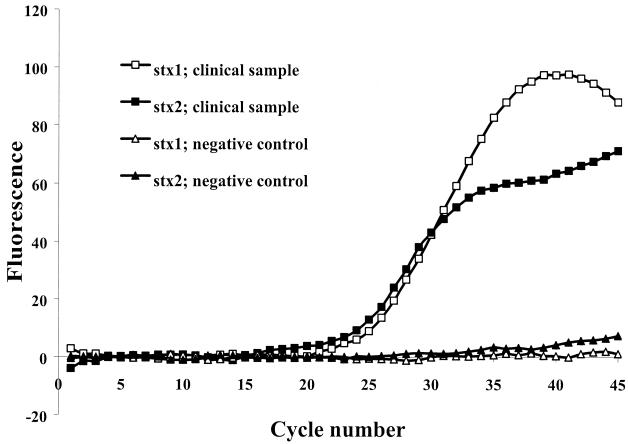
To evaluate the applicability of the assay to the detection of STEC in infected patients, 38 fecal samples from 27 different patients were tested. Twenty-six of these samples were positive by both the PCR assay and the culture-based assay. An example of a representative result for a PCR-positive sample is illustrated in Fig. 2. On the basis of the results of the latex agglutination assay, the STEC isolates found in these samples were all of serotype O157. One of these samples contained a low concentration of STEC, as we observed the growth of only one small non-sorbitol-fermenting colony after 48 h of incubation. However, the STEC in this sample was easily detected by the PCR assay. Of these 26 samples, 25 samples were positive for both stx1 and stx2, while 1 sample was positive for stx2 only. Of the 12 culture-negative samples tested, 1 sample was PCR positive for both stx1 and stx2. Clinical samples received with transport medium were amplified as efficiently as samples received without transport medium, suggesting that the transport medium had no significant influence on the performance of the assay. When the PCR results were compared with the SMAC results, both the sensitivity and the negative predictive value were 100%. The specificity was 92%, and the positive predictive value was 96%.
FIG.2.
Example of real-time detection of STEC in feces from one infected patient by amplification of stx1 and stx2 genes. The fluorescence signals were from the 6-carboxyfluorescein (white dots) or tetrachloro-6-carboxyfluorescein (black dots) dyes of the stx1- or stx2-specific probes.
DISCUSSION
STEC strains have emerged in recent years as important human pathogens implicated in severe complications including hemorrhagic colitis and HUS (3, 12). Thus, the rapid diagnosis of STEC infections in individual patients is crucial to avoid the development of an outbreak and to enable the implementation of control measures to prevent more cases. Despite this, most of the clinical microbiology laboratories are still using time-consuming culture-based methods (5, 7). Moreover, these methods are mostly focused on the detection of E. coli O157:H7, while other serotypes of STEC as well as other Shiga toxin-producing bacteria associated with both sporadic and epidemic cases of disease are neglected. At present, other immunoassays for Shiga toxin detection are available, but they still require an incubation period of about 20 h and are expensive (13). Other PCR assays have been developed for the detection of STEC, but most of them require gel electrophoresis for the analysis of PCR products (1). Moreover, most of these PCR assays require enrichment of the fecal sample to allow the detection of STEC (6, 18). These procedures increase the time to diagnosis and are not as specific as the use of internal hybridization probes to confirm the identity of the amplicon. Recently, other assays that use real-time PCR for the detection of STEC in food or fecal samples have been developed (8, 11, 15, 21, 22). However, none of these assays was used for the detection of STEC in naturally infected clinical fecal samples. Moreover, some of them require an incubation period for enrichment of the sample (8, 21, 22). Some of these assays do not allow the detection of all STEC strains, as they target only stx2 (11, 15) or the rfbE gene, specific for E. coli O157 (8). Thus, the objective of this study was to develop a real-time PCR assay for the rapid detection of all serotypes of STEC and other Shiga toxin-producing bacteria by the direct detection of stx1 and stx2 gene sequences from fecal samples. The ubiquity of the assay was demonstrated by efficient amplification of genomic DNA purified from a variety of STEC strains from diverse geographic origins and of different serotypes. Also, the absence of amplification of DNA from non-STEC strains demonstrated that the assay is specific for the detection of STEC. The assay was sensitive, as it allowed the detection of 10 to 50 genome copies for all 27 STEC and S. dysenteriae strains tested.
The applicability of the PCR assay to the detection of STEC directly from fecal samples was demonstrated by testing samples spiked with different concentrations of STEC cells. One of the major factors in optimizing an assay for the detection of STEC in fecal samples is overcoming the inhibition of the PCR by components found in feces. Following our 10-min sample preparation protocol (2, 14), we were able to detect about 105 bacteria per g of feces by PCR in less than 1 h for all spiked samples tested. These results are comparable to those of four other methods of extraction of STEC from feces evaluated by Holland et al. (9). However, all of the other sample preparation methods require from 1.5 to 4.5 h for completion. The analytical sensitivity of our PCR assay is approximately 1 log better than that of the conventional SMAC, which can detect a minimum of 106 STEC organisms per g of feces due to the high concentration (∼108 per g of feces) of the aerobic coliform bacilli found in the normal flora (23). Moreover, no significant PCR inhibition was observed with any of the fecal samples tested. Although other assays more sensitive than SMAC have been developed for the direct detection of Shiga toxin, such as tissue culture cytotoxicity assays, enzyme-linked immunosorbent assays, and latex agglutination assays (20), they are not routinely used in most clinical laboratories (4, 6).
Thus, the PCR assay was used for the detection of STEC directly from fecal samples obtained from infected patients and compared with the commonly used SMAC method. All clinical samples found to be positive for E. coli O157 by culture methods were also positive by our real-time PCR assay. All except 1 of the 26 SMAC-positive clinical samples tested were positive for both stx1 and stx2; the 1 sample that was the exception was positive only for stx2. The results of a previous study showed that STEC infections occur predominantly in young children (3). Although the sample size in our study was small, the majority of the positive samples were also obtained from young children and teenagers. All PCR results were obtained within 1 h following reception of the samples at the laboratory. This is much more rapid than conventional culture methods, which take at least 24 h. For most positive samples, the load of bacteria carrying the Shiga toxin gene(s) appeared to be relatively high, since the fluorescent signal became positive early in the amplification protocol (at about the 25th cycle). This corresponds to more than 107 bacteria per g of feces, based on testing with purified genomic DNA. By contrast, one clinical sample presented an STEC concentration at the limit of detection by the SMAC method, as it yielded the growth of only a single non-sorbitol-fermenting colony after 48 h of incubation. However, STEC was clearly detected in this sample by our PCR assay with a signal threshold at about the 32nd cycle, thereby confirming that our assay is more sensitive than the culture method. All except one of the clinical samples yielding no growth of O157 were found to be negative for stx genes; one sample, however, was positive for both stx1 and stx2. It is possible that the patient who provided this sample was infected by a non-O157 STEC strain or another Shiga toxin-producing bacterium which is undetectable by classical culture methods, showing the advantage of the PCR assay described here for the detection of any Shiga toxin-producing bacteria, irrespective of the species or serotype. The specificity of our PCR assay was calculated to be 92% when the results were compared to those of a “gold standard” reference test limited to the detection of E. coli O157 (SMAC). However, we cannot exclude the possibility that other Shiga toxin-producing species could have been present in the single PCR-positive sample missed by culture, hence increasing the real specificity and positive predictive values of our PCR assay to 100%.
In summary, our results demonstrate that the PCR-based assay described here, which targets both Shiga toxin genes, stx1 and stx2, is quite promising for direct detection of STEC and other Shiga toxin-producing bacteria from fecal samples in less than 1 h. A larger-scale clinical trial is needed to further validate this test.
Acknowledgments
This study was supported by grant PA-15586 from the Medical Research Council of Canada and Infectio Diagnostic (I.D.I.) Inc. S.D.B. received studentships from the Dr. Georges Phénix Foundation and FCAR-FRSQ Santé.
We thank Louise Coté and Nathalie Boucher of the microbiology laboratory of CHUL for providing the clinical samples. We thank Cepheid for providing the Smart Cycler thermal cycler. We also thank Pierre Harbec (Laboratoire de Santé Publique du Québec), Nancy A. Strockbine (Centers for Disease Control and Prevention), and Helge Karch for providing STEC strains.
REFERENCES
- 1.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, M. G., D. Ke, C. Ménard, F. J. Picard, M. Gagnon, M. Bernier, M. Ouellette, P. H. Roy, S. Marcoux, and W. D. Fraser. 2000. Rapid detection of group B streptococci in pregnant women at delivery. N. Engl. J. Med. 343:175-179. [DOI] [PubMed] [Google Scholar]
- 3.Besser, R. E., P. M. Griffin, and L. Slutsker. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355-367. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim, K. A. 2000. Role of non-O157 VTEC. J. Appl. Microbiol. 88:38S-50S. [DOI] [PubMed] [Google Scholar]
- 5.Bopp, C. A., F. W. Brenner, J. G. Wells, and N. A. Strockbine. 1999. Escherichia, Shigella, and Salmonella, p. 459-474. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 6.Fagan, P. K., M. A. Hornitzky, K. A. Bettelheim, and S. P. Djordjevic. 1999. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 65:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer, J. J., III. 1999. Enterobacteriaceae: introduction and identification, p. 442-458. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 8.Fortin, N. Y., A. Mulchandani, and W. Chen. 2001. Use of real-time polymerase chain reaction and molecular beacons for the detection of Escherichia coli O157:H7. Anal. Biochem. 289:281-288. [DOI] [PubMed] [Google Scholar]
- 9.Holland, J. L., L. Louie, A. E. Simor, and M. Louie. 2000. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 38:4108-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, M. P., R. J. Neill, A. D. O'Brien, R. K. Homes, and J. W. Newland. 1987. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 44:109-114. [DOI] [PubMed] [Google Scholar]
- 11.Kai, E., K. Ikebukuro, S. Hoshina, H. Watanabe, and I. Karube. 2000. Detection of PCR products of Escherichia coli O157:H7 in human stool samples using surface plasmon resonance (SPR). FEMS Immunol. Med. Microbiol. 29:283-288. [DOI] [PubMed] [Google Scholar]
- 12.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karmali, M. A., M. Petric, and M. Bielaszewska. 1999. Evaluation of a microplate latex agglutination method (Verotox-F assay) for detecting and characterizing verotoxins (Shiga toxins) in Escherichia coli. J. Clin. Microbiol. 37:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke, D., C. Ménard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 15.McKillip, J. L., and M. Drake. 2000. Molecular beacon polymerase chain reaction detection of Escherichia coli O157:H7 in milk. J. Food. Prot. 63:855-859. [DOI] [PubMed] [Google Scholar]
- 16.McLigeyo, S. O. 1999. Haemolytic uraemic syndrome: a review. East Afr. Med. J. 76:148-153. [PubMed] [Google Scholar]
- 17.Obrig, T. G. 1998. Interaction of Shiga toxins with endothelial cells, p. 303-311. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing strains. American Society for Microbiology, Washington, D.C.
- 18.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton, A. W., R. Morona, and J. C. Paton. 2000. A new biological agent for treatment of Shiga toxigenic Escherichia coli infections and dysentery in humans. Nat. Med. 6:265-270. [DOI] [PubMed] [Google Scholar]
- 20.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma, V. K., and S. A. Carlson. 2000. Simultaneous detection of Salmonella strains and Escherichia coli O157:H7 with fluorogenic PCR and single-enrichment-broth culture. Appl. Environ. Microbiol. 66:5472-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13:291-302. [DOI] [PubMed] [Google Scholar]
- 23.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]