Abstract
This study is focused on real-time detection of gyrA mutations and of the presence of class I integrons in a panel of 100 veterinary isolates of Salmonella enterica from farm animals. The isolates were selected on the basis of resistance to nalidixic acid, representing a variety of the most prevalent serotypes in England and Wales. In addition, organic solvent (cyclohexane) resistance in these isolates was investigated in an attempt to elucidate the presence of efflux pump mechanisms. The most prevalent mutation among the isolates studied was Asp87-Asn (n = 42), followed by Ser83-Phe (n = 38), Ser83-Tyr (n = 12), Asp87-Tyr (n = 4), and Asp87-Gly (n = 3). Two distinct subpopulations were identified, separated at the 1-mg/liter breakpoint for ciprofloxacin: 86% of isolates with mutations in codon 83 showed MICs of ≥1 mg/liter, while 89.8% of isolates with mutations in codon 87 presented MICs of ≤0.5 mg/liter. Cyclohexane resistance was more prevalent among Ser83 mutants than among Asp87 mutants (34.7 and 4%, respectively), and in 79% of isolates that presented both gyrA mutations and cyclohexane resistance, the level of ciprofloxacin resistance was ≥2.0 mg/liter. Thirty-four isolates contained class I integrons, with 71% of the S. enterica serovar Typhimurium isolates and 6.9% of isolates belonging to other serotypes containing such elements. The methods used represent sensitive ways of investigating the presence of gyrA mutations and of detecting class-I integrons in Salmonella isolates. The results can be obtained in less than 1 h from single colonies without the need for purifying DNA.
The concern about antimicrobial resistance and the problems that it causes in both human and veterinary medicine has grown in recent years. The selective pressure created by widespread use of antimicrobial drugs in animals and humans as therapeutic and prophylactic agents, as well as for growth promotion in animal production, may have contributed to the dissemination of resistant bacterial strains. There is both indirect and direct evidence that resistant bacteria or resistance genes can be exchanged between different reservoirs (soil, water, plants, animals, and humans) (European Commission [http://europa.eu.int/comm/dg24/health/sc/ssc/out50_en.html]). The higher the levels of resistance in one reservoir the more frequently transmission of resistance (bacteria or genes) to other reservoirs will occur. Concern about resistance is reflected in demands for more surveillance, as such studies are vital in the search for information on the epidemiology of resistance in animals and humans. There is a recognized need for simple, rapid, and accurate methods for screening large numbers of isolates for resistance (3) and for building up banks of data useful for future investigations.
Quinolone resistance in field isolates of Salmonella enterica belonging to several serotypes has been reported with increasing frequency in recent years (5, 14, 18, 34). The Veterinary Laboratories Agency (VLA; United Kingdom) surveillance program has shown that 5.3% of all Salmonella strains isolated from animals in 1999 were resistant to nalidixic acid; among the isolates from different animal species, poultry isolates showed the highest percentage of resistance, with 13.4% of these isolates being resistant to nalidixic acid (6). Two main mechanisms of resistance have been identified for gram-negative organisms: mutations in DNA gyrase and reduced intracellular drug accumulation (4, 16). High-level quinolone resistance has been associated with single mutations in the quinolone resistance-determining region (QRDR) of gyrA in Salmonella (4, 10, 13, 15, 31), with the most commonly described mutations being Ser83-to-Phe (TCC→TTC), Asp87-to-Gly (GAC→GGC), and Asp87-to-Asn (GAC→AAC). Mutations of gyrB and parC of Salmonella are not normally found in nalidixic acid-resistant isolates (8, 24, 34) although they may contribute to high-level resistance to fluoroquinolones (27). There are several studies suggesting that fluoroquinolones may have an impaired effect for treating infections due to Salmonella isolates that are resistant to nalidixic acid (20, 30). These isolates normally have decreased susceptibility to fluoroquinolones, and therefore, standard dosages of these drugs may not be sufficient to treat clinical infections.
Integrons are potentially mobile genetic elements that have been identified as loci at which site-specific incorporation and excision of gene cassettes frequently occur. As a result, these elements are able to incorporate a single antibiotic resistance gene or groups of antibiotic resistance genes by site-specific recombination (17). These elements may be found in both chromosomal and extrachromosomal DNA (11, 29), and their presence is often indicative of multiple resistance to unrelated antimicrobials.
In a previous paper, we reported the use of real-time methods to assess the frequency of gyrA mutations in S. enterica serovar Typhimurium isolates from humans and animals (31). In the present paper, we describe a study of a wider panel of veterinary isolates from some of the common serotypes in England and Wales. This study focused on the detection of mutations in gyrA and of the presence of class I integrons. In addition, organic solvent (cyclohexane) resistance in these isolates was investigated in an attempt to elucidate the presence of efflux pump mechanisms.
MATERIALS AND METHODS
Bacterial strains.
One hundred S. enterica isolates from pigs (n = 20), cattle (n = 16), chickens (n = 34), and turkeys (n = 30) isolated in England and Wales between 1998 and 2000 were selected for the study on the basis of resistance to nalidixic acid. The isolates represented a variety of some of the most prevalent serotypes in animals (S. enterica serovar Typhimurium [n = 42], S. enterica serovar Newport [n = 12], S. enterica serovar Hadar [n = 10], S. enterica serovar Montevideo [n = 10], S. enterica serovar Senftenberg [n = 7], S. enterica serovar Dublin [n = 4], S. enterica serovar Kedougou [n = 4], S. enterica serovar Mbandaka [n = 3], S. enterica serovar Enteritidis [n = 2], S. enterica serovar Goldcoast [n = 2], S. enterica serovar Choleraesuis [n = 1], S. enterica serovar Virchow [n = 1], S. enterica serovar Indiana [n = 1], and S. enterica serovar Saintpaul [n = 1]). Isolates were serotyped following a microagglutination method (28) and, where appropriate, were phage typed (32) at the Veterinary Laboratories Agency (Weybridge).
Determination of antibiotic resistance.
Isolates were screened for susceptibility to a panel of 16 antibiotics on iso-sensitest agar (Oxoid CM471) by a disk diffusion method similar to that previously described (23). The following disks (Oxoid, Basingstoke, Hampshire, United Kingdom) were used: amikacin (10 μg), amoxicillin/clavulanic acid (30 μg), ampicillin (10 μg), apramycin (15 μg), chloramphenicol (10 μg), cefoperazone (30 μg), cefuroxime (30 μg), colistin (25 μg), furazolidone (15 μg), gentamicin (20 μg), nalidixic acid (30 μg), neomycin (10 μg), streptomycin (25 μg), sulfamethoxazole/trimethoprim (25 μg), tetracycline (10 μg), and triple sulfonamide (300 μg). Organisms with a zone diameter of less than 13 mm were classified as resistant. The Escherichia coli strain (NCTC 10418) sensitive to all antibiotics was used on the plates for quality control purposes.
MICs of ciprofloxacin.
MICs were determined by an agar doubling dilution method similar to that described by the National Committee of Clinical Laboratory Standards (NCCLS) (22), with the main exception that iso-sensitest agar (Oxoid CM471) rather than Mueller Hinton agar (Oxoid CM337) was used. Bacteria grown overnight at 37°C in peptone water were diluted 1/10 in normal saline and inoculated using a multi-point inoculator onto the agar with suitable dilutions of ciprofloxacin (kindly donated by Bayer, Newbury, Berkshire, United Kingdom). Plates were incubated overnight at 37°C, and the MIC was recorded as the lowest concentration of ciprofloxacin inhibiting growth.
Cyclohexane resistance.
Cyclohexane resistance was determined by the method of Asako et al. (2). Briefly, bacteria grown overnight at 30°C were inoculated (using a multipoint inoculator) onto duplicate solid agar media in glass petri dishes. Each plate was flooded to a depth of 3 mm with hexane (control) or cyclohexane, sealed with Nescofilm, and incubated overnight at 30°C. Strains that grew in the presence of cyclohexane were deemed cyclohexane resistant. E. coli K-12 strains AG100 and AG102 (a marR mutant of AG100) (7, 26) were included as controls in the plates.
DNA preparation.
Bacteria were grown overnight at 37°C on blood agar base (Oxoid) plates. A single colony was emulsified in 100 μl of water and heated in a boiling water bath for 10 min, the suspension was centrifuged at 11,000 × g for 5 min in a microcentrifuge (Biofuge pico; Hareaus), and the supernatant was stored at −20°C until tested.
LightCycler gyrA mutation assay.
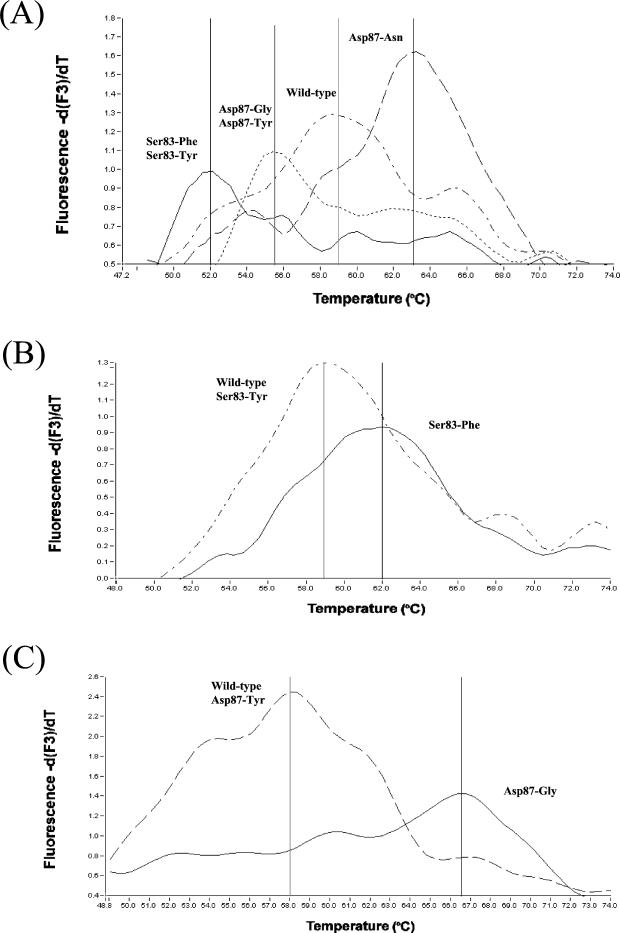
Isolates were tested following a modification of the original protocol described by Walker et al. (31). Reactions were performed in a LightCycler instrument (Roche Diagnostics UK Ltd., Lewes, United Kingdom) in glass capillaries containing 20 μl with 1× LightCycler DNA Master SYBR Green (Roche Molecular Biochemicals), 10 pmol of gyrAF, 4 pmol of gyrAR, 10 pmol of one of the specific probes (gyrAPI, gyrAP2, or gyrAPIII), 5 mM MgCl2 (final concentration), and 2 μl of bacterial DNA template. The PCR run consisted of denaturation at 95°C for 30 s, followed by an amplification protocol of 60 cycles of denaturation at 95°C for 0 s, annealing at 55°C for 10 s, and elongation at 72°C for 10 s. The temperature transition rates were of 20°C/s for all the steps. A melting protocol was included at the end of the PCR run, consisting of 95°C for 0 s followed by 45°C for 15 s, both at a transition rate of 20°C/s, and finally increasing the temperature at a 0.1°C/s rate to reach 95°C. During this last step, fluorescence was detected in a continuous mode in channel 3. Control DNAs (strains P3801900 [Asp87-to-Asn], P3749380 [Ser83-to-Phe], P3424780 [Asp87-to-Gly], and 42R500 [wild type]) were included in each run. Oligonucleotide probe gyrAPI was always used as a screening probe, based on previous reports suggesting that Asp87-to-Asn is one of the most commonly found mutations and due to the clear differences in melting temperatures (Tm's) achieved for the different mutations of interest. Subsequently to the screening reactions, if a Tm similar to the Ser83-to-Phe or Asp97-to-Gly control mutants was detected, DNAs were again tested with the specific probes (gyrAPII or gyrAPIII). Isolates that could not be confirmed with any of the three probes were subjected to sequencing of the gyrA fragment. Figure 1 shows the expected Tm's for the different gyrA mutants after testing with the three oligonucleotide probes.
FIG. 1.
gyrA mutation assay analysis of Salmonella control strains. (A) DNA from strains P3801900 (Asp87-Asn), 42R500 (wild type), P3424780 (Asp87-Gly), and P3749380 (Ser83-Phe) were tested with the screening oligonucleotide probe gyrAPI yielded Tm's of 63, 59, 55.5, and 51.8°C, respectively. (B) DNA from strains P3749380 and 42R500 tested with confirmatory oligonucleotide probe gyrAPII yielded Tm's of 62 and 59°C, respectively. (C) DNA from strains P3424780 and 42R500 tested with confirmatory probe gyrAPIII yielded Tm values of 66.5 and 58°C, respectively.
Detection of integrons by real-time PCR.
Amplification was performed in a LightCycler instrument (Roche Diagnostics UK Ltd.) in glass capillaries containing 20 μl with 1× LightCycler DNA Master SYBR Green (Roche Molecular Biochemicals), L2 and L3 primers (17) at 6.6 pmol each, 4 mM MgCl2 (final concentration), and 2 μl of bacterial DNA template. The PCR run was as described by Maguire et al. (17). The nature of the amplicon was determined by melting analysis over a temperature range from 70 to 97°C with a transition rate of 0.1°C/s and continuous detection of fluorescence in channel 1. The Tm for the PCR product was 90°C. The size of the product was 300 bp, and this was verified by agarose gel electrophoresis of the positive samples.
DNA sequencing.
DNA sequencing was performed by amplifying a fragment of the gyrA gene as follows: 25-μl reaction mixtures were prepared with 10 pmol of both primers gyrAF and gyrAR, 200 μM deoxynucleoside triphosphates (dNTPs) (each), 2.5 mM MgCl2, 0.5 U of platinum Taq, 1× PCR buffer (Life Technologies, Paislay, United Kingdom), and 5 μl of template DNA. PCR was performed in a GeneAmp PCR system 9700 (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). The PCR run comprised denaturation at 94°C for 10 min, followed by 33 cycles each consisting of 94°C for 10 s, 60°C for 1 min and 72°C for 1 min. PCR products were purified prior to sequencing by using QIAQuick purification columns (QIAgen Ltd., Crawley, United Kingdom). Products were sequenced in both directions by MWG Biotech by using the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer Applied Biosystems) on an ABI 3700 sequencer.
RESULTS
Antibiotic resistance profiles.
Table 1 summarizes the antibiotic resistance profiles for 100 salmonella strains selected on the basis of resistance to nalidixic acid. Sixty-three of the isolates were resistant to three or more antimicrobials. All but a single isolate (n = 99) showed at least decreased susceptibility to ciprofloxacin (MICs of ≥0.25 mg/liter). As expected, the selection of nalidixic acid-resistant isolates provided a good sensitivity and specificity for detection of isolates with resistance to ciprofloxacin at a MIC of ≥0.125 μg/ml (12).
TABLE 1.
Antibiotic resistance profiles for 100 Salmonella strains selected on the basis of resistance to nalidixic acid
| Salmonella serotype | No. of isolates with antibiotic resistance profile:
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI | XVII | XVIII | XIX | XX | XXI | XXII | |
| Choleraesuis (n = 1) | 1 | |||||||||||||||||||||
| Dublin (n = 4) | 4 | |||||||||||||||||||||
| Enteritidis (n = 2) | 2 | |||||||||||||||||||||
| Gold Coast (n = 2) | 2 | |||||||||||||||||||||
| Hadar (n = 10) | 3 | 4 | 1 | 1 | 1 | |||||||||||||||||
| Indiana (n = 1) | 1 | |||||||||||||||||||||
| Kedougou (n = 4) | 1 | 3 | ||||||||||||||||||||
| Mbandaka (n = 3) | 1 | 2 | ||||||||||||||||||||
| Montevideo (n = 10) | 10 | |||||||||||||||||||||
| Newport (n = 12) | 1 | 4 | 1 | 1 | 1 | 4 | ||||||||||||||||
| Saintpaul (n = 1) | 1 | |||||||||||||||||||||
| Senftenberg (n = 7) | 1 | 6 | ||||||||||||||||||||
| Typhimurium (n = 42) | 1 | 17 | 1 | 1 | 1 | 7 | 1 | 2 | 8 | 1 | 2 | |||||||||||
| Virchow (n = 1) | 1 | |||||||||||||||||||||
| Total (n = 100) | 3 | 17 | 2 | 1 | 1 | 7 | 1 | 2 | 1 | 1 | 1 | 4 | 9 | 1 | 4 | 5 | 1 | 1 | 1 | 2 | 1 | 34 |
Profiles indicate the antimicrobial(s) to which each isolate was resistant: I, ANxSSuTTm; II, ACNxSSuT; III, ACNxSSuTTm; IV, ACGNxSSuT; V, ACNNxSSuT; VI, ACCzNxSSuT; VII, ACCzNNxSSuT; VIII, ACNxSuTTm; IX, CNxSuTTm; X, ACNxSSu; XI, ApGNxSu; XII, ACzNxST; XIII, NxSSuT; XIV, ANxSSu; XV, ANxST; XVI, NxSSu; XVII, NxST; XVIII, AFNx; XIX, ANxT; XX, NxS; XXI, ANx; XXII, Nx. Antimicrobial abbreviations: A, ampicillin; Ap, apramycin; C, chloramphenicol; Cs, colistin sulfate; Cz, cefoperazone; F, furazolidone; G, gentamicin; N, neomycin; Nx, nalidixic acid; S, streptomycin; Su, sulfonamides; T, tetracyclines; Tm, trimethoprim.
GyrA mutations in Salmonella serotypes and correlation with ciprofloxacin MICs.
Table 2 depicts mutations encountered for the 100 Salmonella isolates in the study. All but one of the isolates tested (n = 99) presented mutations in the gyrA gene. The most prevalent mutation among the isolates studied was Asp87-Asn (n = 42), followed by Ser83-Phe (n = 38). Three of the isolates presented an Asp87-Gly mutation, and finally, 16 of the isolates presented mutations different from the classical ones (12 Ser83-Tyr and 4 Asp87-Tyr). Within the group of S. enterica serovar Typhimurium isolates, 40.5% presented an Asp87-Asn mutation, 54.8% presented a Ser83-Phe mutation, and 4.8% presented Ser83-Tyr mutations. Within the group of isolates from other serotypes, 43% presented an Asp87-Asn mutation, 25.9% presented a Ser83-Phe mutation, 17.3% presented a Ser83-Tyr mutation, 6.9% presented a Asp87-Tyr mutation, and finally, 5.2% presented a Asp87-Gly mutation.
TABLE 2.
Distribution of gyrA mutations within Salmonella serotypes
| Serotype | No. of isolates with mutation:
|
No. of wild type | ||||
|---|---|---|---|---|---|---|
| Asp87- Asn | Ser83- Phe | Asp87- Gly | Asp87- Tyr | Ser83- Tyr | ||
| Choleraesuis (n = 1) | 1 | |||||
| Dublin (n = 4) | 1 | 2 | 1 | |||
| Enteritidis (n = 2) | 1 | 1 | ||||
| Gold Coast (n = 2) | 2 | |||||
| Hadar (n = 10) | 8 | 1 | 1 | |||
| Indiana (n = 1) | 1 | |||||
| Kedougou (n = 4) | 4 | |||||
| Mbandaka (n = 3) | 3 | |||||
| Montevideo (n = 10) | 10 | |||||
| Newport (n = 12) | 5 | 1 | 6 | |||
| Saintpaul (n = 1) | 1 | |||||
| Senftenberg (n = 7) | 3 | 1 | 3 | |||
| Typhimurium (n = 42) | 17 | 23 | 2 | |||
| Virchow (n = 1) | 1 | |||||
| Total (n = 100) | 42 | 38 | 3 | 4 | 12 | 1 |
Table 3 shows correlation between ciprofloxacin MICs and gyrA mutations. Two distinct subpopulations seem to coexist, separated at the 1-mg/liter breakpoint for ciprofloxacin. Eighty-six percent of isolates with mutations in codon 83 showed MICs of ≥1 mg/liter, while 89.8% of isolates with mutations in codon 87 presented MICs of ≤0.5 mg/liter. The isolate showing a wild-type gene was fully sensitive to ciprofloxacin (MIC < 0.06 mg/liter).
TABLE 3.
Correlation between ciprofloxacin MIC and gyrA mutations
| gyrA mutation | No. of isolates inhibited by ciprofloxacin at MIC (mg/liter) of:
|
|||||
|---|---|---|---|---|---|---|
| <0.06 | 0.25 | 0.5 | 1.0 | 2.0 | 4.0 | |
| Asp87-Asn (n = 42) | 9 | 30 | 3 | |||
| Ser83-Phe (n = 38) | 4 | 24 | 3 | 7 | ||
| Asp87-Gly (n = 3) | 1 | 1 | 1 | |||
| Asp87-Tyr (n = 4) | 1 | 3 | ||||
| Ser83-Tyr (n = 12) | 3 | 4 | 2 | 3 | ||
| Wild type (n = 1) | 1 | |||||
| Total (n = 100) | 1 | 11 | 40 | 32 | 6 | 10 |
Cyclohexane resistance in fluoroquinolone-resistant isolates.
Table 4 shows the relationship between presence of gyrA mutations and/or cyclohexane resistance and MIC level for ciprofloxacin. Cyclohexane resistance was found to be more prevalent among Ser83 mutants than among Asp87 mutants (34.7 and 4%, respectively). In 79% of the isolates that presented both gyrA mutations and cyclohexane resistance, the level of ciprofloxacin resistance was ≥2.0 mg/liter. The only exceptions were four isolates with gyrA mutations coupled with cyclohexane resistance that had MICs of ≤1.0 mg/liter. Only a single isolate that had a substitution in codon 87 and was sensitive to cyclohexane had a MIC of ≥2.0 mg/liter.
TABLE 4.
Cyclohexane resistance in nalidixic acid-resistant isolates
| Substitution at codon/cyclohexane tolerance | No. of isolates inhibited by ciprofloxacin at MIC (mg/liter) of:
|
|
|---|---|---|
| ≤1.0 | ≥2.0 | |
| Ser83/sensitive | 32 | 0 |
| Ser83/resistant | 2 | 15 |
| Asp87/sensitive | 47 | 1 |
| Asp87/resistant | 2 | 0 |
Frequency of class I integrons.
Of the 100 isolates, 34 contained class I integrons, as determined by the LightCycler method. All of these isolates presented a phenotype of resistance to sulfonamides. Seventy-one percent of the S. enterica serovar Typhimurium isolates showed the presence of class I integrons, while only 6.9% (two Goldcoast, one Saintpaul, and one Virchow) of the isolates belonging to serotypes other than Typhimurium presented these elements and were resistant to three or more antibiotics. The integron-free S. enterica serovar Typhimurium isolates (29%) were from a variety of phage types (DT193 [n = 4], U302 [n = 2], U308 [n = 2], U288 [n = 1], DT104 [n = 1], and untypeable [n = 2]), and all but two of these isolates showed resistance to three or more antibiotics.
DISCUSSION
According to the NCCLS guidelines, MICs of ≤1 and ≥4 μg/ml are regarded as the respective breakpoints for susceptibility and resistance to fluoroquinolones. However, these are not universally accepted, and several reports have emphasized the importance of detection of Salmonella strains with decreased susceptibility (MIC ≥ 0.125 μg/ml) to this group of antimicrobials. New rapid technologies have made possible the investigation of the mechanisms of quinolone resistance in a real-time cost-efficient way, which presents advantages compared to the classical phenotypic tests (31). Our study was aimed at analyzing a variety of Salmonella serotypes from different animal species and investigating not only the occurrence of changes at codons 83 and 87 but giving specific information on the mutation present at those sites. The information generated is useful per se for studies on fluoroquinolone resistance in Salmonella. In addition, sequence variations in the gyrA gene have been used previously for the verification of an epidemiological relationship between isolates (21). In order to use these variations, more information is needed on the frequency and locations of these changes in isolates from different serotypes and origins. Several studies have addressed this question for human isolates, but limited comprehensive information is available for animal strains. Griggs et al. (10) studied a panel of animal isolates from the United Kingdom, but they did not provide data on the specific substitution at codons 83 or 87. In a later study (24), a panel of 108 animal isolates from the United Kingdom was analyzed by single-stranded conformational polymorphism (SSCP), but no information on the presence of specific mutations in the different serotypes was presented in the study. A more recent study gave data on 37 Danish isolates from serotypes Typhimurium, Enteritidis, and Dublin (34). In our present study, Ser83-Phe (38%) and Asp87-Asn (42%) were found to be the most common mutations. These results contrast with those generated by Griggs et al. (10), in which 95.4% of animal Salmonella isolates were Ser83 mutants. This discrepancy could be due to the high number of S. enterica serovar Newport isolates (79%) within the population of that study. Our results also differ from those of Piddock et al. (24), in which 76% of animal Salmonella isolates were Asp87-Gly mutants; in our study, only two S. Dublin and one S. Newport isolates presented this substitution. In our previous study with S. Typhimurium DT104 (31), only 4.6% of the animal isolates presented Ser83-Tyr substitutions and this mutation was not identified in any human strains. In the present study, this mutation was not uncommon in animal isolates, with 12% presenting Ser83-Tyr mutations. This mutation was found in S. enterica serovar Typhimurium (4.7%), S. enterica serovar Senftenberg (43%), S. enterica serovar Enteritidis (50%), and S. enterica serovar Newport (50%) isolates. The only other report on gyrA mutations in S. enterica serovar Newport (10) did not provide sequence information on the nature of the change in codon 83.
In other gram-negative bacteria, high-level fluoroquinolone resistance has always been associated with the presence of multiple mutations in the different QRDRs. However, recent studies have shown that, for S. enterica, fluoroquinolone resistance is not very well correlated with the presence of mutations in gyrase B (gyrB) and topoisomerase IV (parC and parE) genes and that highly resistant mutants may present only single mutations in gyrA (8). These studies have shown that changes in the genes that code for the target proteins of fluoroquinolones (i.e., gyrase) could not totally account for the resistance phenotypes of S. enterica serovar Typhimurium mutants (8, 9). In our paper, we have investigated the presence of mutations in gyrA and the resistance to the organic solvent cyclohexane. Organic solvent tolerance in E. coli has been shown to be linked to mutations in marR, to overexpression of marA, soxS, and rob, to upregulation of acrAB, and to increased resistance to antibiotics (1, 2, 33). There is also evidence that cyclohexane resistance in Salmonella may be induced via a mar-dependent pathway (25) and that cyclohexane resistance in Salmonella may be associated with active efflux pump mechanisms (26). We have demonstrated that 93.7% of isolates with a ciprofloxacin MIC of ≥2 mg/liter showed a resistant phenotype to cyclohexane, and this could be due to enhanced active efflux mechanisms. Although we cannot be certain that these isolates do not possess additional mutations in other QRDRs, based on previous reports, it seems unlikely that nalidixic acid-resistant Salmonella isolates present such mutations (24, 34). There is some suggestion that these efflux mechanisms may appear early compared to mutation in gyrA and thus could be responsible for the first decrease in susceptibility to fluoroquinolones (9) and that further selective pressure by the use of antibiotics as therapeutic agents may trigger the selection of gyrA mutants and the appearance of fully resistant phenotypes. However, from our data, it seems clear that isolates with only single mutations in the gyrA-QRDR show low-level resistance (MIC ≤ 1 mg/liter), while gyrA mutants exhibiting cyclohexane resistance (possibly related to the presence of efflux pump mechanisms) show increased resistance levels (MIC ≥ 2 mg/liter). In addition, cyclohexane resistance phenotypes seemed more likely to be present in isolates showing Ser83 mutations than in isolates with Asp87 substitutions.
A high proportion (60%) of the turkey isolates showed ciprofloxacin MICs of ≥1 mg/liter. There has been published evidence that management practices within the turkey industry are associated with increase medication using enrofloxacin (5), and this may have selected populations of bacteria with increased resistance to fluoroquinolones.
A majority (63%) of the isolates in the study showed multiple resistance to three or more antimicrobials. It is well established that the dissemination of antimicrobial resistance is often plasmid and/or transposon mediated and that the presence of these elements is often indicative of multi-resistance to several antibiotics. There is evidence that integron transfer between bacteria may not be as common as was previously thought (19). Therefore, screening for these elements may be also a useful tool for epidemiological investigations. As was expected, all the S. enterica serovar Typhimurium DT104 resistant to three or more antibiotics included in the study presented class I integrons. A total of 10 S. enterica serovar Typhimurium resistant to three or more antibiotics were found to be integron-free by the light-cycler method. These isolates were from several phage types (DT193, U302, U308, and U288). Further studies are in process to investigate the mechanisms of resistance in these isolates.
The Light-Cycler methods used in this study allowed a rapid and sensitive way of investigating the presence of gyrA mutations and of detecting the presence of class I integrons in Salmonella isolates. The results can be obtained from single colonies without the need for purifying DNA. This approach offers great potential for screening large numbers of isolates in a cost-efficient manner.
Acknowledgments
This work was supported by a grant from the Seedcorn Program (SC0096) Veterinary Laboratories Agency, Weybridge, United Kingdom.
REFERENCES
- 1.Asako, H., H. Nakajima, K. Kobayshi, M. Kobayshi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 64:1428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asako, H., K. Kobayashi, and R. Aono. 1999. Organic solvent tolerance of Escherichia coli is independent of OmpF levels in the membrane. Appl. Environ. Microbiol. 65:294-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron, M. G., and M. Ouellete. 1998. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J. Clin. Microbiol. 36:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cambau, E., and L. Gutmann. 1993. Mechanisms of resistance to quionolones. Drugs 45(Suppl. 3):15-23. [DOI] [PubMed] [Google Scholar]
- 5.Davies, R. H., C. J. Teale, C. Wray, I. M. McLaren, Y. E. Jones, S. Chappell, and S. Kidd. 1999. Nalidixic acid resistance in salmonellae isolated from turkeys and other livestock in Great Britain. Vet. Rec. 20:320-322. [DOI] [PubMed] [Google Scholar]
- 6.Evans, S., and S. Kidd. 1999. Salmonella in livestock production, p. 127-133. Veterinary Laboratories Agency, Ministry of Agriculture, Fisheries and Food, London, United Kingdom.
- 7.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giraud, E., A. Brisadois, J. L. Martel, and E. Chaslus-Dancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43:2131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griggs, D. J., K. Gensberg, and L. J. V. Piddock. 1996. Mutations in gyrA gene of quinolone-resistant Salmonella serotypes isolated from humans and animals. Antimicrob. Agents Chemother. 40:1009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra, B., S. M. Soto, J. M. Arguelles, and M. C. Mendoza. 2001. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella serotype [4,5,12:I:-]. Antimicrob. Agents Chemother. 45:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakanen, A., P. Kotilainen, J. Jalava, A. Siitonen, and P. Huovinen. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J. Clin. Microbiol. 37:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heising, P., B. Kratz, E. Halle, Y. Graser, M. Altwegg, W. Rabsch, and J. P. Faber. 1995. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella typhimurium from men and cattle in Germany. Microb. Drug Resist. 1:211-218. [DOI] [PubMed] [Google Scholar]
- 14.Herikstad, H., P. Hayes, M. Mokhtar, M. L. Fracaro, E. J. Threlfall, and F. J. Angulo. 1997. Emerging quinolone resistant Salmonella in the United States. Emerg. Infect. Dis. 3:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heurtin-Le Corre, C., P. Y. Donnio, M. Perrin, M. F. Travert, and J. L. Avril. 1999. Increasing incidence and comparison of nalidixic acid-resistant Salmonella enterica subs. enterica serotype typhimurium isolates from human and animals. J. Clin. Microbiol. 37:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, D. C., J. S. Wolfson, M. A. Mozza, and E. Y. Ng. 1992. Genetics and regulation of outer membrane protein expression by quinolone resistance loci nfxB, nfxC, and cfxB. Antimicrob. Agents Chemother. 36:1151-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire, A. J., D. F. J. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malorny, B., A. Schroeter, and R. Helmuth. 1999. Incidence of quinolone resistance over the period 1986 to 1998 in veterinary isolates from Germany. Antimicro. Agents Chemother. 43:2278-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, J. Verhoef, and M. E. Jones. 1999. Many class 1 integrons comprise distinct stable structures occuring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarron, B., and W.C. Love. 1997. Acalculous nontyphoidal salmonella cholecystitis requiring surgical intervention despite ciprofloxacin therapy: report of three cases. Clin. Infect. Dis. 24:707-709. [DOI] [PubMed] [Google Scholar]
- 21.Molbak, K., D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multiresistant quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 22.National Committee of Clinical Laboratory Standards. 1998. Methods for dilution antimicrobial sensitivity testing for bacteria that grow aerobically. National Committee of Clinical Laboratory Standards, Wayne, Pa.
- 23.Phillips, I. 1991. A report of the working party on antibiotic sensitivity of the British Society for Antimicrobial Chemotherapy London. London Academic Press, London, United Kingdom.
- 24.Piddock, L. J. V., V. Ricchi, I. McLaren, and D. J. Griggs. 1998. Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41:635-641. [DOI] [PubMed] [Google Scholar]
- 25.Randall, L. P., and M. J. Woodward. 2001. Multiple antibiotic resistance (mar) locus in Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 67:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall, L. P., S. W. Cooles, R. Sayers, and M. J. Woodward. 2001. Association between cyclohexane resistance in Salmonella of different serovars and increased resistance to multiple antibiotics, disinfectants and dyes. J. Med. Microbiol. 50:919-924. [DOI] [PubMed] [Google Scholar]
- 27.Reyna, F., M. Huesca, V. Gonzalez, and L. Y. Fuchs. 1995. Salmonella Typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 39:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shipp, C. R., and B. Rowe. 1980. A mechanical microtechnique for Salmonella serotyping. J. Clin. Pathol. 33:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes, H., and R. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 30.Vasallo, F. J., P. Martin-Rabadan, L. Alcala, J. M. Garcia-Lechuz, M. Rodriguez-Creimems, and E. Bouza. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 31.Walker, R. A., N. Saunders, A. J. Lawson, E. A. Lindsay, M. Dassama, L. R. Ward, M. J. Woodward, R. H. Davies, E. Liebana, and E. J. Threlfall. 2001. Use of LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J. Clin. Microbiol. 39:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, L. D., J. D. H. De Sa, and B. Rowe. 1987. A phage typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiuff, C., M. Madsen, D. L. Baggesen, and F. M. Aarestrup. 2000. Quinolone resistance among Salmonella enterica from cattle, broilers, and swine in Denmark. Microb. Drug Res. 6:11-17. [DOI] [PubMed] [Google Scholar]