Abstract
Identification of Nocardia to the species level is useful for predicting antimicrobial susceptibility patterns and defining the pathogenicity and geographic distribution of these organisms. We sought to develop an identification method which was accurate, timely, and employed tests which would be readily available in most clinical laboratories. We evaluated the API 20C AUX yeast identification system as well as several biochemical tests and Kirby-Bauer susceptibility patterns for the identification of 75 isolates encompassing the 8 medically relevant Nocardia species. There were few biochemical reactions that were sufficiently unique for species identification; of note, N. nova were positive for arylsulfatase, N. farcinica were positive for opacification of Middlebrook 7H11 agar, and N. brasiliensis and N. pseudobrasiliensis were the only species capable of liquefying gelatin. API 20C sugar assimilation patterns were unique for N. transvalensis, N. asteroides IV, and N. brevicatena. There was overlap among the assimilation patterns for the other species. Species-specific patterns of susceptibility to gentamicin, tobramycin, amikacin, and erythromycin were obtained for N. nova, N. farcinica, and N. brevicatena, while there was overlap among the susceptibility patterns for the other isolates. No single method could identify all Nocardia isolates to the species level; therefore, a combination of methods was necessary. An algorithm utilizing antibiotic susceptibility patterns, citrate utilization, acetamide utilization, and assimilation of inositol and adonitol accurately identified all isolates. The algorithm was expanded to include infrequent drug susceptibility patterns which have been reported in the literature but which were not seen in this study.
The genus Nocardia has undergone a revolution in its taxonomy in recent years. Fewer than 10 years ago, only three species of pathogenic Nocardia were defined based on the hydrolysis of casein, tyrosine, and xanthine substrates (8). Even at that time, it was apparent that there was a great deal of heterogeneity within the most common species, N. asteroides. Subsequent studies of antibiotic susceptibility patterns revealed six types within the N. asteroides complex (16). With the advent of molecular analyses, these drug pattern types were confirmed as distinct species based on PCR-restriction fragment length polymorphism (RFLP) analysis (5, 12). Accurate identification has become increasingly important as differences among species have emerged in terms of epidemiology, virulence, and antibiotic susceptibility (9, 10). In addition, a timely species identification by the laboratory can have an immediate impact on therapy. Molecular methods such as PCR-RFLP and 16S ribosomal DNA sequencing can accurately identify all medically relevant Nocardia isolates to the species level; however, not all laboratories have the capability to perform these tests. Also, biochemical characterization of species has been time consuming and problematic.
In an effort to develop a timely and technically simple identification method, several investigators have examined the utility of commercially available kits for the identification of Nocardia (1-3, 7, 11). Although several kits proved to be useful for identification (1, 11), additional species of Nocardia have been recognized since the publication of these studies (4). Although a number of phenotypic characteristics for each Nocardia species can be found in the literature, these have not been applied in a systematic manner for routine identification of Nocardia in the clinical laboratory (6, 13-15, 17; B. A. Brown, R. W. Wilson, V. A. Steingrube, Z. Blacklock, and R. J. Wallace, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. C-65, 1997). Therefore, we sought to develop an identification method which was accurate, timely, and employed tests which would be readily available in most laboratories, taking into account phenotypic characteristics previously reported. We evaluated the API 20C AUX yeast identification system as well as several biochemical tests and Kirby-Bauer susceptibility patterns for the identification of 75 isolates encompassing the 8 medically relevant Nocardia species.
MATERIALS AND METHODS
Study strains.
A total of 75 Nocardia isolates, representing 8 species, were used in this study. These isolates were obtained from the culture collections of SUNY Upstate Medical University, Syracuse, N.Y.; Good Samaritan Regional Medical Center, Phoenix, Ariz.; University of North Carolina Hospitals, Chapel Hill, N.C.; and University of Texas Health Center, Tyler, Tex. The species were as follows: N. nova (n = 19), N. farcinica (n = 16), N. asteroides VI (n = 10), N. asteroides I (n = 8), N. otitidiscaviarum (n = 8), N. brasiliensis (n = 6), N. pseudobrasiliensis (n = 5), N. asteroides IV (n = 1), N. transvalensis (n = 1), and N. brevicatena (n = 1). The following ATCC type strains are included in the numbers above: N. nova ATCC 33726, N. farcinica ATCC 3318, N. asteroides VI ATCC 14759, N. asteroides I ATCC 23824, N. otitidiscaviarum ATCC 14629, N. brasiliensis ATCC 19296, N. pseudobrasiliensis ATCC 51511 and ATCC 51512, N. asteroides IV ATCC 49872, N. transvalensis ATCC 29982, N. brevicatena ATCC 15333. Two criteria for inclusion of clinical isolates in the study were the presence of aerial hyphae and resistance to lysozyme (as described below). All isolates were initially identified to the species level using hsp65 RFLP analysis as described previously (12). For final identification purposes, the N. asteroides species were defined as follows. N. asteroides I and N. asteroides VI were considered the same species (N. asteroides sensu stricto) (4). N. asteroides IV is a member of the N. transvalensis complex but was considered a separate species from N. transvalensis based upon previous analyses (17). All isolates were stored in 15% glycerol-tryptic soy broth at −70°C during the course of the study. Isolates were subcultured twice to tryptic soy agar with 10% sheep blood (SBA) and incubated at 35°C in air for 48 to 72 h prior to testing.
Biochemical identification.
A 0.5 McFarland suspension of each organism was prepared in sterile distilled water for use in the biochemical and disk diffusion susceptibility tests. Glass beads (1 mm) were added to each tube to facilitate suspension of the organisms during vortexing. Most isolates gave a fine particulate suspension; however, N. pseudobrasiliensis strains and some N. asteroides I/VI strains were very difficult to resuspend. In these cases, a 0.5 McFarland standard was approximated visually. This did not affect the results of biochemical and susceptibility testing, since all isolates demonstrated reactions characteristic for their species.
Lysozyme resistance was determined by inoculating 4 drops of the standardized organism suspension into glycerol broth with and without lysozyme (50 μg/ml) (BBL; Becton Dickinson Microbiology Systems, Cockeysville, Md.). Tubes were incubated at 35°C in air and examined daily for up to 7 days. Lysozyme resistance was defined as equivalent growth in the lysozyme and the glycerol control tubes. Utilization of acetamide was determined by inoculation of 2 drops of the standardized organism suspension to acetamide agar slants (BBL), which were incubated at 35°C in air for up to 7 days. The test was considered positive if the entire slant turned from pale yellow-orange to bright pink. Citrate utilization was determined by inoculating a loopful of organism to citrate agar slants (BBL), which were incubated at 35°C in air for up to 7 days. The test was considered positive if a blue color developed in the slant. Gelatin hydrolysis (liquefaction) was determined by stabbing a loopful of growth ∼1 cm into a nutrient gelatin tube (BBL). The tube was incubated at 35°C in air for up to 7 days. Detection of hydrolysis was accomplished by placing the tubes at 4°C for 15 min and then tilting each tube at a 45o angle. The test was considered positive if the gelatin was liquefied. Equivalent growth at 35 and 45°C was determined by inoculating duplicate SBA plates with 2 drops of the standardized organism suspension, streaking for isolation, and incubating the plates at the respective temperatures for 3 days. This test was considered positive if the growth at 35°C was identical to that at 45°C. Production of arylsulfatase was assessed by heavily inoculating a tube of 0.003 M arylsulfatase broth (BBL) with several loopfuls of organism. The tubes were incubated at 37°C in air for 7 days, and then 6 drops of 2 N sodium carbonate (Remel, Lenexa, Kans.) was added to test for a color change. The test was considered positive if any amount of pink color developed throughout the broth. Opacification of Middlebrook 7H11 agar (BBL) was determined by inoculating 2 drops of the standardized organism suspension onto the plate and streaking for isolation (6). The plates were incubated at 35°C in air for up to 7 days. The test was considered positive if any amount of milky-white opacification developed in the agar. This was best observed by viewing from the bottom of the plate.
Antibiotic susceptibility testing.
The 0.5 McFarland suspension described above was used to inoculate Mueller-Hinton (MH) agar for disk diffusion susceptibility testing. Large MH plates (diameter, 14 cm) were used because of the large zone sizes produced with some of the antibiotics. The antibiotic disks used were gentamicin (10 μg), tobramycin (10 μg), amikacin (30 μg), erythromycin (15 μg), and ciprofloxacin (5 μg). Plates were incubated at 35°C in air for 72 h. Zone diameters were measured, and resistance to the drugs was interpreted as follows: gentamicin, ⩽15 mm (4); tobramycin, <20 mm (4); amikacin, ⩽20 mm (17); erythromycin, <30 mm (4); and ciprofloxacin, <35 mm (13).
API 20C AUX system.
API 20C AUX strips (bioMerieux, Hazelwood, Mo.) contain 19 substrates for carbohydrate assimilation testing. These include glucose, glycerol, 2-keto-d-gluconate, l-arabinose, d-xylose, adonitol, xylitol, galactose, inositol, sorbitol, α-methyl-d-glucoside, N-acetyl-d-glucosamine, cellobiose, lactose, maltose, sucrose, trehalose, melezitose, and raffinose. The test was set up according to the manufacturer's instructions using a 2 McFarland standard as an inoculum. The inoculum was prepared in sterile distilled water with glass beads (1 mm) added to the tube to facilitate suspension. As described above, with some strains only an approximate 2 McFarland standard could be prepared due to clumping of the organisms. The strips were incubated for up to 7 days at 35°C in air. To keep the substrates from dehydrating, the strips were placed in a partially sealed plastic bag with paper towels moistened with sterile distilled water. Reactions were read at 4 and 7 days by comparing turbidity in the carbohydrate wells against that of the control well.
RESULTS
The biochemical results for the isolates are shown in Table 1. The reaction endpoints for all tests were clear and easy to read. Reactions that were characteristic for particular species included the arylsulfatase test, which was positive for 17 of 19 (89%) N. nova isolates and negative for all other species. It was critical that a heavy inoculum be used in this test, since several strains gave less than a 1+ reaction in 7 days compared with arylsulfatase quantitative standards. The two strains that were negative for arylsulfatase were incubated for an additional 7 days (total, 14 days) but still remained negative. Opacification of 7H11 agar was demonstrated for 15 of 16 (94%) N. farcinica isolates; the test was negative for all other species. Most isolates demonstrated opacity within 3 to 5 days, although a few isolates required 7 days. The single isolate that had a negative opacity reaction was ATCC strain 3318, which demonstrated opacity in a previous study (6); prolonged storage of this isolate may account for this discrepancy. Acetamide utilization was positive for all N. farcinica isolates tested, a characteristic shared with only 5 of 18 (28%) N. asteroides I/VI isolates. Few species demonstrated equivalent growth at 35 and 45°C. These included all N. farcinica isolates, 5 of 10 (50%) N. asteroides I/VI isolates, 1 of 8 (12%) N. otitidiscaviarum isolates, and 1 of 19 (5%) N. nova isolates. The latter isolate was 1 of 2 N. nova strains negative for arylsulfatase. N. brasiliensis and N. pseudobrasiliensis were distinguished from all other species by their ability to liquefy gelatin; 5 of 6 (83%) N. brasiliensis and 5 of 5 (100%) N. pseudobrasiliensis isolates were positive. Although most biochemical tests required up to 7 days' incubation, reproducible results could be obtained with acetamide in 3 days.
TABLE 1.
Biochemical reactions of Nocardia species
| Species (no. of isolates) | Arylsulfatase (7 d)a | Acetamide utilization (3 d) | Gelatin liquefaction (7 d) | Equivalent growth at 35°/45°C (3 d) | Opacification of 7H11 agar (7 d) | API 20C assimilation results (4-7 d)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| glub | gly | gal | nag | ino | ado | tre | ||||||
| N. nova (19) | + (89)c | − | − | − (95) | − | + | − | − | − | − | − | − |
| N. asteroides I (8) | − | − (88) | − | − | − | + | + | − | + (63) | − | − | − |
| N. asteroides VI (10) | − | − (60) | − | + (60) | − | + | + (90) | − | − (90) | − | − | − (80) |
| N. asteroides IV (1) | − | − | − | − | − | + | + | + | − | − | − | + |
| N. farcinica (16) | − | + | − | + | + (94)e | + | +d | +d (63) | − | − | − | − (94) |
| N. otitidiscaviarum (8) | − | − | − | − (88) | − | + | + | − (75) | + | + | − | + (50) |
| N. brasiliensis (6) | − | − | + (83) | − | − | + | + | + | + | + | − | + |
| N. pseudobrasiliensis (5) | − | − | + | − | − | + | + | + | + | + | − | + |
| N. transvalensis (1) | − | − | − | − | − | + | + | + | + | − | + | −f (+) |
| N. brevicatena (1) | − | − | − | − | − | − | − | − | − | − | − | + |
Incubaton time required for result is indicated in parentheses in boxheads. d, days.
Abbreviations: glu, glucose; gly, glycerol; gal, galactose; nag, N-acetyl-d-glucosamine; ino, inositol; ado, adonitol; tre, trehalose.
Numbers in parentheses are percentages of isolates with the indicated reactions; 100% unless specified.
Some weak reactions observed.
ATCC 3318 was negative.
Result of ATCC 29982; 90% of clinical isolates are positive (17).
The API 20C assimilation results were sometimes variable among isolates of the same species, making a definitive identification by biocode somewhat difficult. Therefore, the results for a select group of carbohydrates are shown in Table 1. This combination of carbohydrates gave the best discriminatory power among species; however, it was not 100% specific. N. nova was easily distinguished by its assimilation pattern. This species was positive for glucose and negative for all other carbohydrates on the reaction strip. Only one other isolate, an N. asteroides VI strain, demonstrated this pattern. Some N. farcinica strains (7) gave weak reactions with glycerol and galactose which could result in identification of N. nova if growth was not carefully compared to the carbohydrate control. N. brevicatena was the only species negative for glucose. N. transvalensis gave a characteristic reaction pattern, this species being the only one positive for adonitol. N. brasiliensis and N. pseudobrasiliensis showed identical reaction patterns and thus could not be separated, but they could be easily distinguished from the majority of isolates. There were only two strains of N. otitidiscaviarum which gave a reaction pattern similar to the pattern of the former two species. N. asteroides IV and N. asteroides I/VI could be confused with N. farcinica on some occasions, but most of the isolates gave characteristic patterns.
The patterns of sensitivity to gentamicin, tobramycin, amikacin, and erythromycin delineated three species of Nocardia, N. brevicatena, N. nova, and N. farcinica (Table 2). The other species were divided into three groups: one consisting of the amikacin-resistant species, N. asteroides IV and N. transvalensis, another which included some N. otitidiscaviarum isolates and one isolate of N. asteroides I, and a large heterogenous group which included N. otitidiscaviarum, N. brasiliensis, N. pseudobrasiliensis, and N. asteroides I/VI.
TABLE 2.
Antibiotic susceptibility patterns of Nocardia species
| Sensitivity patterna | Speciesb |
|---|---|
| SSSS | N. nova |
| SRSS | N. nova |
| RRSS | N. nova |
| N. farcinica | |
| RSSR | N. brevicatena |
| RRSR | N. farcinica |
| N. asteroides IV | |
| SSSR | N. brasiliensis |
| N. pseudobrasiliensis | |
| N. asteroides I/VI | |
| N. otitidiscaviarum | |
| SRSR | N. otitidiscaviarum |
| N. asteroides I | |
| N. asteroides IV | |
| RRRR | N. transvalensis |
| N. asteroides IV | |
| SRRR | N. transvalensis |
| N. asteroides IV |
Sensitivity (S) or resistance (R) to gentamicin, tobramycin, amikacin, and erythromycin, respectively, as determined by Kirby-Bauer disk diffusion. See Materials and Methods for disk diffusion zone sizes.
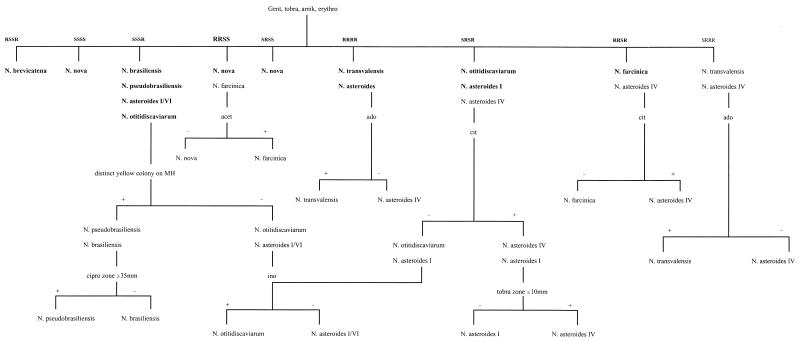
It was evident from the results that a single method could not identify all Nocardia isolates to the species level; therefore, a combination of methods was necessary. Table 3 presents additional tests that could be used in conjunction with API 20C to achieve an accurate species identification. Basically, results of a Kirby-Bauer susceptibility test could distinguish species with similar API 20C patterns. For N. brasiliensis and N. pseudobrasiliensis, production of bright yellow colony growth on MH agar could distinguish these two species from N. otitidiscaviarum. Additional supplemental tests included gelatin liquefaction, acetamide, 7H11 opacification, and arylsulfatase, but most of these reactions required up to 7 days for an accurate result. Figure 1 presents an algorithm using a minimal number of tests that demonstrated an accuracy of 100% for species identification of Nocardia. The sensitivity patterns shown in bold type represent those seen in this study and are the most common patterns reported in the literature. The additional patterns added to the algorithm are ones which have been reported previously among larger collections of isolates (15, 17). The algorithm employs the sensitivity pattern for gentamicin, tobramycin, amikacin, erythromycin, and ciprofloxacin in addition to colony pigment, citrate utilization, acetamide utilization, and results of adonitol and inositol assimilation from API 20C. An attractive feature of this identification algorithm is that most results were available within 3 to 4 days. A total of 7 days was required for distinguishing N. transvalensis and N. asteroides IV by adonitol assimilation. Separation of these species is probably not critical from a clinical standpoint, since both of these species demonstrate similar sensitivity patterns: of note, resistance to aminoglycosides (17).
TABLE 3.
Supplemental tests needed to resolve overlap of API 20C assimilation patterns
| Identification | Supplemental testsa |
|---|---|
| N. nova vs.: | |
| N. asteroides VI | Sensi or AS |
| N. farcinica | Sensi or AS or acetamide or 7H11 |
| N. farcinica vs.: | |
| N. asteroides IV | Sensi or 7H11 or acetamide |
| N. asteroides I | Sensi or 7H11 |
| N. asteroides VI | Sensi or 7H11 |
| N. otitidiscaviarum vs.: | |
| N. pseudobrasiliensis | Pigment or gelatin |
| N. brasiliensis | Pigment or gelatin |
| N. brasiliensis vs.: | |
| N. pseudobrasiliensis | Sensi |
definitions: sensi, sensitivity pattern to gentamicin, tobramycin, amikacin, erythromycin, ciprofloxacin; AS, arylsulfatase; 7H11, 7H11 opacification; pigment, distinct yellow colony on Mueller-Hinton agar.
FIG. 1.
Algorithm for identification of Nocardia species. Identification using sensitivity (S) or resistance (R) to gentamicin (gent), tobramycin (tobra), amikacin (amik), ciprofloxacin (cipro), and erythromycin (erythro) coupled with acetamide (acet), adonitol (ado), inositol (ino), citrate (cit), and pigment on MH agar. Sensitivity patterns to gentamicin, tobramycin, amikacin, and erythromycin in bold indicate reactions seen in this study; others are reported from previous studies (15, 17).
DISCUSSION
This study demonstrated the difficulty encountered with species identification of Nocardia. There were few unique biochemical reactions among the species tested, and there was overlap in the susceptibility patterns and API 20C results. However, a combination of susceptibility pattern, colony pigment, citrate utilization, acetamide utilization, and adonitol and inositol assimilation accurately identified all isolates to the species level, with most identifications complete in 4 days.
The percentage of species in this study that were either positive or negative for a particular biochemical reaction was consistent with previous reports of studies which used a larger number of strains (13-15, 17; Brown et al., 97th Gen. Meet. Am. Soc. Microbiol.). This is an important point, since only single isolates of N. brevicatena, N. transvalensis, and N. asteroides IV were used in this study due to the difficulty in acquiring these infrequent isolates. The consistency of the results suggested that the biochemicals chosen for the algorithm would provide accurate identification if applied to a larger group of isolates. In this study and other reports, the biochemicals chosen for the algorithm were highly specific, demonstrating a 100% positive or negative result. The only exception was acetamide utilization. All isolates of N. farcinica in this study were positive for acetamide, but a previous report found a positivity rate of 85% among 40 isolates (15). Only 16 isolates were tested in this study, so perhaps the number was too small to detect a strain(s) with a negative reaction. Also, differences in the acetamide formulation may explain the results. In the algorithm, acetamide is used to distinguish isolates of N. farcinica which demonstrate a susceptibility pattern similar to that of N. nova (15). These isolates are rare, and none were seen in this study. Growth temperature may be helpful in this regard, since all N. farcinica isolates and only 5% of N. nova isolates demonstrate equivalent growth at 35 and 45°C (this study and reference 15).
Several studies have examined commercially available kits for their usefulness in identifying Nocardia. Biehle et al. evaluated the MicroScan Rapid Anaerobe Identification panel, which uses chromogenic substrates for the detection of preformed enzymes (1). A combination of three substrates (p-nitrophenyl-α-d-mannopyranoside, indoxyl phosphate, and l-pyrrolidonyl-β-napthylamide) and equivalent growth at 45°C allowed identification of all species in 3 days. N. pseudobrasiliensis, N. brevicatena, N. transvalensis, and N. asteroides IV were not evaluated, since they were not yet described or were just newly recognized at that time. These species were easily distinguished by their sensitivity patterns in this study. It would seem likely that the addition of a sensitivity pattern to the above scheme coupled with p-nitrophenyl-α-d-mannopyranoside results for N. asteroides IV and N. transvalensis (17) would allow differentiation of these additional species. Muir et al. used the bioMerieux ID 32C yeast identification system for identification of Nocardia in 7 days (11). This system is similar to the API 20C system but includes additional substrates. As with our study, the assimilation results were variable among isolates of the same species; however, accurate species identification was obtained by using the last four digits of the profile. This suggested that certain substrates were more discriminatory in delineating species, as we discovered with API 20C. N. pseudobrasiliensis, N. brevicatena, and N. asteroides IV were not evaluated with the ID 32C system. N. brevicatena would likely give a unique profile, as this is the only species unable to assimilate glucose. N. pseudobrasiliensis is very similar to N. brasiliensis biochemically and would likely give a similar profile, but as noted above, the sensitivity pattern could reliably distinguish these species. N. asteroides IV would likely be distinguished by this system due to the inclusion of erythritol and rhamnose. Application of either of the two systems presented in the above-described studies, along with the suggested additional testing, would likely result in accurate identification of all eight medically relevant species in 3 to 7 days.
In the present study, API 20C assimilation patterns were for the most part characteristic for a particular species, but definitive identification required additional tests. An accurate identification could be obtained by using results from API 20C coupled with susceptibility testing. The only drawback to this method was that some isolates required 7 days for an API 20C result due to poor growth. The algorithm was devised in an effort to shorten this identification time. In the algorithm, only the inositol and adonitol results from the API 20C were used. In terms of cost and simplicity, it would be better to have individual reactions for inositol and adonitol rather than setting up an API 20C strip solely for this purpose. An attempt was made to use commercially prepared CTA media with these substrates, but the results were negative for isolates which demonstrated positive reactions with API 20C. Use of other media/reagents is being explored.
As a final note, it is critical that the algorithm be applied to isolates which demonstrate lysozyme resistance and aerial hyphae, since other genera of aerobic actinomycetes may demonstrate similar reactions to Nocardia. For some Nocardia species, particularly N. farcinica, aerial hyphae were best discerned using a dissecting microscope. Some of the thermophilic actinomycetes may demonstrate aerial hyphae and lysozyme resistance; however, these isolates would rarely be encountered in the clinical laboratory. Growth at 50°C would distinguish these genera from Nocardia (4).
In conclusion, accurate identification of Nocardia isolates was achieved in a timely manner using commercially available tests and media. The algorithm presented here should allow most clinical laboratories to routinely identify Nocardia isolates to the species level.
Acknowledgments
We thank the following individuals for providing isolates used in this study: Roy Hopfer, University of North Carolina Hospitals; Michael Saboulle and Den Sussland, Good Samaritan Regional Medical Center; and Richard Wallace and Barbara Brown-Elliot, University of Texas Health Center. Also, we gratefully acknowledge the technical advice provided by Barbara Brown-Elliot.
REFERENCES
- 1.Biehle, J. R., S. J. Cavalieri, T. Felland, and B. L. Zimmer. 1996. Novel method for rapid identification of Nocardia species by detection of preformed enzymes. J. Clin. Microbiol. 34:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bizet, C., C. Barreau, C. Harmant, M. Nowakowski, and A. Pietfroid. 1997. Identification of Rhodococcus, Gordona, and Dietzia species using carbon source utilization tests (“Biotype- 100” strips). Res. Microbiol. 148:799-809. [DOI] [PubMed] [Google Scholar]
- 3.Boiron, P., and F. Provost. 1990. Enzymatic characterization of Nocardia spp. and related bacteria by API ZYM profile. Mycopathologia 110:51-56. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. M., M. M. McNeil, and E. P. Desmond. 1999. Nocardia, Rhodococcus, Gordona. Actinomadura, Streptomyces, and other actinomycetes of medical importance, p. 370-398. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of Clinical Microbiology, 7th ed. ASM Press, Washington, D.C.
- 5.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores, M., and E. Desmond. 1993. Opacification of Middlebrook agar as an aid in identification of Nocardia farcinica. J. Clin. Microbiol. 31:3040-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilian, M. 1978. Rapid identification of Actinomycetaceae and related bacteria. J. Clin. Microbiol. 8:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Land, G., M. R. McGinnis, J. Staneck, and A. Gatson. 1991. Aerobic pathogenic Actinomycetales, p. 340-359. In A. Balows, W. J. Hausler, K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. (ed.), Manual of clinical microbiology, 5th ed. ASM Press, Washington, D.C.
- 9.Lerner, P. I. 1996. Nocardiosis. Clin. Infect. Dis. 22:891-905. [DOI] [PubMed] [Google Scholar]
- 10.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muir, D. B., and R. C. Pritchard. 1997. Use of the BioMerieux ID 32C yeast identification system for identification of aerobic actinomycetes of medical importance. J. Clin. Microbiol. 35:3240-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Z. Blacklock, J. L. Gibson, and R. J. Wallace. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace, R. J., B. A. Brown, Z. Blacklock, R. Ulrich, K. Jost, J. M. Brown, M. M. McNeil, G. Onyi, V. A. Steingrube, and J. Gibson. 1995. New Nocardia taxon among isolates of Nocardia brasiliensis associated with invasive disease. J. Clin. Microbiol. 33:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace, R. J., B. A. Brown, M. Tsukamura, J. M. Brown, and G. O. Onyi. 1991. Clinical and laboratory features of Nocardia nova. J. Clin. Microbiol. 29:2407-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace, R. J., M. Tsukamura, B. A. Brown, J. Brown, V. A. Steingrube, Y. Zhang, and D. R. Nash. 1990. Cefotaxime-resistant Nocardia asteroides strains are isolates of the controversial species Nocardia farcinica. J. Clin. Microbiol. 28:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. J. Clin. Microbiol. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson, R. W., V. A. Steingrube, B. A. Brown, Z. Blacklock, K. C. Jost, A. McNabb, W. D. Colby, J. R. Biehle, J. L. Gibson, and R. J. Wallace. 1997. Recognition of a Nocardia transvalensis complex by resistance to aminoglycosides, including amikacin, and PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]