Abstract
The phylogeny of 20 Actinobacillus actinomycetemcomitans strains isolated from patients with localized juvenile periodontitis (LJP) was investigated by using partial sequence analysis of 16S rRNA genes, arbitrarily primed PCR (AP-PCR), and four additional PCR assays that amplified polymorphic regions in the leukotoxin (lkt), cytolethal distending toxin (cdt), major fimbrial subunit (flp-1), and serotype-specific O polysaccharide gene clusters. Our analysis also included four strains isolated from healthy subjects and nine reference strains. We found that A. actinomycetemcomitans strains comprised three major phylogenetic lineages. One lineage consisted of serotype b strains, a second lineage consisted of serotype c strains, and a third lineage consisted of serotype a, d, e, and f strains. 16S rRNA sequences within each lineage were highly conserved (<1% base substitutions), whereas sequences between lineages were exceptionally divergent (1.9 to 5.0% substitutions). Two strains exhibited 16S rRNA sequences that were even more distantly related to those of the three major lineages (2.7 to 6.7% substitutions), indicating that additional minor lineages or variants exist. The distribution of 16S rRNA sequences and lkt, cdt, flp-1, and AP-PCR genotypes was consistent with a clonal population structure, with little evidence of assortative recombination between strains of different serotypes. Strains from all three major lineages were recovered from LJP patients, suggesting that phylogenetically diverse strains of A. actinomycetemcomitans carry pathogenic potential.
Actinobacillus actinomycetemcomitans is a gram-negative, capnophilic coccobacillus that colonizes the human oral cavity (24). A. actinomycetemcomitans is implicated in the etiology of localized juvenile periodontitis (LJP), a severe and rapid form of periodontal disease that affects more than 70,000 predominantly African-Americans in the United States annually (42). Infrequently, A. actinomycetemcomitans can enter the submucosa and cause extraoral infections including endocarditis, bacteremias, and abscesses (21). A. actinomycetemcomitans produces adhesive pili which cause tight adherence to surfaces such as glass, plastic, and saliva-coated hydroxyapatite (8, 20), a property that has been shown to play an important role in the ability of A. actinomycetemcomitans to colonize the mouths of rats (10). A. actinomycetemcomitans also produces several potential virulence factors, including leukotoxin, a 116-kDa secreted lipoprotein that specifically kills human polymorphonuclear leukocytes and macrophages (26), and cytolethal distending toxin, an immunosuppressive factor which is homologous to a family of toxins expressed by several other gram-negative bacteria (37). Little is known about the role of these and other potential virulence factors in the pathogenesis of LJP (11).
Numerous studies on the genetic diversity of A. actinomycetemcomitans strains isolated in Europe, Japan, and the United States have been reported (1, 3, 6, 12, 14-16, 28, 29, 34, 40, 43). These studies showed that there is considerable genetic variation among natural isolates of A. actinomycetemcomitans and suggested that variation in the virulence potential of strains may exist. For example, studies on the distribution of A. actinomycetemcomitans serotype a to e strains showed that serotype a and b strains are frequently isolated from LJP patients, serotype c strains are more common in nonoral infections and in healthy people, and serotype d and e strains are rare in all populations (1, 15, 28, 43). Studies on the leukotoxin gene showed that strains carrying a 530-bp deletion in the leukotoxin gene promoter that results in 10- to 20-fold-higher levels of leukotoxin production are almost exclusively isolated from LJP patients, suggesting that these strains may have an increased virulence potential (3, 12, 14-16). Other studies have shown an association between disease and certain restriction fragment length polymorphism genotypes (6) or arbitrarily primed PCR (AP-PCR) genotypes (1, 28).
The serologic specificity of A. actinomycetemcomitans serotype a to e strains is defined by five structurally and antigenically distinct O polysaccharide (O-PS) components of their respective lipopolysaccharide molecules (27, 31, 32). We recently identified a sixth A. actinomycetemcomitans serotype, designated serotype f, which synthesizes an O-PS molecule that is structurally distinct from those of serotype a to e strains but which shows immunological cross-reactivity with the O-PS molecule of serotype b strains (22). We also analyzed the DNA sequence of the gene cluster responsible for the synthesis of serotype f O-PS and showed that it is homologous to the serotype b, c, and e O-PS gene clusters (22). The aim of the present study was to clarify the genetic relationship between serotype f strains and the five previously identified A. actinomycetemcomitans serotypes. This report describes our analysis of 33 A. actinomycetemcomitans clinical isolates and reference strains, including strains representing all six of the currently known serotypes, by using four genotype-specific PCR assays and a comparison of partial 16S rRNA sequences.
MATERIALS AND METHODS
A. actinomycetemcomitans strains.
The strains analyzed in this study are listed in Table 1. Strains designated NJ, DF, and CU were isolated from cheek, tongue, saliva, or subgingival plaque samples obtained from subjects in New York City and Newark, N.J., between 1992 and 2000. All subjects were unrelated except for one pair of half siblings (NJ5500 and NJ5800). LJP was diagnosed according to the criteria described by Baer (2). Strains were identified as A. actinomycetemcomitans by the following criteria: (i) characteristic star-positive colony morphology on AAGMBV selective medium (9); (ii) CO2 required for growth; (iii) catalase positive; (iv) tight adherence to surfaces when grown in broth (9); (v) PCR amplification products with primers that hybridize to lktCA of strain JP2 (25), cdtABC of strain Y4 (37), and flp-1 or flp-2 of strain CU1000 (20); and (vi) amplification of a diagnostic PCR product in an A. actinomycetemcomitans serotype-specific PCR assay (22). Haemophilus aphrophilus strains NJ8500 and NJ8700 were distinguished from A. actinomycetemcomitans by the following criteria: (i) yellowish colony on AAGMBV selective medium; (ii) catalase negative; (iii) absence of PCR products with lktCA, cdtABC, and flp-1/flp-2 primers; (iv) absence of a PCR product in the A. actinomycetemcomitans serotype-specific PCR assay; (v) 16S rRNA sequence >97% identical to that of H. aphrophilus ATCC 33389 (5) (see Fig. 2); and (vi) presence of an fnr homologue >97% identical to that of H. aphrophilus ATCC 13252 (13). Strains were preserved as −70°C frozen stocks in 10% dimethyl sulfoxide and grown in Trypticase soy broth supplemented with yeast extract and glucose as previously described (9).
TABLE 1.
A. actinomycetemcomitans strains analyzed in this work
| Straina | Subjectb
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Serotype | 1kt | flp-1 | cdt | AP-PCR | 16S rRNA type | GenBank accession no. | Source or reference | Age (yr) | Sex | Race | LJP |
| DF2200 | a | 652 | o | 2 | 1 | I | AF355071 | This work | 27 | F | Afr | + |
| NJ3800 | a | 652 | 1 | 2 | 2 | II | AF355072 | This work | 7 | F | Cau | + |
| NJ4600 | a | 652 | o | 2 | 1 | I | AF355078 | This work | 15 | M | Afr | + |
| NJ5000 | a | 652 | 2 | 2 | 1 | I | AF355079 | This work | 22 | F | Afr | + |
| NJ6000 | a | 652 | o | 2 | 1 | I | AF355080 | This work | 13 | F | Afr | − |
| ATCC 29523 | a | 652 | 2 | 2 | 1 | I | M75038 | ATCC | ? | M | ? | ? |
| SUNYab75 | a | 652 | o | o | 1 | I | AF355067 | SUNY | 17 | F | Cau | + |
| NJ1000 | b | JP2 | 1 | 1 | 3 | II | AF355081 | This work | 18 | F | Afr | + |
| NJ3500 | b | JP2 | 1 | 2 | 3 | II | AF355082 | This work | 22 | F | Afr | + |
| NJ4500 | b | JP2 | 1 | 2 | 3 | II | AF355083 | This work | 19 | F | Afr | + |
| NJ5400 | b | JP2 | 1 | 2 | 3 | II | AF355084 | This work | 11 | F | Afr | + |
| NJ8800 | b | JP2 | 1 | 2 | 3 | II | AF355085 | This work | 6 | F | Afr | + |
| NJ5800 | b | JP2 | 1 | 2 | 3 | II | AF355086 | This work | 13 | M | Afr | − |
| JP2 | b | JP2 | 1 | 2 | 4 | II | AF355087 | Penn | 25 | M | Afr | + |
| NK1651 | b | JP2 | 1 | 2 | 3 | II | AGSP | SUNY | 18 | M | Afr | + |
| ATCC 29524 | b | 652 | 1 | o | 3 | II | M75037 | ATCC | 50 | F | Afr | ? |
| Y4 | b | 652 | 1 | 1 | 3 | II | M75035 | ATCC | ? | ? | ? | + |
| DF2000 | c | 652 | o | 4 | 3 | III | AF355076 | This work | 8 | F | Afr | + |
| DF2300 | c | 652 | 2 | o | 3 | III | AF355077 | This work | 40 | M | Afr | + |
| NJ2300 | c | 652 | 2 | 4 | 5 | III | AF355088 | This work | 30 | M | Cau | + |
| NJ2700 | c | 652 | 2 | 4 | 3 | III | AF355068 | This work | 18 | M | Afr | + |
| NJ6700 | c | 652 | 2 | 3 | 3 | IV | AF355089 | This work | 18 | F | Afr | + |
| NJ7200 | c | 652 | 2 | 4 | 3 | III | AF355090 | This work | 19 | F | Afr | + |
| NJ7500 | c | 652 | 2 | 3 | 5 | III | AF355091 | This work | 30 | M | Afr | + |
| NJ9300 | c | 652 | 2 | o | 3 | III | AF355092 | This work | 14 | F | Afr | + |
| NJ8400 | c | 652 | 2 | o | 3 | III | AF355093 | This work | 22 | M | Cau | − |
| Aa307 | c | 652 | 2 | nt | nt | III | AF355094 | SUNY | 60 | M | Cau | − |
| IDH781 | d | 652 | 2 | nt | nt | I | AF355069 | SUNY | 10 | M | Cau | − |
| NJ9500 | e | 652 | o | nt | 6 | V | AF355073 | This work | 22 | M | Cau | − |
| IDH1705 | e | 652 | 2 | nt | nt | I | AF355066 | SUNY | 32 | M | Cau | − |
| CU1000 | f | 652 | o | 3 | 1 | I | AF355070 | 22 | 13 | F | Afr | + |
| NJ9100 | f | 652 | o | 3 | 1 | I | AF355074 | This work | 21 | M | His | + |
| NJ9200 | f | 652 | 2 | 3 | 1 | I | AF355075 | This work | 23 | M | Asi | + |
Boldfacing denotes the 20 LJP strains isolated in our laboratory. The serotype, 1kt, flp-1, cdt, and AP-PCR columns show the results of PCR assays described in the text. o, no PCR product; nt, not tested. 16S rRNA types correspond to those shown in Fig. 2. The GenBank accession number is for the 16S rRNA sequence. AGSP, Actinobacillus Genome Sequencing Project (www.genome.ou.edu/act.html). Source: ATCC, American Type Culture Collection; SUNY, J. Zambon and H. Reynolds, State University of New York at Buffalo; Penn, E. T. Lally, University of Pennsylvania, Philadelphia.
Race: Cau, Caucasian; Afr, African or African-American; His, Hispanic; Asi, Asian. LJP: +, LJP; −, periodontally healthy. ?, Not known.
FIG. 2.
Polymorphisms in 16S rRNA sequences. Numbers on top correspond to coordinates of E. coli 16S rRNA sequence (5). Letters in parentheses indicate serotype. Bases occupying the indicated positions in the A. actinomycetemcomitans DF2200 sequence are shown. Gaps (indicated by hyphens) were inserted after E. coli positions 77, 93, 250, 455, 469, and 477. A dot indicates that the base is identical to that in DF2200. Roman numerals indicate five 16S rRNA types discussed in the text. The GenBank accession numbers for strains not listed in Table 1 are as follows: A. actinomycetemcomitans ATCC 29522, M75036; NCTC9710, M75039; H. aphrophilus NJ8700, AF355095; NJ8300, AF355096; ATCC 33389, M75041; H. paraphrophilus, M75042; H. influenzae, M35019; P. multocida, M35018; and H. segnis, M75043.
PCR assays.
Genomic DNA was prepared from test strains by using a DNeasy tissue kit (Qiagen, Valencia, Calif.) according to the instructions provided by the manufacturer. PCR primers were purchased from Biosource International (Camarillo, Calif.) or Integrated DNA Technologies (Coralville, Iowa). Unless otherwise indicated, PCR assays were performed in 0.5-ml thin-walled tubes containing 10 μl of 10× PCR buffer (500 mM KCl; 100 mM Tris-HCl, pH 8.3; 150 mM MgCl2), 1 mM deoxynucleoside triphosphates, 2.5 U of Taq DNA polymerase (Applied Biosystems, Foster City, Calif.), and 5 ng of genomic DNA as a target in a 100-μl reaction volume overlaid with 100 μl of mineral oil. A 10-μl aliquot of each PCR was electrophoresed through a 1.0 or 1.8% agarose gel in 1× TAE buffer or a 5% acrylamide-0.17% bisacrylamide gel in 1× Tris-borate-EDTA buffer, and the PCR products were visualized by staining with ethidium bromide (36). The specific primers and reaction conditions for the PCR assays were as follows.
(i) Serotype assay.
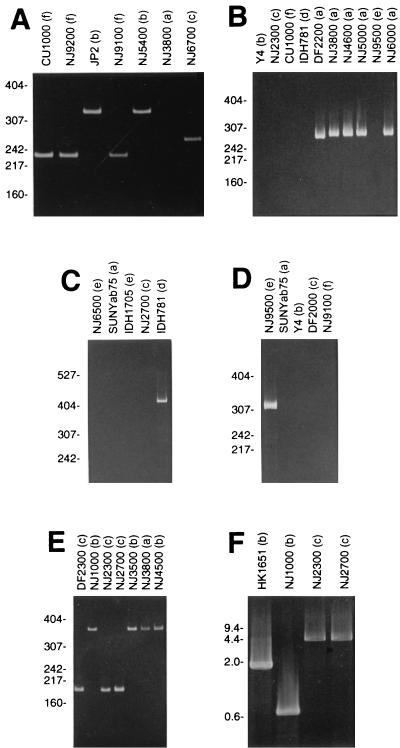
Four PCRs were used to amplify unique sequences in the gene clusters responsible for the synthesis of serotype a-, b-, c-, d-, e-, and f-specific O-PS molecules as previously described (22). The first PCR utilized four primers that hybridized to the serotype b-, c-, and f-specific O-PS gene clusters, resulting in the amplification of 333-, 268-, and 232-bp PCR products for serotype b, c, and f strains, respectively (Fig. 1A). The remaining three PCRs amplified sequences in the serotype a-, d-, and e-specific O-PS gene clusters individually, resulting in the synthesis of 293-, 411-, and 311-bp PCR products for serotype a, d, and e strains, respectively (Fig. 1B to D). Assays were validated with DNAs from strains SUNYab75, Y4, NJ2700, IDH781, NJ9500, and CU1000 (serotypes a to f, respectively). Seroclassification of serotype a, b, c, and f strains was confirmed by enzyme-linked immunosorbent assay with anti-serotype a-, b-, c-, and f-specific rabbit antisera as previously described (22).
FIG. 1.
Examples of PCR assays utilized in this study. Lane labels indicate strain name with serotype in parentheses. (A) Serotype b-, c-, and f-specific quadriplex reaction; (B) serotype a-specific reaction; (C) serotype d-specific reaction; (D) serotype e-specific reaction; (E) flp-1-specific reaction; (F) cdt-specific reaction. Panel F shows the PCR products amplified with primer pair P1. The sizes of molecular weight standards run in an adjacent lane are shown on the left. Molecular weight standard sizes are given in base pairs for panels A to E and in kilobase pairs for panel F. Panels A to E show 5% acrylamide gels; panel F shows a 1.0% agarose gel.
(ii) lkt assay.
The characteristic 530-bp deletion present in the leukotoxin promoter of highly leukotoxic strains was detected by using a PCR assay as previously described (16). PCR products were analyzed on 5% acrylamide gels as described above. Strains that amplified a 504-bp PCR product were classified as strain JP2-like (highly leukotoxic); those that amplified a 1,034-bp PCR product were classified as strain 652-like (minimally leukotoxic).
(iii) flp-1 assay.
PCR primers 5-AGCCACTTTCATCTGCGCTGG-3 and 5-ATTGCATCTAGTCTCTTTCG-3 were used to amplify a region located ca. 0.5 kb upstream from the flp-1 start codon (corresponding to bp 1448 to 1626 in GenBank accession no. AB005741). Thirty PCR cycles of 94°C, 55°C, and 72°C (1 min each) were performed. Strains that amplified a 363-bp PCR product were classified as type 1 (strain NK1651-like); those that amplified a 179-bp PCR product were classified as type 2 (strain 304-like) (Fig. 1E).
(iv) cdt assay.
PCR primer pairs P1 (5-GTCAACGAAGCTCCCAAGAACGCT-3 and 5-TGTACCTCTCCTTAGATCCATCCT-3), P2 (5-ACGTTCACCACCCAGTAACAGGAT-3 and 5-TTCGCCATAACGTCAACGTAGTAA-3), and P3 (5-ATCCCGGGAAACGGGTAACGG-3 and 5-ACAACAAGCAACGTTAGGTCTGTG-3) were used to amplify three regions upstream from the cdtABC cytolethal distending toxin gene cluster. PCR primer pair P1 amplified the region corresponding to bp 1 to 667 of the cdtABC sequence of strain Y4 reported by Sugai et al. (37). PCR primer pair P1 amplified a 1,893-bp PCR product in strain NK1651, due to a 1,226-bp sequence located 607-bp upstream from the cdtA start codon in strain NK1651 (Actinobacillus Genome Sequencing Project, www.genome.ou.edu/act.html) that is absent in strain Y4 (37). PCR primer pairs P2 and P3 hybridized within the 1,226-bp fragment present in strain NK1651. PCRs were incubated at 94°C for 5 min, followed by 30 cycles of PCR at 94°C, 55°C, and 72°C (1 min each), with a final elongation step of 72°C for 5 min. When primer pairs P2 and P3 were used, the annealing temperature was changed from 55 to 60°C. Four cdt genotypes (designated 1 to 4 in Table 1) were defined as follows: strains that amplified a 667-bp PCR product with primer pair P1 were classified as type 1; those that amplified a 1,893-bp PCR product with primer pair P1, a 1,272-bp PCR product with primer pair P2, and a 932-bp PCR product with primer pair P3 were classified as type 2; those that amplified a 932-bp PCR product with primer pair P3 were classified as type 3; those that amplified a 4.4-kb PCR product with primer pair P1, a 1,272-bp PCR product with primer pair P2, and a 932-bp PCR product with primer pair P3 were classified as type 4 (Fig. 1F).
(v) AP-PCR assay.
AP-PCR (41) was performed by using Ready-To-Go RAPD Analysis Beads (Amersham Pharmacia, Piscataway, N.J.) according to the instructions supplied by the manufacturer. AP-PCR products were analyzed on 5% polyacrylamide gels as described above. Three of the six AP-PCR primers supplied with the kit (RAPD analysis primers 2 [5-GTTTCGCTCC-3], 3 [5-GTAGACCCGT-3], and 4 [5-AAGAGCCCGT-3]) amplified AP-PCR products that were clearly and reproducibly polymorphic in the strains tested. These included 780- and 470-bp AP-PCR products amplified by RAPD analysis primer 2, a 625-bp AP-PCR product amplified by RAPD analysis primer 3, and a 530-bp AP-PCR product amplified by RAPD analysis primer 4. Six AP-PCR genotypes (designated 1 through 6 in Table 1) were defined as follows: AP-PCR genotype 1 amplified the 625- and 780-bp AP-PCR products; AP-PCR genotype 2 amplified all four AP-PCR products; AP-PCR genotype 3 amplified the 470- and 530-bp AP-PCR products; AP-PCR genotype 4 amplified the 530-bp AP-PCR product; AP-PCR genotype 5 amplified the 470-, 530-, and 780-bp AP-PCR products; and AP-PCR genotype 6 amplified the 470-bp AP-PCR product.
16S rRNA sequencing.
PCR primers 5-GCTTAACACATGCAAGTCGG-3 and 5-TGCTGGCACGGAGTTAGCCG-3 were used to amplify a 478-bp region of the A. actinomycetemcomitans 16S rRNA gene corresponding to coordinates 46 to 523 of the Escherichia coli 16S rRNA sequence (5). The ends of the PCR products were made flush by using E. coli DNA polymerase (Klenow fragment) and ligated into the EcoRV site of plasmid LITMUS28 (New England Biolabs, Beverly, Mass.) as described previously (36). Plasmid DNAs were prepared by using a QIAprep Spin Miniprep kit (Qiagen) and subjected to DNA sequence analysis on an ABI model 377 PRISM automated sequencer. Sequences were aligned by using CLUSTALW 1.8 (19), and the alignment was manually adjusted to minimize the number of multistate positions.
Phylogenetic analysis.
Phylogenetic trees were generated by using the software package PAUP 4.0b4a (Phylogenetic Analysis Using Parsimony), developed by D. L. Swofford (Sinauer Associates, Sunderland, Mass.), by using the heuristic search algorithm with default options. Attributes included those listed in Table 1, as well as the genotype of the flp-1 structural gene as determined by DNA sequence analysis (J. B. Kaplan, unpublished data). The results of lkt, flp-1, and cdt PCR assays which failed to produce a PCR product were considered unknown. Strains carrying the JP2-like leukotoxin promoter were topologically constrained to a single clade, since these strains have previously been shown to be monophyletic (12, 14, 16). The phylogram was rooted between the serotype {b,c} and {a,d,e,f} clusters, since these two groups have previously been shown to be genotypically distinct based on genomic DNA fingerprinting, restriction fragment length polymorphisms, and multilocus enzyme electrophoresis (15, 16, 34). Rooting the tree with a hypothetical distant outgroup placed the root on branch 4 of the phylogram (Fig. 2), which did not alter the conclusions drawn from the analysis. Because of insufficient data or a high level of divergence or assortative recombination, strains NJ3800 and NJ9500 could not be confidently positioned in the phylogram and were therefore excluded from the analysis.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the 16S rRNA sequences reported in this study are listed in Table 1 and in the legend of Fig. 2.
RESULTS
A. actinomycetemcomitans strains.
Table 1 lists the 33 A. actinomycetemcomitans strains analyzed in this study. A total of 20 of these strains were isolated in our laboratory from specimens obtained from localized juvenile periodontitis patients (referred to as the 20 LJP strains and indicated by boldface in Table 1). Our analysis also included four A. actinomycetemcomitans strains isolated from the oral cavities of healthy subjects (NJ5800, NJ6000, NJ8400, and NJ9500) and nine reference strains (ATCC29523, SUNYab75, ATCC29524, Y4, JP2, NK1651, Aa307, IDH781, and IDH1705).
Serotypes.
All 33 A. actinomycetemcomitans strains amplified one of the six O-PS gene clusters in the serotype-specific PCR assay (Table 1). The serotypes of the nine reference strains were consistent with the reported seroclassifications of these strains (7, 34, 35, 43). Among the 20 LJP strains, four were serotype a (20%), five were serotype b (25%), eight were serotype c (40%), and three were serotype f (15%). The four strains isolated from healthy subjects included one each of serotypes a, b, c, and e.
Leukotoxin promoter.
All 33 A. actinomycetemcomitans strains amplified a PCR product in the leukotoxin-specific PCR assay. Of the 33 strains, 8 carried the 530-bp deletion in the leukotoxin promoter (highly leukotoxic), including 5 of the 20 LJP strains, 1 strain isolated from a healthy subject (NJ5800), and reference strains JP2 and NK1651. All highly leukotoxic strains were serotype b, but two serotype b reference strains (ATCC 29524 and Y4) contained the minimally leukotoxic promoter. It has been shown that highly leukotoxic strains of A. actinomycetemcomitans are frequently isolated from LJP patients (12, 14), Africans and African-Americans (3, 12), and younger subjects (12). In our study, highly leukotoxic strains comprised 25% of the isolates from LJP patients (5 of 20) and 25% of the isolates from healthy subjects (1 of 4). All six of these highly leukotoxic strains were isolated from African-Americans. Subjects harboring highly leukotoxic strains were younger than those harboring minimally leukotoxic strains (14.8 versus 20.3 years old), but the difference was not statistically significant (P = 0.18, unpaired two-tailed t test).
flp-1 upstream region.
Comparison of the DNA sequence upstream from flp-1 in strain NK1651 (serotype b) (Actinobacillus Genome Sequencing Project [www.genome.ou.edu/act.html]) with the sequence upstream from the flp-1 gene of strain 304 (serotype a) (18) revealed a 184-bp sequence located 0.5 kb upstream from the flp-1 start codon in strain NK1651 that was absent in strain 304. The effect of this 184-bp sequence on the expression of flp-1 or other genes is not known. We used a PCR assay to detect the presence of this 184-bp sequence in all 33 test strains (Table 1). Eight strains failed to produce a PCR product in this assay. The structure of the flp-1 upstream regions in these eight strains in not known. Of the remaining 25 strains, 11 contained the 184-bp sequence (designated type 1 in Table 1), including all 10 of the serotype b strains and 1 serotype a strain (NJ3800). The remaining serotype a strains and all serotype c, d, e, and f strains lacked the 184-bp sequence (type 2 in Table 1). Among the 20 LJP strains, subjects harboring strains containing the 184-bp sequence (type 1) were significantly younger than those harboring strains with the strain 304-like flp-1 upstream region (type 2) (13.7 ± 6.2 versus 23.8 ± 8.1 years old; P = 0.02).
cdt upstream region.
Twenty-four strains could be classified as cdt genotypes 1 to 4 based on the cdt-specific PCR assay (Table 1). The distribution of the four cdt genotypes correlated with serotypes: cdt genotype 1 was found only in serotype b strains; cdt genotype 2 was found only in serotype a and b strains; cdt genotype 3 was found only in serotype c and f strains; and cdt genotype 4 was found only in serotype c strains. Over one half of the strains belonged to the cdt genotype 2. The structure of the cdtABC upstream region in the five strains that failed to amplify a PCR product in this assay is not known.
AP-PCR.
Of the 33 test strains, 30 were analyzed by using AP-PCR genotyping. The distribution of the six detected AP-PCR genotypes correlated with serotypes: AP-PCR genotypes 1 and 2 were found only in serotype a and f strains, and AP-PCR genotypes 3, 4 and 5 were found only in serotype b and c strains. AP-PCR genotype 6 was found in only one serotype e strain. A total of 25 of the 30 strains tested (83%) belonged to AP-PCR genotypes 1 and 3.
16S rRNA sequencing.
Figure 2 shows the nucleotide polymorphisms between coordinates 46 and 523 in the 16S rRNA sequences of 35 A. actinomycetemcomitans strains and seven strains of closely related bacteria. This section of the 16S rRNA molecule is the most informative for determining the relatedness of bacteria because it contains several variable regions (30, 39). The 35 A. actinomycetemcomitans strains include the 33 strains listed in Table 1, as well as reference strains ATCC29522 (serotype b) and NCTC9710 (serotype c). Nucleotide sequences were derived from published sources (5) or were determined in our laboratory. The nucleotide sequence of the 16S rRNA gene from strain NK1651 was obtained from the Actinobacillus Genome Sequencing Project (www.genome.ou.edu/act.html). There are 92 sites that are polymorphic in at least one of the 42 strains shown in Fig. 2, and 52 sites that are polymorphic in at least one of the 35 A. actinomycetemcomitans strains. Three major A. actinomycetemcomitans 16S rRNA types were observed (labeled I to III in Fig. 2). Nucleotide sequences were highly conserved within each 16S rRNA type (0.0 to 0.8% nucleotide substitutions) but highly divergent between types (1.9 to 5.0% substitutions). In addition, strains NJ6700 (serotype c) and NJ9500 (serotype e) contained 16S rRNA sequences (designated types IV and V in Fig. 2, respectively) that were more divergent (2.7 to 6.7% substitutions) from those of the three major types. The distribution of the three major 16S rRNA types was serotype specific: type I included all serotype a, d, e, and f strains; type II included all serotype b strains and one serotype a strain (NJ3800); and type III included all serotype c strains. The sequence of the type III 16S rRNA contained a large region (coordinates 77 to 226 in Fig. 2) that was homologous to the corresponding region of the 16S rRNAs of H. aphrophilus and H. paraphrophilus. This region corresponds to the stem regions designated helices 6 to 18 of the 16S rRNA molecule (helix numbering according to Van de Peer et al. [39]). The 16S rRNA sequence from strain NJ6700 contained this same region, but it also contained a sequence homologous to helix 18 of the H. segnis 16S rRNA (coordinates 455 to 490). The 16S rRNA sequence from strain NJ9500 contained a sequence homologous to helix 6 of the H. aphrophilus and H. paraphrophilus 16S rRNAs (coordinates 77 to 92) and a sequence homologous to helix 18 of the Pasteurella multocida 16S rRNA (coordinates 455 to 478). These interspecific homologous regions may have resulted from retention of homologous sequences in some strains with concomitant DNA sequence evolution in other strains or from horizontal gene transfer between A. actinomycetemcomitans and other closely related oral bacteria (38).
Phylogenetic analysis of A. actinomycetemcomitans strains.
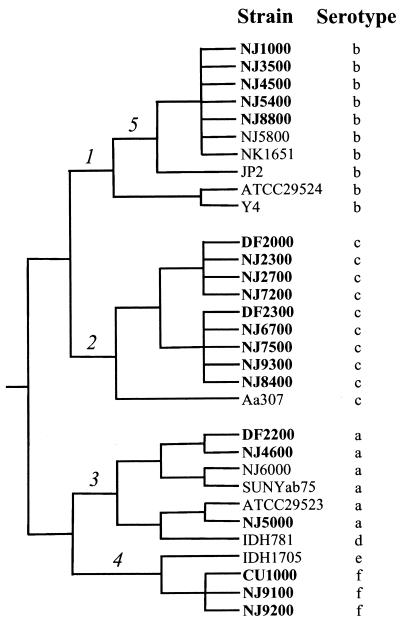
Phylogenetic analysis of A. actinomycetemcomitans strains based on the six attributes listed in Table 1 (serotype, 16S rRNA type, and lkt, flp-1, cdt, and AP-PCR genotypes) and the genotype of the flp-1 structural gene (Kaplan, unpublished) was performed, and three clusters were observed (Fig. 3). One cluster consisted of serotype b strains, a second cluster consisted of serotype c strains, and a third cluster consisted of serotype a, d, e, and f strains. Serotype b and c strains formed distinct, monophyletic groups within the serotype {b,c} cluster (branches 1 and 2, respectively, in Fig. 3). Our analysis also revealed that serotype f strains were closely related to serotypes a, d, and e, confirming that serotype f strains constitute a unique A. actinomycetemcomitans serotype distinct from serotype b (22). The tree shown in Fig. 3 also suggests that the phylogenetic relatedness of strains within the serotype {a,d,e,f} cluster is {a,d}{e,f} (branches 3 and 4, respectively), although more data are needed to confidently resolve the branching pattern in this part of the tree. Highly leukotoxic strains of A. actinomycetemcomitans that carry the 530-bp deletion in the leukotoxin promoter (JP2-like strains in Table 1) have previously been shown to comprise a monophyletic group based on multilocus enzyme electrophoresis and genomic DNA fingerprinting (16), DNA sequence analysis (14), and AP-PCR genotyping (12). Our data suggest that the 530-bp deletion in the leukotoxin promoter occurred in a branch within the serotype b cluster (branch 5 in Fig. 3). Our findings are in good agreement with previous phylogenetic analyses of A. actinomycetemcomitans (15, 34).
FIG. 3.
Phylogeny of A. actinomycetemcomitans. Boldface type indicates strains isolated from LJP patients in this study. Numbers 1 to 5 denote branches discussed in the text. Horizontal branch lengths are not to scale and do not metrically represent evolutionary change.
DISCUSSION
The distribution of PCR genotypes among the strains analyzed in our study was consistent with previous population genetic analyses of A. actinomycetemcomitans which showed considerable genetic diversity both between and within serotype a to e strains, with little evidence of assortative recombination between strains of different serotypes (1, 7, 15, 28, 34). Our data revealed five types of A. actinomycetemcomitans 16S rRNA sequences which displayed 0.0 to 6.7% sequence divergence, higher than the amount of interspecific 16S rRNA divergence observed many other bacteria. 16S rRNA sequence divergence of 1 to 2% corresponds to a species-level difference for most bacteria (30). We recently identified a strain isolated from the gingival pocket of a rapidly progressive periodontitis patient in Japan (strain OEN12-5, serotype e) that contains a 16S rRNA sequence identical to that of strain NJ9500 (also serotype e) (J. B. Kaplan and S. Kokeguchi, unpublished data), suggesting that 16S rRNA type V constitutes a distinct and geographically diverse A. actinomycetemcomitans clade. Because of the small number of strains analyzed to date, it is possible that other A. actinomycetemcomitans 16S rRNA types exist.
Three of the five A. actinomycetemcomitans 16S rRNA types identified in our study (types III to V) contained sequences that were homologous to sequences in the 16S rRNAs of H. aphrophilus and H. paraphrophilus, a finding consistent with the close evolutionary relationship of A. actinomycetemcomitans to these species (4, 5, 17, 33). Our PCR genotyping data, however, indicated that A. actinomycetemcomitans 16S rRNA types III to V were more closely related to other A. actinomycetemcomitans strains than to H. aphrophilus or H. paraphrophilus. 16S rRNA type III strains include type strain NCTC9710, which has also been shown to be more closely related to other A. actinomycetemcomitans strains than to H. aphrophilus or H. paraphrophilus, based on DNA-DNA hybridization (4, 33) and partial infB sequence comparisons (17). These data support the classification of 16S rRNA types III to IV as A. actinomycetemcomitans.
Previous studies showed that highly leukotoxic (JP2-like) strains are isolated almost exclusively from LJP patients (12, 14), suggesting that these strains may have an increased virulence potential. Highly leukotoxic strains also comprise approximately one-half of the strains isolated from Africans and African-Americans but only 0 to 2% of strains isolated from Europeans and Asians (3, 12, 14, 15). These findings led to the hypothesis that LJP may have two different etiologies and epidemiologies: in Caucasians LJP may be associated with diverse A. actinomycetemcomitans clones acting as opportunistic pathogens, whereas in some Africans and African-Americans LJP may be associated with JP2-like strains acting as exogenous pathogens (23). It is also possible that JP2-like strains are acting as opportunistic pathogens that are simply more prevalent in Africans and African-Americans, who have an increased susceptibility to LJP for other reasons, perhaps related to host determinants. In the present study, all six JP2-like strains were isolated from African-Americans, but JP2-like strains constituted less than one-third (5 of 16) of the strains isolated from African-American LJP patients. The fact that phylogenetically diverse strains were recovered from LJP patients in our study supports the hypothesis that A. actinomycetemcomitans plays the role of an opportunistic pathogen (14, 15, 34). Since all data presented to date have been cross-sectional in nature, it is impossible to determine the temporal relationship of JP2-like strains to disease. This question can only be resolved in a prospective longitudinal study that measures the frequency of conversion from health to disease in a sufficient number of healthy subjects who carry JP2-like and non-JP2-like strains. Resolution of this question could have implications for treatment strategies (23).
Acknowledgments
We thank Paul Planet for critically reviewing the manuscript; Homer Reynolds, Sigmund Socransky, Ben Hammond, Edward Lally, and Sirkka Asikainen for providing A. actinomycetemcomitans reference strains and patient histories; and Malvin Janal for helpful discussions.
REFERENCES
- 1.Asikainen, S., C. Chen, and J. Slots. 1995. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol. Immunol. 10:65-68. [DOI] [PubMed] [Google Scholar]
- 2.Baer, P. N. 1971. The case for periodontitis as a clinical entity. J. Periodontol. 42:516-519. [DOI] [PubMed] [Google Scholar]
- 3.Contreras, A., T. Rusitanonta, C. Chen, W. G. Wagner, B. S. Michalowicz, and J. Slots. 2000. Frequency of 530-bp deletion in Actinobacillus actinomycetemcomitans leukotoxin promoter region. Oral Microbiol. Immunol. 15:338-340. [DOI] [PubMed] [Google Scholar]
- 4.Coykendall, A. L., J. Setterfiels, and J. Slots. 1983. Deoxyribonucleic acid relatedness among Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, and other Actinobacillus species. Int. J. Syst. Bacteriol. 33:422-424. [Google Scholar]
- 5.Dewhirst, F. E., B. J. Paster, I. Olsen, and G. J. Fraser. 1992. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J. Bacteriol. 174:2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiRienzo, J. M., J. Slots, M. Sixou, M.-A. Sol, R. Harmon, and T. L. McKay. 1994. Specific genetic variants of Actinobacillus actinomycetemcomitans correlate with disease and health in a regional population of families with localized juvenile periodontitis. Infect. Immun. 62:3058-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dogan, B., M. Saarela, and S. Asikainen. 1999. Genotyping of Actinobacillus actinomycetemcomitans serotype d isolates based on polymerase chain reaction. Oral Microbiol. Immunol. 14:387-390. [DOI] [PubMed] [Google Scholar]
- 8.Fine, D. H., D. Furgang, J. B. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 9.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 10.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 46:1065-1078. [DOI] [PubMed] [Google Scholar]
- 11.Fives-Taylor, P. M., D. M. Meyer, K. P. Mintz, and C. Brissette. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20:136-167. [DOI] [PubMed] [Google Scholar]
- 12.Haraszthy, V. I., G. Hariharan, E. M. B. Tinoco, J. R. Cortelli, E. T. Lally, E. Davis, and J. J. Zambon. 2000. Evidence for the role of highly leukotoxic Actinobacillus actinomycetemcomitans in the pathogenesis of localized juvenile and other forms of early-onset periodontitis. J. Periodontol. 71:912-922. [DOI] [PubMed] [Google Scholar]
- 13.Hattori, T., K. Takahishi, T. Nakanishi, H. Ohta, K. Fukui, S. Taniguchi, and M. Takigawa. 1996. Novel FNR homologues identified in four representative oral facultative anaerobes: Capnocytophaga ochracea, Capnocytophaga sputigena, Haemophilus aphrophilus, and Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 137:213-220. [DOI] [PubMed] [Google Scholar]
- 14.Haubek, D., J. M. DiRienzo, E. M. B. Tinoco, J. Westergaard, N. J. Lopez, C.-P. Chung, K. Poulsen, and M. Kilian. 1997. Racial tropism in a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 35:3037-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haubek, D., K. Poulsen, S. Asikainen, and M. Kilian. 1995. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 33:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haubek, D., K. Poulsen, J. Westergaard, G. Dahlèn, and M. Kilian. 1996. Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J. Clin. Microbiol. 34:1576-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedegaard, J., H. Okkels, B. Bruun, M. Kilian, K. K. Mortensen, and N. Nørskov-Lauritsen. 2001. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology 147:2599-2609. [DOI] [PubMed] [Google Scholar]
- 18.Inoue, T., I. Tanimoto, H. Ohta, K. Kato, Y. Murayama, and K. Fukui. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42:253-258. [DOI] [PubMed] [Google Scholar]
- 19.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 20.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, first representative of a new pilin gene subfamily, is required for nonspecific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, A. H., D. J. Weber, E. Z. Oddone, and J. R. Perfect. 1989. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev. Infect. Dis. 11:46-63. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilian, M. 1998. Clonal basis of bacterial virulence, p. 131-142. In B. Guggenheim and S. Shapiro (ed.), Oral biology at the turn of the century. S. Karger AG, Basel, Switzerland.
- 24.King, E. O., and H. W. Tatum. 1962. Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J. Infect. Dis. 111:85-94. [DOI] [PubMed] [Google Scholar]
- 25.Kraig, E., T. Dailey, and D. Kolodrubetz. 1990. Nucleotide sequence of the leukotoxin gene from Actinobacillus actinomycetemcomitans: homology to the alpha-hemolysin/leukotoxin gene family. Infect. Immun. 58:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodrubetz, D., T. Dailey, J. Ebersole, and E. Kraig. 1989. Cloning and expression of the leukotoxin gene from Actinobacillus actinomycetemcomitans. Infect. Immun. 57:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page, R. C., T. J. Sims, L. D. Engel, B. J. Moncla, B. Bainbridge, J. Stray, and R. P. Darveau. 1991. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect. Immun. 59:3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paju, S., P. Carlson, H. Jousimies-Somer, and S. Asikainen. 2000. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotypes, genotype, and antimicrobial susceptibility. J. Clin. Microbiol. 38:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paju, S., M. Saarela, S. Alaluusua, P. Fives-Taylor, and S. Asikainen. 1998. Characterization of serologically nontypeable Actinobacillus actinomycetemcomitans isolates. J. Clin. Microbiol. 36:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry, M. B., L. L. MacLean, J.-R. Brisson, and M. E. Wilson. 1996. Structures of the antigenic O-polysaccharides of lipopolysaccharides produced by Actinobacillus actinomycetemcomitans serotypes a, c, d and e. Eur. J. Biochem. 242:682-688. [DOI] [PubMed] [Google Scholar]
- 32.Perry, M. B., L. L. MacLean, R. Gmür, and M. E. Wilson. 1996. Characterization of the O-polysaccharide structure of lipopolysaccharide from Actinobacillus actinomycetemcomitans serotype b. Infect. Immun. 64:1215-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potts, T. V., T. Mitra, T. O'Keefe, J. J. Zambon, and R. J. Genco. 1986. Relationships among isolates of oral haemophili as determined by DNA-DNA hybridization. Arch. Microbiol. 145:136-141. [DOI] [PubMed] [Google Scholar]
- 34.Poulsen, K., E. Theilade, E. T. Lally, D. R. Demuth, and M. Kilian. 1994. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology 140:2049-2060. [DOI] [PubMed] [Google Scholar]
- 35.Saarela, M., S. Asikainen, S. Alaluusua, L. Pyhälä, C.-H. Lai, and H. Jousimies-Somer. 1992. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol. Immunol. 7:277-279. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sugai, M., T. Kawamoto, S. Y. Peres, Y. Ueno, H. Komatsuzawa, T. Fujiwara, H. Kurihara, H. Suginaka, and E. Oswald. 1998. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 66:5008-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity in 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van de Peer, Y., S. Chapelle, and R. De Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Steenbergen, T. J. M., C. J. Bosch-Tijhoh, A. J. van Winkelhoff, R. Gmür, and J. de Graaff. 1994. Comparison of six typing methods for Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 32:2769-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, J. G. K., A. R. Kubelik, J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 22:6531-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 43.Zambon, J. J., J. Slots, and R. J. Genco. 1983. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect. Immun. 41:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]