Abstract
A triplex PCR targeting the 16S rRNA, mecA, and nuc genes was developed for identification of staphylococci and detection of methicillin resistance. After validation of the assay with a collection of strains of staphylococci and enterococci (n = 169), the assay was evaluated with cultures of blood with gram-positive cocci from 40 patients. Accurate results were obtained for 59 (98%) of 61 cultures within 6 h of growth detection.
Methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci(MRCoNS) are important causes of nosocomial infections (12, 15, 20, 22). As methicillin resistance mediated by PBP 2a is often heterogeneously expressed in staphylococci (2, 7, 24), PCR detection of mecA gene is the “gold standard” for the detection of methicillin resistance (1, 2, 10, 16, 21). In the case of severe infections, it is clinically useful to provide rapid identification and antimicrobial susceptibility test results (5). Many multiplex PCR techniques have been described for the characterization of staphylococci (4, 13, 14, 25, 27). Most of these are performed with genomic DNA extracts from bacterial colonies or broth culture lysates. Evaluation of mecA detection by PCR with simulated positive blood cultures, with blood samples spiked with staphylococci (4, 27), or from blood cultures directly has also been reported (6, 11; S. M. Benson, G. W. Coombs, and I. D. Kay, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 877, 1999). This report describes a PCR assay that targets three genes: mecA, a determinant of methicillin resistance; nuc, which encodes the S. aureus-specific region of the thermonuclease gene; and a genus-specific 16S rRNA sequence used as an internal amplification control for staphylococcal DNA. The expected sizes of the amplified DNA fragments are 533, 279, and 750 bp, respectively (Fig. 1). The objectives of this study were to evaluate the accuracy of characterization of staphylococci by the 16S rRNA-mecA-nuc triplex PCR directly from blood cultures and to compare its diagnostic performance with that of conventional methods in terms of specificity, sensitivity, and detection time.
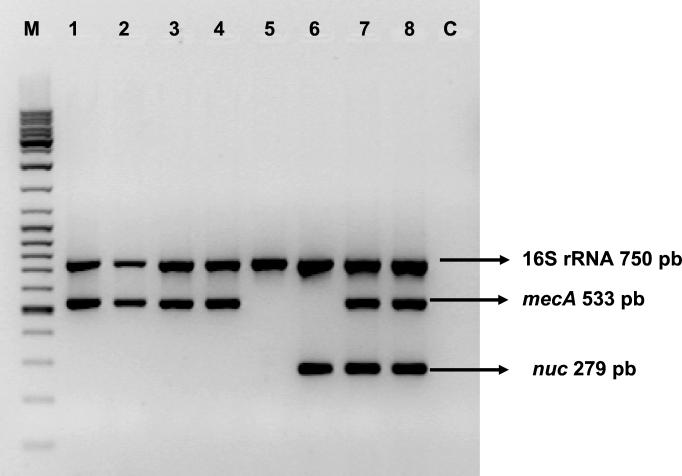
FIG. 1.
Interpretation of PCR results after electrophoresis on an agarose gel. Lane M, molecular size marker (MBI Fermentas ladder mix); lanes 1 to 4, MRCoNS; lane 5, MSCoNS; lane 6, MSSA; lanes 7 and 8, MRSA; lane C, negative control; pb, base pairs.
Bacterial strains were phenotypically characterized by determination of coagulase activity and an agglutination test with a Pastorex Staph Plus kit (Sanofi Diagnostics Pasteur, Marnes la Coquette, France), and coagulase-negative staphylococci (CoNS) were identified with the ID 32 Staph system (Biomérieux, Marcy l'Etoile, France). The strains were tested for oxacillin susceptibility by use of the Oxacillin Agar Screen (Becton Dickinson, Sparks, Md.) and the rapid ATB Staph system (Biomérieux) for S. aureus and by a 1-μg disk (Rosco, Taastrup, Denmark) diffusion test for CoNS. Oxacillin MICs were determined by either the E-test (AB Biodisk, Solna, Sweden) for the reference strains of CoNS or the agar dilution method according to the guidelines of NCCLS (19) for the clinical isolates of CoNS selected for assay validation. The sets of primers were previously reported by Murakami et al. (18), Brakstad et al. (3), and Jaffe et al. (11), respectively.
In the first part of the study, the amplification conditions were optimized and validated with 169 well-characterized clinical isolates and reference strains (Table 1). The clinical isolates were blood isolates of MRSA (n = 88) and methicillin-susceptible S. aureus (MSSA; n = 24) selected from a multicenter survey of S. aureus bacteremia in Belgian hospitals conducted in 1999 and 2000 and blood isolates of CoNS (n = 25) obtained from a prospective study of bacteremia in patients admitted to Erasme Hospital during 1998 and 1999 (9). The Staphylococcus and Enterococcus isolates were grown on Columbia agar with 5% sheep blood (Biomérieux) for 24 h at 37°C. DNA was extracted from the colonies as described by Unal et al. (26). The 50-μl PCR mixture contained 1× PCR buffer (Perkin-Elmer Applied Biosystems, Foster City, Calif.), 2 mM MgCl2, 16S rRNA-specific primers (0.6 μM) and mecA- and nuc-specific primers (0.4 μM each) (Amersham Pharmacia Biotech, Roosendaal, The Netherlands), deoxynucleoside triphosphates (250 μM each; Promega, Madison, Wis.), and AmpliTaq Gold DNA polymerase (2 U; Perkin-Elmer Applied Biosystems). A DNA sample of 5 μl was used as the target in the PCR. Amplification conditions consisted of 10 min at 94°C, followed by 23 cycles of 1 min at 94°C, 1 min at 51°C, and 2 min at 72°C, with a final step of 5 min at 72°C. The DNA fragments were separated by electrophoresis on a 1.5% agarose (Invitrogen, Merelbeke, Belgium) gel stained with ethidium bromide.
TABLE 1.
Bacterial strains used for preliminary validation of 16S rRNA-mecA-nuc triplex PCR
| Strain(s) | No. of isolates | PCR resultsa
|
Oxacillin resistance
|
|||
|---|---|---|---|---|---|---|
| 16S rRNA | mecA | nuc | Diffusion testb | MIC or MIC range (μg/ml) | ||
| MRSA clinical isolates | 88 | + | + | + | R | ND |
| MSSA clinical isolates | 24 | + | 0 | + | S | ND |
| MRSA reference strains ATCC 43300, ATCC 33592, CCM 885 | 3 | + | + | + | R | ND |
| MSSA reference strain ATCC 29213 | 1 | + | 0 | + | S | ND |
| Staphylococcus epidermidis clinical isolates | 18 | + | + | 0 | R | 0.5->128 |
| Staphylococcus haemolyticus clinical isolates | 3 | + | + | 0 | R | 64-128 |
| Staphylococcus hominis clinical isolate | 1 | + | + | 0 | R | >128 |
| Staphylococcus lugdunensis clinical isolates | 2 | + | 0 | 0 | S | 0.25 |
| S. lugdunensis clinical isolate | 1 | + | 0 | 0 | R | 0.5 |
| Staphylococcus auricularis ATCC 33753 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus capitis capitis CCM 2734 | 1 | + | 0 | 0 | S | ND |
| S. capitis ureolyticus ATCC 49326 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus caprae CCM 3573 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus cohnii cohnii CCM 2736 | 1 | + | 0 | 0 | R | 0.75 |
| S. cohnii urealyticum ATCC 49330 | 1 | + | 0 | 0 | R | 0.75 |
| S. epidermidis CCM 2124 | 1 | + | 0 | 0 | S | ND |
| S. haemolyticus CCM 2737 | 1 | + | 0 | 0 | S | ND |
| S. hominis hominis DSM 20328 | 1 | + | 0 | 0 | S | ND |
| S. lugdunensis ATCC 43809 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus pasteuri ATCC 51129 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus saprophyticus CCM 883 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus schleiferi coagulans GA 211 | 1 | + | 0 | 0 | S | ND |
| S. schleiferi schleiferi ATCC 43808 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus simulans ATCC 27848 | 1 | w | 0 | 0 | S | ND |
| Staphylococcus wameri CCM 2730 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus xylosus ATCC 29971 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus intermedius CCM 5739 | 1 | + | 0 | 0 | S | ND |
| Staphylococcus sciuri sciuri ATCC 29062 | 1 | 0 | w | 0 | R | 0.75 |
| S. sciuri rodentium ATCC 700061 | 1 | 0 | 0 | 0 | R | 0.75 |
| S. sciuri camaticus ATCC 700058 | 1 | 0 | w | 0 | R | 0.75 |
| S. hominis novobiosepticus ATCC 700236 | 1 | + | + | 0 | R | >256 |
| Enterococcus faecium lowa 1 | 1 | 0 | 0 | 0 | ND | ND |
| Enterococcus faecalis ATCC 51299, 29212 | 2 | 0 | 0 | 0 | ND | ND |
| Enterococcus casseliflavus ATCC 25788 | 1 | 0 | 0 | 0 | ND | ND |
| Enterococcus flavescens CCM 439 | 1 | 0 | 0 | 0 | ND | ND |
| Enterococcus gallinarum BM 4174 | 1 | 0 | 0 | 0 | ND | ND |
+, positive result; 0, negative result; w, weakly positive results; ND, not determined.
R, resistant; S, sensitive.
The PCR results for nuc and mecA detection were in agreement with those of conventional methods for 100 and 98% of strains, respectively. The staphylococcal 16S rRNA sequence was amplified for 98% of the strains. Discrepancies between phenotypic resistance in the absence of an amplified mecA gene fragment was observed with Staphylococcus sciuri subsp. rodentium, Staphylococcus cohnii, and Staphylococcus lugdunensis (Table 1). A weak mecA PCR signal was noted with S. sciuri subsp. sciuri and S. sciuri carnaticus strains, but no signal appeared with S. sciuri subsp. rodentium. Most of the S. sciuri strains carry a genetic element closely related to the mecA gene (8), and some S. sciuri isolates harbor a second copy of the gene identical to mecA (8). Therefore, our PCR results suggest that sequence variations in the S. sciuri mecA gene are probably localized in the region recognized by the mecA-specific primers. mecA failed to be amplified from isolates of S. cohnii and S. lugdunensis for which oxacillin MICs were 0.75 and 0.5 μg/ml, indicating that this borderline resistance is mediated by a mechanism other than mecA. The 16S rRNA signal was weak with Staphylococcus simulans and was absent with S. sciuri strains. The lack of a complete 16S rRNA sequence for these two species precluded a comparison of the region recognized by the primers.
In the second part of the study, the triplex PCR assay was prospectively evaluated directly with cultures of blood specimens from 40 patients for whom cultures of at least two blood samples showed the growth of gram-positive cocci in clusters. A total of 61 BACTEC-Plus (Becton Dickinson) blood culture broths were analyzed, as were colonies obtained by subculture of the broth cultures on Columbia agar with 5% sheep blood for 24 h at 37°C. Blood culture broths were processed for the PCR assay between 1 and 12 h after being flagged as positive by the instrument. A 100-μl aliquot was diluted 1:100 in brain heart infusion broth (Becton Dickinson) and incubated at 35°C for 45 min. After centrifugation at 2,000 × g for 5 min to remove blood cells, followed by centrifugation at 13,000 × g for 5 min, the supernatant was discarded. The cell pellet was washed twice with 1 ml of sterile water to remove inhibitors. DNA extraction and amplification conditions were as described above, except that 5 μl of each of the DNA extracts was tested nondiluted and diluted 10-fold in distilled water. Isolates recovered from the subcultures of the blood cultures were identified by the conventional techniques described above as MRCoNS (n = 22), methicillin-susceptible coagulase-negative staphylococci (MSCoNS; n = 4), MSSA (n = 10), MRSA (n = 4), and a Stomatococcus sp. (n = 1). CoNS were identified as Staphylococcus epidermidis (n = 22), Staphylococcus hominis (n = 2), Staphylococcus warneri (n = 1), and Staphylococcus capitis (n = 1).
Staphylococcal bacteremia and methicillin susceptibility were accurately detected by PCR from blood culture broths for 59 of 61 samples within 6 h. In general, a bacterial cell pellet was visible after centrifugation of the brain heart infusion broth subculture. For two specimens, specific DNA fragments were correctly detected by PCR only when the test was repeated with a sample from the blood culture bottle. Identical results were obtained by the PCR performed with samples from the blood culture bottles and overnight colonies. In one mixed culture containing both MRSA and MRCoNS, the amplification signal for nuc with a sample of the blood culture broth was weak. The subculture showed a predominance of MRCoNS, which could explain the weak nuc signal in comparison with those for 16S rRNA and mecA. In one case, amplification occurred only with the diluted extract from the blood culture, suggesting the presence of inhibitors in the undiluted sample.
Our results are in agreement with those of recent studies of PCR detection of MRSA from blood. In a clinical evaluation of a mecA PCR assay with 181 BACTEC blood cultures, Carroll et al. (6) found that the results had a 99% correlation with the results of standard susceptibility tests. Their assay had a turnaround time of less than 3 h but could not differentiate S. aureus from CoNS. Benson et al. (39th ICAAC) reported mecA-nuc duplex PCR results in total concordance with phenotypic characterization for 184 BacTAlert blood cultures. However, no internal amplification control was included in the two assays described above. The region of 16S rRNA selected used as a positive control in the PCR assay described here was adequate for the detection of the most important species of staphylococci encountered in humans.
The PCR assay described here provided results, on average, 18 to 42 h faster than the conventional identification and susceptibility testing methods used in our laboratory, which required 24 to 48 h after detection of colonies on blood cultures, whereas the PCR required 6 h. Other investigators have evaluated rapid phenotypic methods directly with blood culture specimens, such as the coagulase test for identification of staphylococci (23) and the BBL Crystal MRSA ID system (Becton Dickinson) for detection of methicillin resistance (17). The coagulase test was found to be fairly sensitive (92%), with results obtained within 2 h. Lower sensitivities (84 and 54% for S. aureus and CoNS, respectively) were found for the BBL Crystal MRSA ID test, with results obtained within 6 h.
In conclusion, the 16S rRNA-mecA-nuc triplex PCR is a good tool for rapid characterization of staphylococci in positive blood cultures. To confirm the specificity of this test, more samples from patients with Enterococcus and Streptococcus sp. bacteremias need to be studied. Further evaluations of the clinical impact and the cost-effectiveness of this rapid diagnostic test on the management of staphylococcal bacteremia are under way.
REFERENCES
- 1.Barski, P., L. Piechowicz, J. Galinski, and J. Kur. 1996. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol. Cell. Probes 10:471-475. [DOI] [PubMed] [Google Scholar]
- 2.Bignardi, G. E., N. Woodford, A. Chapman, A. P. Johnson, and D. C. Speller. 1996. Detection of the mec-A gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J. Antimicrob. Chemother. 37:53-63. [DOI] [PubMed] [Google Scholar]
- 3.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brakstad, O. G., J. A. Maeland., and Y. Tveten. 1993. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS 101:681-688. [DOI] [PubMed] [Google Scholar]
- 5.Byl, B., P. Clevenbergh, F. Jacobs, M. J. Struelens, F. Zech, A. Kentos, and J. P. Thys. 1999. Impact of infectious diseases specialists and microbiological data on the appropriateness of antimicrobial therapy for bacteremia. Clin. Infect. Dis. 29:60-66. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, K. C., R. B. Leonard, P. L. Newcomb-Gayman, and D. R. Hillyard. 1996. Rapid detection of the staphylococcal mecA gene from BACTEC blood culture bottles by the polymerase chain reaction. Am. J. Clin. Pathol. 106:600-605. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, H. F. 1988. Methicillin-resistant staphylococci. Clin. Microbiol. Rev. 1:173-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couto, I., I. S. Sanches, R. Sa-Leao, and H. de Lencastre. 2000. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J. Clin. Microbiol. 38:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 10.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffe, R. I., J. D. Lane, S. V. Albury, and D. M. Niemeyer. 2000. Rapid extraction from and direct identification in clinical samples of methicillin-resistant staphylococci using the PCR. J. Clin. Microbiol. 38:3407-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karchmer, A. W. 2000. Nosocomial bloodstream infections: organisms, risk factors, and implications. Clin. Infect. Dis. 31(Suppl. 4):S139-S143. [DOI] [PubMed] [Google Scholar]
- 13.Kearns, A. M., P. R. Seiders, J. Wheeler, R. Freeman, and M. Steward. 1999. Rapid detection of methicillin-resistant staphylococci by multiplex PCR. J. Hosp. Infect. 43:33-37. [DOI] [PubMed] [Google Scholar]
- 14.Kizaki, M., Y. Kobayashi, and Y. Ikeda. 1994. Rapid and sensitive detection of the femA gene in staphylococci by enzymatic detection of polymerase chain reaction (ED-PCR): comparison with standard PCR analysis. J. Hosp. Infect. 28:287-295. [DOI] [PubMed] [Google Scholar]
- 15.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolbert, C. P., J. Arruda, P. Varga-Delmore, X. Zheng, M. Lewis, J. Kolberg, and D. H. Persing. 1998. Branched-DNA assay for detection of the mecA gene in oxacillin-resistant and oxacillin-sensitive staphylococci. J. Clin. Microbiol. 36:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubina, M., B. Jaulhac, X. Delabranche, C. Lindenmann, Y. Piemont, and H. Monteil. 1999. Oxacillin susceptibility testing of staphylococci directly from BACTEC Plus blood cultures by the BBL Crystal MRSA ID system. J. Clin. Microbiol. 37:2034-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M2 A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Refsahl, K., and B. M. Andersen. 1992. Clinically significant coagulase-negative staphylococci: identification and resistance patterns. J. Hosp. Infect. 22:19-31. [DOI] [PubMed] [Google Scholar]
- 21.Salisbury, S. M., L. M Sabatini, and C. A. Spiegel. 1997. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Am. J. Clin. Pathol. 107:368-373. [DOI] [PubMed] [Google Scholar]
- 22.Silva, H. L., T. M. Strabelli, E. R. Cunha, S. F. Neres, L. F. Camargo, and D. E. Uip. 2000. Nosocomial coagulase negative staphylococci bacteremia: five year prospective data collection. J. Infect. Dis. 4:271-274. [PubMed] [Google Scholar]
- 23.Speers, D. J., T. R. Olma., and G. L. Gilbert. 1998. Evaluation of four methods for rapid identification of Staphylococcus aureus from blood cultures. J. Clin. Microbiol. 36:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Towner, K. J., D. C. S. Alobot, R. Curran, C. A. Webster, and H. Humphreys. 1998. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 47:607-613. [DOI] [PubMed] [Google Scholar]
- 26.Unal, S., J. Hoskins, J. E. Flokowitsch, C. Y. Wu, D. A. Preston, and P. L. Skatrud. 1992. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J. Clin. Microbiol. 30:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vannuffel, P., J. Gigi, H. Ezzedine, B. Vandercam, M. Delmee, G. Wauters, and J.-L. Gala. 1995. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J. Clin. Microbiol. 33:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]