Abstract
The complete sequence of rpoB, the gene encoding the beta subunit of RNA polymerase was determined for Staphylococcus saccharolyticus, Staphylococcus lugdunensis, S taphylococcus caprae, and Staphylococcus intermedius and partial sequences were obtained for an additional 27 Staphylococcus species. The complete rpoB sequences varied in length from 3,452 to 3,845 bp and had a 36.8 to 39.2% GC content. The partial sequences had 71.6 to 93.6% interspecies homology and exhibited a 0.08 to 0.8% intraspecific divergence. With a few exceptions, the phylogenetic relationships inferred from the partial rpoB sequences were in agreement with those previously derived from DNA-DNA hybridization studies and analyses of 16S ribosomal DNA gene sequences and partial HSP60 gene sequences. The staphylococcal rpoB sequence database we established enabled us to develop a molecular method for identifying Staphylococcus isolates by PCR followed by direct sequencing of the 751-bp amplicon. In blind tests, this method correctly identified 10 Staphylococcus isolates, and no positive results were obtained with 10 non-Staphylococcus gram-positive and gram-negative bacterial isolates. We propose partial sequencing of the rpoB gene as a new tool for the accurate identification of Staphylococcus isolates.
The genus Staphylococcus comprises 36 species of gram-positive, catalase-positive cocciform bacteria, nine of which also contain subdivisions with subspecies designations (11, 25). Only a few species are coagulase positive, namely, Staphylococcus aureus, Staphylococcus intermedius, Staphylococcus delphini, Staphylococcus schleiferi subsp. coagulans, and some strains of Staphylococcus hyicus (22). These species are readily isolated, and cultures may be obtained from environmental, animal, and human clinical samples (24). S. aureus is well documented as an opportunistic pathogen in humans and is responsible for staphylococcal toxin-mediated food poisoning, toxic shock syndrome, and pyogenic infections characterized by frequent metastatic spread (22). Also, nosocomial infections due to methicillin-resistant S. aureus have emerged as major clinical and epidemiological problems (22). The coagulase-negative staphylococcal species constitute a major component of the normal microflora of humans. Their role in nosocomial infections has been well documented and has increased dramatically within the past 20 years (21). Precise delineation of the clinical disease produced by this group of bacteria is based on accurate species identification (21), aiding clinicians in the choice of appropriate antibiotic (13). Therefore, there is a pressing need for the rapid and accurate identification of isolates of clinically important coagulase-negative staphylococci. Such a goal remains difficult.
Currently, conventional benchtop phenotypic characterization is used for the identification of Staphylococcus species. For the reference identification of staphylococci the Centers for Disease Control and Prevention has developed a numerical code system based on a panel of 18 selected conventional biochemical reactions. Several biochemical-based commercial kits and automated systems (23) are also available for this purpose. In clinical studies, however, various degrees of accuracy have been achieved with these different phenotypic formats (12, 33, 35). For example, commercial identification systems have been shown to commonly misidentify Staphylococcus hominis and Staphylococcus warneri with error rates as high as 36 and 27%, respectively (16, 17). S. schleiferi may also be misidentified by automated systems (5). Phenotypic characterizations thus have limitations that are partly due to variable expression of characters and ambiguity in the interpretation of end point reactions (3).
The use of nucleic acid targets with their high sensitivity and specificity may provide an alternative means of accurately identifying Staphylococcus species. Several molecular targets have been exploited for the molecular identification of Staphylococcus species, including the 16S rRNA gene (2, 8), the tRNA gene intergenic spacer (27), the heat shock protein 60 (HSP60) gene (14, 15, 26), and the femA gene (44). These targets, however, have been exploited through the technology of molecular probe hybridization, and therefore, they are useful only in laboratories that have the complete panel of probes and then only for identifying recognized Staphylococcus species. Further molecular targets that have been identified include the nuc gene, which occurs only in S. aureus (4), and a chromosomal DNA fragment specific for Staphylococcus epidermidis (29). There is, therefore, still a need for a simple and reliable molecular test for the identification of all Staphylococcus species (19).
rpoB, the gene encoding the highly conserved β subunit of the bacterial RNA polymerase, has previously been demonstrated to be a suitable target on which to base the identification of enteric bacteria (31), spirochetes (37), bartonellas (36), and rickettsias (10). The gene has been shown to be more discriminative than the 16S ribosomal DNA (rDNA) gene (31), which has also been used for identifying enteric bacteria. In this report we describe the molecular identification of Staphylococcus isolates using a PCR-sequencing approach based on the sequence of staphylococcal rpoB.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. They were grown on blood agar at 37°C under aerobic conditions except for Staphylococcus aureus subsp. anaerobius and Staphylococcus saccharolyticus, which were grown under anaerobic conditions. All isolates were streaked on blood agar plates to determine the purity of each of the cultures by macroscopic examination of colonies and microscopic examination of gram-stained preparations. Colonies were scraped from the plates and boiled for 15 min in Chelex 100 (39) and 1% sodium dodecyl sulfate solution (Bio-Rad Laboratories, Hercules, Calif.) before genomic DNA was extracted and purified with QIAamp DNA minikits (Qiagen GmbH, Hilden, Germany).
TABLE 1.
List of 31 Staphylococcus species investigated by rpoB sequencing
| Species or subspecies | GenBank accession no. | Straina |
|---|---|---|
| S. caprae | AF325868 | CIP 104000T |
| S. gallinarum | AF325890 | CIP 103504T |
| S. aureus subsp. anaerobius | AF325894 | CIP 103780T |
| S. aureus subsp. aureus | X64172 | CIP 103428 |
| S. epidermidis | AF325872 | CIP 81.55T |
| S. haemolyticus | AF325888 | CIP 81.56T |
| S. intermedius | AF325869 | CIP 81.60T |
| S. lugdunensis | AF325870 | CIP 103642T |
| S. saccharolyticus | AF325871 | CIP 103275T |
| S. schleiferi subsp. schleiferi | AF325886 | CIP 103643T |
| S. xylosus | AF325883 | CIP 81.66T |
| S. capitis subsp. capitis | AF325885 | ATCC 27840T |
| S. arlettae | AF325874 | ATCC 43957T |
| S. warneri | AF325887 | ATCC 27836T |
| S. hominis | AF325875 | ATCC 27844T |
| S. simulans | AF325877 | ATCC 27848T |
| S. saprophyticus | AF325873 | ATCC 15305T |
| S. equorum | AF325882 | ATCC 43958T |
| S. cohnii subsp. cohnii | AF325893 | ATCC 29974T |
| S. auricularis | AF325889 | ATCC 33753T |
| S. carnosus subsp. carnosus | AF325880 | ATCC 51365T |
| S. kloosii | AF325891 | ATCC 43959T |
| S. chromogenes | AF325892 | ATCC 43764T |
| S. hyicus subsp. hyicus | AF325876 | ATCC 11249T |
| S. pulvereri | AF325879 | CCUG 33938T |
| S. felis | AF325878 | CIP 103366T |
| S. lentus | AF 036973 | ATCC 49574 |
| S. muscae | AF325884 | CIP 1035641T |
| S. sciuri | AF325881 | ATCC 29062T |
ATCC, American Type Culture Collection; CCUG, Culture Collection of the University of Göteborg, Goteborg, Sweden; CIP, Institut Pasteur Collection, Paris, France.
Construction of a Staphylococcus rpoB database.
Consensus rpoB PCR primers were designed after the alignment of rpoB genes of S. aureus (GenBank accession number X64172) and Bacillus subtilis (GenBank accession number L43593) and numbered on the basis of the S. aureus rpoB sequence. Primer pair 1302F-2309R was used to amplify a 1,007-bp fragment in S. saccharolyticus, Staphylococcus lugdunensis, Staphylococcus caprae, and S. intermedius. These species were chosen as representatives of the major phyla from the 16S rDNA phylogenetic tree. Additional oligonucleotides were selected on the basis of data obtained from ongoing base sequence determinations (Table 2). The alignment of the rpoB sequences of S. aureus, S. lugdunensis, S. intermedius, S. saccharolyticus, and S. caprae enabled us to design consensus PCR primers 2491F (5′-AACCAATTCCGTATIGGTTT-3′; base positions 2491 to 2511) and 3554R (5′-CCGTCCCAAGTCATGAAAC-3′; base positions 3554 to 3573) to amplify a 1,081-bp variable fragment in 27 additional Staphylococcus species under investigation. All PCR mixtures contained 2.5 × 10−2 U of Taq polymerase per μl, 1× Taq buffer, 1.8 mM MgCl2 (Gibco BRL, Life Technologies, Cergy Pontoise, France), 200 μM concentrations of dATP, dTTP, dGTP, and dCTP (Boehringer Manheim GmbH, Hilden, Germany), and 0.2 μM concentrationsof each primer (Eurogentec, Seraing, Belgium). PCR mixtures were subjected to 35 cycles of denaturation at 94°C for 30 s, primer annealing at 52°C for 30 s, and de novo DNA extension at 72°C for 60 s. Every amplification program began with a denaturation step of 95°C for 2 min and ended with a final elongation step of 72°C for 5 min. Amplicons were purified for sequencing using a QIAquick spin PCR purification kit (Qiagen) following the protocol of the supplier. Partial reverse and forward (bidirectional) sequencing of a 598-bp fragment was obtained by using internal primers at positions 2643 (2643F: 5′-CAA TTC ATG GAC CAA GC-3′) and 3241 (3241R: 5′-GCI ACI TGI TCC ATA CCT GT-3′). Sequencing reactions were carried out with the reagents of the ABI Prism dRhodamine dye terminator cycle sequencing ready reaction kit (Perkin Elmer Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions and with the following program: 30 cycles of denaturation at 94°C for 10 s, primer annealing at 50°C for 10 s, and extension at 60°C for 2 min. Products of sequencing reactions were separated by electrophoresis on a 0.2-mm 6% polyacrylamide denaturing gel and recorded with an ABI Prism 377 DNA sequencer (Perkin Elmer Applied Biosystems) following the standard protocol of the supplier. The results were processed into sequence data with Sequence Analysis software (Applied Biosystems), and partial sequences were combined into a single consensus sequence with Sequence Assembler software (Applied Biosystems). The rpoB sequences were aligned by using the multisequence alignment program of Sequence Assembler software (Applied Biosystems). Pairwise sequence comparisons for nucleic acid or peptidic sequence homology were determined with PC Gene software (Intelligenetics, Mountain View, Calif.).
TABLE 2.
Primers used in the sequencing of the entire rpoB gene in S. lugdunensis, S. intermedius, S. saccharolyticus, and S. caprae
| Primer name | Primer sequence (5′-3′) | Positiona | Tm (°C) |
|---|---|---|---|
| 30F | GGTTTAGGATTAAAAGATGC | 30-50 | 41 |
| 192F | GAAGAAGTTGGAGCTACTG | 192-211 | 44 |
| 806F | AATAAGAGCAGGGAAAGAAAC | 806-827 | 43 |
| 920F | AAAGAAAAGAATGAATGAACTT | 920-942 | 39 |
| 1165F | TATGCTTATGGTATTTAGCTA | 1165-1186 | 39 |
| 1302F | AAACTTAATAGAAATTCAAACTAAA | 1302-1327 | 58 |
| 1450F | GTTCAAACGATAAATAGAGAA | 1450-1471 | 39 |
| 1741F | GAAACAGATGCTAAAGATGT | 1741-1761 | 41 |
| 1850F | CCATATACTGCGAGTGGGAA | 1850-1870 | 47 |
| 2245F | TAGAAATTCAATCAATTAAGTATATG | 2245-2271 | 62 |
| 2309F | TTGGTAATGCTTTACCAGAT | 2309-2329 | 41 |
| 2334F | TGCATTACACCAGCAGATATCATTG | 2334-2359 | 70 |
| 2412F | GATGATATTGACCATTTAGG | 2412-243 | 41 |
| 2534F | TGAAAGAATGTCAATTCAAGA | 2534-2555 | 39 |
| 2663F | AAACCCATTAGCTGAGTT | 2663-268 | 38 |
| 2995F | TGGTCGTTTCATGGATGATGAAGTTG | 2995-3119 | 74 |
| 2924F | AAGATAGCTATGTTGTAGCA | 2924-2944 | 41 |
| 3200F | CTTAGAGAACGATGACTCTAA | 3200-3221 | 43 |
| 3498F | TAGTTGGTTTCATGACTTGGGA | 3498-3520 | 46 |
| 3550F | TTGAAAGTCCAACAAAGCAA | 3550-3570 | 38 |
| 3843F | GGTAAAGTAACGCCTAAAGGT | 3843-3864 | 45 |
| 4494F | TGGAGGTATGGGCACTTGAA | 4494-4514 | 47 |
| 1759R | ACATCTTTMGCATCTGTTTC | 1779-1759 | 48 |
| 1460R | ATCGTTTGAACGCCACTCTT | 1480-1460 | 45 |
| 1910R | TCATAGTAAGTTTGCGCCAT | 1930-1910 | 43 |
| 2309R | ATCTGGTAAAGCATTACCAA | 2329-2309 | 41 |
| 2334R | CAATGATATCTGCTGGTGTAATGCA | 2354-2334 | 68 |
| 2432R | CCTAAATGGTCAATATCATC | 2452-2432 | 41 |
| 2573R | CGAATATTAATTAATTGTTG | 2593-2573 | 34 |
| 2892R | GTGATAGCATGTGTATCTAAATCA | 2912-2892 | 64 |
| 2915R | TAACTATCTTCTTCATCAGC | 2935-2915 | 41 |
| 2924R | TGCTACAACATAGCTATCTT | 2944-2924 | 41 |
| 2995R | CAACTTCATCATCCATGAAACGACCA | 3015-2995 | 74 |
| Cm32b | ATGCAACGTCAGGCCGTTCCG | 3211-3191 | 64 |
| 3321R | AGACGACGAACAGAATTTCA | 3341-3321 | 56 |
| 3610R | GCTCGAATGATAACGTGATT | 3630-3610 | 43 |
| 4139R | ACTTGTCCrATGTTCATACG | 4159-4139 | 44 |
| 4502R | CATATGCTTCAAGTGCCCATA | 4523-4502 | 45 |
| 4508R | CCAAGTGGTTGTTGTGTAAC | 2428-4508 | 45 |
| 4871R | TTTAGAGCTTTCACTGTTTG | 4891-4871 | 41 |
| 5000R | CACCATATGACCAAGAACGAA | 5021-5000 | 45 |
| 5018R | CAATCAAGGAGCCTACCTCCTT | 5040-5018 | 50 |
| 5030R | GAAATTATTTACATCAATCAA | 5051-5030 | 36 |
| 5041R | TAACTATCTTCTTCATCAGC | 5061-5041 | 41 |
| 5085R | CCCAGTCTTTTGTAGGTCCG | 5105-5085 | 49 |
| 5188R | CCCATTCTTTCACGACGTAC | 5208-5188 | 47 |
Position relative to the S. aureus rpoB sequence.
Phylogenetic comparisons.
Phylogenetic trees were constructed based on rpoB sequences and compared with the phylogenetic tree based on 16S rDNA sequence data. The nucleotide sequences were aligned using the ClustalW program (42) supported in the Bisance workstation (9). Phylogenetic trees were inferred with the PHYLIP software package (12) supported in the Bisance workstation. Evolutionary matrices, generated by DNADIST, were constructed by the method of Kimura (18). The matrices were used to infer dendrograms using the neighbor-joining method (38). Bootstrap values were obtained for a consensus based on 100 randomly generated trees using Seqboot and Consense in the PHYLIP package. Tree figures were generated with the Tree View program, version 1.61 (32). Sequence homology comparisons were made with the GAP program of the Genetics Computer Group (Madison, Wis.) package, again supported in the Bisance workstation.
rpoB sequence-based identification blind testing.
The rpoB-based system we developed to identify staphylococci was first applied to a collection of clinical isolates in order to assess intraspecific sequence variation. This collection comprised five S. aureus isolates, five S. epidermidis isolates, three S. saprophyticus isolates, and three S. haemolyticus isolates. Blind tests were then performed on a collection of 20 bacterial isolates, including rifampin-susceptible S. aureus, rifampin-resistant S. aureus, S. epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus, S. intermedius, Staphylococcus equorum, S. schleiferi, S. lugdunensis, Staphylococcus gallinarum, Escherichia coli, Pseudomonas aeruginosa, Streptococcus pneumoniae, Enterococcus faecalis, Streptococcus pyogenes, Corynebacterium amycolatum, Gemella morbillorum, Acinetobacter anitratus, Micrococcus luteus, and Propionibacterium acnes. After the isolates were coded, extraction of bacterial DNA and PCR incorporating primer pair 2491F-3241R were performed as described above. The presence of 751-bp amplicons was revealed by 1% agarose gel electrophoresis, and amplicons were sequenced with sequencing primers 2643F and 3241F as described above. Sequences were aligned by using FASTA with the Staphylococcus rpoB database in Infobiogen (http://www.infobiogen.fr/services/analyseq/cgi-bin/fasta_in.pl), and identification was assessed by >97% similarity with one of the database sequences.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the staphylococcal rpoB sequences determined in this study are AF325870 for the S. lugdunensis complete rpoB sequence, AF325869 for the S. intermedius complete rpoB sequence, AF325871 for the S. saccharolyticus complete rpoB sequence, AF325868 for the S. caprae complete rpoB sequence, AF325875 for the S. hominis partial rpoB sequence, AF325876 for the Staphylococcus hyicus subsp. hyicus partial rpoB sequence, AF325880 for the Staphylococcus carnosus partial rpoB sequence, AF325881 for the Staphylococcus sciuri partial rpoB sequence, AF325885 for the Staphylococcus capitis partial rpoB sequence, AF325890 for the S. gallinarum partial rpoB sequence, AF325894 for the S. aureus subsp. anaerobius partial rpoB sequence, X64172 for the Staphylococcus aureus subsp. aureus partial rpoB sequence, AF325872 for the S. epidermidis partial rpoB sequence, AF325888 for the S. haemolyticus partial rpoB sequence, AF325886 for the Staphylococcus schleiferi subsp. schleiferi partial rpoB sequence, AF325883 for the Staphylococcus xylosus partial rpoB sequence, AF325885 for the Staphylococcus capitis subsp. capitis partial rpoB sequence, AF325874 for the S. arlettae partial rpoB sequence, AF325887 for the S. warneri partial rpoB sequence, AF325877 for the Staphylococcus simulans partial rpoB sequence, AF036973 for the Staphylococcus lentus partial rpoB sequence, AF325873 for the S. saprophyticus partial rpoB sequence, AF325882 for the S. equorum partial rpoB sequence, AF325893 for the Staphylococcus cohnii subsp. cohnii partial rpoB sequence, AF325889 for the Staphylococcus auricularis partial rpoB sequence, AF325880 for the Staphylococcus carnosus subsp. carnosus partial rpoB sequence, AF325891 for the Staphylococcus kloosii partial rpoB sequence, AF325892 for the Staphylococcus chromogenes partial rpoB sequence, AF325879 for the Staphylococcus pulvereri partial rpoB sequence, AF325878 for the Staphylococcus felis partial rpoB sequence, and AF325884 for the Staphylococcus muscae partial rpoB sequence.
RESULTS
Determination of rpoB sequences in Staphylococcus species.
Consensus primers 1302F and 2309R enabled the amplification of a 1,107-bp rpoB fragment and consensus primers 3843F and 5041R enabled the amplification of a 1,198-bp rpoB fragment in S. intermedius. Alignment of these two fragment sequences along with that of S. aureus rpoB enabled us to design the additional primers 2245F, 4139F and 4139R, 4508F and 4502R, and 4494F and 1751R, which allowed us to determine the complete sequence of rpoB of S. intermedius. The complete sequence for S. caprae was obtained with the same primers, whereas the additional primers 2924F, 2924R, 3200F, and 3050R were required for S. lugdunensis and the additional primers 5000R, 2915R, and 2663F were required for S. saccharolyticus. The rpoB sequence varied in length, being 3,845 bp in S. intermedius, 3,461 bp in S. caprae, 3,452 bp in S. lugdunensis, and 3,461 bp in S. saccharolyticus. The GC contents were 39.2, 37.4, 38.4, and 36.8% in the above species, respectively. Based on the alignment of these four sequences with that of S. aureus rpoB in GenBank, the most variable region flanked by conserved regions was chosen as a target for PCR-sequencing-based molecular identification of staphylococci. Consensus PCR primers 2491F and 3241R were designed for the amplification of the region, and one additional primer, 2643F, was designed for sequencing. An rpoB partial sequence database was done for an additional 27 Staphylococcus species under investigation.
Phylogenetic trees.
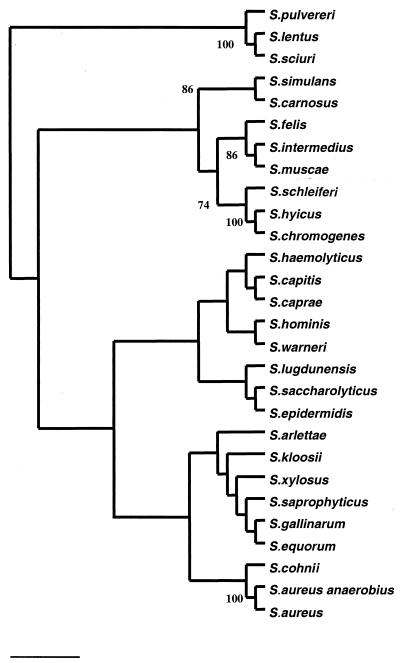
The rpoB sequence similarity matrix tabulated after ClustalW alignment of 29 staphylococcal rpoB partial sequences showed interspecific similarity values of 71.6 to 93.6%, the most closely related sequences being those of S. aureus subsp. aureus and S. aureus subsp. anaerobius. The phylogenetic tree (Fig. 1) inferred from neighbor joining showed nine clusters which were characterized by a sequence similarity level of >85% within the cluster. The nine clusters were the S. aureus group, comprising S. aureus subsp. aureus, S. aureus subsp. anaerobius, and S. cohnii; the S. saprophyticus group, comprising S. saprophyticus, S. xylosus, S. kloosii, S. equorum, S. gallinarum, and S. arlettae; the S. haemolyticus group, comprising S. haemolyticus, S. capitis, and S. caprae; the S. sciuri group, comprising S. sciuri, S. pulvereri, and S. lentus; the S. epidermidis group, comprising S. epidermidis, S. saccharolyticus, and S. lugdunensis; the S. simulans group, comprising S. simulans and S. carnosus; and the S. hyicus group, comprising S. hyicus, S. schleiferi, and S. chromogenes. Bootstrap values of >70% were, however, noted only for five nodes, namely, the S. aureus group, the S. hyicus group, the S. intermedius group, the S. simulans group and the S. sciuri group.
FIG. 1.
Unrooted neighbor-joining tree based on partial rpoB gene sequences showing the phylogenetic relationships among 29 species of the genus Staphylococcus. The numbers are the estimated confidence levels, expressed as percentages, for the positions of the branches, determined by bootstrap analysis; only bootstraps values of >70% are indicated. The scale bar indicates the evolutionary distance between sequences, determined by measuring the lengths of the horizontal lines connecting two organisms (0.01 substitution/site).
Results of blind identification testing.
A 751-bp amplicon was obtained in 16 of 16 Staphylococcus sp. isolates previously identified at the species level and belonging to four different species. In this collection, intraspecies sequence divergence was 0.5% among five S. aureus isolates, 0.8% among five S. epidermidis isolates, and 0.08% among three S. saprophyticus isolates as well as among three S. haemolyticus isolates. Sequence analysis assigned every one of these 16 isolates to the correct species. A 751-bp amplicon was further obtained for 10 of 20 bacterial isolates tested after PCR with the primer pair 2491F-3241R. The sequences of these 10 amplicons had >99% homology with our reference staphylococcal sequences, and an exact identification was obtained for all 10. None of the other species used in the trial were amplified in the PCRs.
DISCUSSION
The data we present show that PCR amplification of a 751-bp rpoB gene fragment using primer pair 2491F-3241R followed by sequence analysis is a suitable molecular approach for the identification of Staphylococcus isolates at the species level. Moreover, these primer pairs were shown to be specific for Staphylococcus, since no amplification products were obtained from species in 10 other bacterial genera.
To date, the primary method for the molecular identification of Staphylococcus species has been the analysis of genomic DNA restriction fragment length patterns on Southern blots probed with labeled rRNA genes (2, 6-8, 43, 45). PCR and sequencing of rRNA genes, however, have been found to have limited discriminating power for Staphylococcus species, since the 16S rDNA sequence similarity has been shown to be very high, 90 to 99%, in 29 Staphylococcus species (26). By contrast, the sequence similarity of the hsp60 gene was 74 to 93% in 23 Staphylococcus species (26), and for the partial rpoB gene sequence in our study the similarity was even less, 71.6 to 93.6% in 29 Staphylococcus species. The rpoB gene, then, clearly has the most discriminative power, and while S. caprae and S. capitis cannot be distinguished by their 16S rDNA gene sequences (41), the partial rpoB sequences of these two species have only 84.5% homology. Similarly, some Staphylococcus taxa have the same 16S rDNA gene sequences in variable regions V1, V3, V7, and V9 (34) with identical sequences occurring in S. pulvereri and Staphylococcus vitulinus; in S. saccharolyticus, S. capitis subsp. urealyticus, and S. caprae; in the two subspecies of S. aureus; and in the two subspecies of S. cohnii (41). The rpoB partial sequence similarity, however, was only 87.3% between the two subspecies of S. aureus and 84.5 to 89% in the S. saccharolyticus, S. capitis, and S. caprae group. Also, we determined that intraspecific sequence variation was 0.08 to 0.5%, further indicating that this rpoB fragment was an efficient tool for species identification.
The gene encoding chaperonin 60 has also been proposed as a suitable target for the molecular identification of Staphylococcus species (14, 15). A unique pair of degenerate PCR primers which amplify a 600-bp region of hsp60 have been described, and the sequences of this region have been reported for S. aureus, S. epidermidis, S. haemolyticus, S. lugdunensis, S. saprophyticus, S. schleiferi (14), and S. intermedius, S. delphini, and four S. hyicus strains (15). It should be noted, however, that the cloned partial HSP60 gene DNA sequences of nine isolates of S. aureus showed a mean variability of 2% (40). Also, cross-hybridization occurred between cloned partial HSP60 gene DNAs from the two S. schleiferi subspecies and between the DNAs from S. intermedius and S. delphini (15). Also, the DNA from isolates identified as S. hyicus failed to hybridize. Other suitable targets for the molecular identification of Staphylococcus species that have been proposed include the femA gene, which was used in a multiplex PCR-reverse hybridization approach to identify 55 clinical isolates (44). These, however, included only five Staphylococcus species, namely S. aureus, S. epidermidis, S. hominis, S. saprophyticus, and S. simulans. Discrepant results were found in the identification of S. aureus using tRNA gene intergenic spacer length polymorphism (27), but restriction fragment length polymorphism analysis of the PCR-amplified 16S-23S rDNA intergenic spacer proved to be effective in the identification of a collection of 221 isolates from 31 Staphylococcus species (30). Some intraspecific heterogeneity was shown for Staphylococcus saprophyticus subsp. saprophyticus and for S. aureus, with five fragment length patterns being observed in eight isolates. Also, PCR-RFLP analysis of the gla gene has been carried out, but only with 12 species of Staphylococcus, with some degree of intraspecific heterogeneity being observed in three species (47). Finally, molecular identification methods for the identification of one or only a few Staphylococcus species have been reported for S. saprophyticus (28), S. aureus (1), and S. epidermidis (29, 46). In our study, we used PCR sequencing of the rpoB gene to effectively identify staphylococcal species. In contrast to the probe hybridization technique and the RFLP approach, sequencing enables any isolate to be characterized, including new species by their phylogenetic relationships.
The phylogenetic relationships among Staphylococcus species we derived in our study of rpoB sequences were limited, as some species and subspecies (11) were not available for study and only partial rpoB gene sequences were analyzed. The rpoB sequence-based relationships we found, however, were in accordance with those published previously. Earlier studies on the taxonomy of Staphylococcus species based on DNA-DNA reassociation indicated that in the genus there were nine distinct species groups, represented by S. epidermidis, S. saprophyticus, S. simulans, S. intermedius, S. hyicus, S. sciuri, S. auricularis, S . aureus, and Staphylococcus caseolyticus (23, 24). The same groups were identified in a study using hsp60 gene sequence analysis (26). The 16S rDNA sequence-derived trees (40, 41) gave different results, however, with 12 genogroups being identified. In fact, it has been shown that S. caseolyticus should be reclassified as Macrococcus caseolyticus (20), thus limiting the number of 16S rDNA gene sequence-derived genogroups.
In our rpoB-derived data, we found that S. lugdunensis is related to S. epidermidis, as was also found by DNA hybridization. The position of the organism should be regarded as uncertain, however, as low bootstrap values (≤50%) were obtained in studies of the bacteria using 16S rDNA (41) and hsp60 (20) sequence data. The rpoB-derived data indicating that S. cohnii is outside the S. saprophyticus group and closely related to S. aureus are in disagreement with previously published data and may be incorrect. Similarly, the positions of S. caprae and S. capitis within the S. haemolyticus group and outside the S. epidermidis group appear to be incorrect. Complete rpoB sequence analysis of these species may be helpful in resolving these questions.
Acknowledgments
We acknowledge Patrick Kelly for reviewing the manuscript.
REFERENCES
- 1.Benito, M. J., M. M. Rodriguez, M. G. Cordoba, E. Aranda, and J. J. Cordoba. 2000. Rapid differentiation of Staphylococcus aureus from Staphylococcus spp. by arbitrarily primed-polymerase chain reaction. Lett. Appl. Microbiol. 31:368-373. [DOI] [PubMed] [Google Scholar]
- 2.Bialkowska-Hobrzanska, H., H. V. Harry, D. Jaskot, and O. Hammerberg. 1990. Typing of coagulase-negative staphylococci by Southern hybridization of chromosomal DNA fingerprints using a ribosomal RNA probe. Eur. J. Microbiol. Infect. Dis. 9:588-594. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum, D., M. Kelly, and A. W. Chow. 1991. Epidemiologic typing systems for coagulase-negative staphylococci. Infect. Control Hosp. Epidemiol. 12:319-326. [DOI] [PubMed] [Google Scholar]
- 4.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo, J., J. L. Hernandez, M. C. Farinas, D. Garcia-Palomo, and J. Aguero. 2000. Osteomyelitis caused by Staphylococcus schleiferi and evidence for misidentification of this Staphylococcus species by an automated identification system. J. Clin. Microbiol. 38:3887-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesneau, O., A. Morvan, F. Grimont, H. Labischinski, and N. El Solh. 1993. Staphylococcus pasteuri sp. nov., isolation from human, animal, and food specimens. Int. J. Syst. Bacteriol. 43:237-244. [DOI] [PubMed] [Google Scholar]
- 7.De Buyser, M. L., A. Morvan, S. Aubert, F. Dilasser, and N. El Solh. 1989. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 135:989-991. [DOI] [PubMed] [Google Scholar]
- 8.De Buyser, M. L., A. Morvan, S. Aubert, F. Dilasser, and N. El Solh. 1992. Evaluation of ribosomal RNA gene probe for the identification of species and sub-species within the genus Staphylococcus. J. Gen. Microbiol. 138:889-899. [DOI] [PubMed] [Google Scholar]
- 9.Dessen, P., C. Fondat, C. Valencien, and G. Meunier. 1990. BISANCE: a French service for access to biomolecular sequences databases. Comput. Appl. Biol. Sci. 6:355-356. [DOI] [PubMed] [Google Scholar]
- 10.Drancourt, M., and D. Raoult. 1999. Characterization of mutations in the rpoB gene in naturally rifampin-resistant Rickettsia species. Antimicrob. Agents Chemother. 43:2400-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Euzéby, J. P. 1997. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int. J. Syst. Bacteriol. 47:590-592. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 13.Fernandes, A. P., T. M. Perl, and L. M. Herwaldt. 1996. Staphylococcus cohnii: a case report on an unusual pathogen. Clin. Performance Qual. Health Care 4:107-109. [PubMed] [Google Scholar]
- 14.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, C. E., D. L. Sewell, M. Pfaller, R. V. Bumgardner, and J. A. Williams. 1994. Evaluation of two commercial systems for identification of coagulase-negative staphylococci to species level. Diagn. Microbiol. Infect. Dis. 18:1-5. [DOI] [PubMed] [Google Scholar]
- 17.Ieven, M., J. Verhoeven, S. R. Pattyn, and H. Goossens. 1995. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 33:1060-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, M. 1980. A simple method for estimating evolutional rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. E 16:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Kleeman, K. T., T. L. Bannerman, and W. E. Kloos. 1993. Species distribution of coagulase-negative staphylococcal isolates at a community hospital and implications for selection of staphylococcal identification procedures. J. Clin. Microbiol. 31:1318-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloos, W. E., D. N. Ballard, C. G. George, J. A. Webster, R. J. Hubner, W. Ludwig, K.-H. Schleifer, F. Fiedler, and K. Schubert. 1998. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. nov. and Macrococcus carouselicus sp. nov. Int. J. Syst. Bacteriol. 48:859-877. [DOI] [PubMed] [Google Scholar]
- 21.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kloos, W. E., and T. L. Bannerman. 1995. Staphylococcus and Micrococcus, p. 282-298. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 23.Kloos, W. E., and C. G. George. 1991. Identification of Staphylococcus species and subspecies with the Microscan Pos ID and rapid Pos ID panel systems. J. Clin. Microbiol. 29:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kloos, W. E., and K.-H. Schleifer. 1986. Genus IV. Staphylococcus, p. 1013-1035. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 25.Kloos, W. E., K.-H. Schleifer, and R. Götz. 1991. The genus Staphylococcus, p.1369-1420. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed., vol. 2. Springer, New York, N.Y.
- 26.Kwok, A. Y., S. C. Su, R. P. Reynolds, S. J. Bay, Y. Av-Gay, N. J. Dovichi, and A. W. Chow. 1999. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int. J. Syst. Bacteriol. 49:1181-1192. [DOI] [PubMed] [Google Scholar]
- 27.Maes, N., Y. De Gheldre, R. DeRyck, M. Vaneechoutte, H. Meugnier, J. Etienne, and M. J. Struelens. 1997. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J. Clin. Microbiol. 35:2477-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martineau, F., F. J. Picard, C. Menard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Development of a rapid PCR assay specific for Staphylococcus saprophyticus and application to direct detection from urine samples. J. Clin. Microbiol. 38:3280-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1996. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J. Clin. Microbiol. 34:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza, M., H. Meugnier, M. Bes, J. Etienne, and J. Freney. 1998. Identification of Staphylococcus species by 16-23S rDNA intergenic spacer PCR analysis. Int. J. Syst. Bacteriol. 48:1049-1055. [DOI] [PubMed] [Google Scholar]
- 31.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB gene sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 32.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 33.Perl, T. M., P. R. Rhomberg, M. J. Bale, P. C. Fuchs, R. N. Jones, F. P. Koontz, and M. A. Pfaller. 1994. Comparison of identification systems for Staphylococcus epidermidis and other coagulase-negative Staphylococcus species. Diagn. Microbiol. Infect. Dis. 18:151-155. [DOI] [PubMed] [Google Scholar]
- 34.Raué, H. A., W. Musters, C. A. Rutgers, J. Van't-Riet, and R. J. Planta. 1990. rRNA: from structure to function, p. 217-235. In W. E. Hill, P. B. Moore, A. Dahlberg, D. Schlessinger, R. A. Garrett, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 35.Refshal, K., and B. M. Andersen. 1992. Clinical relevant coagulase-negative staphylococci: identification and resistance pattern. J. Hosp. Infect. 22:19-31. [DOI] [PubMed] [Google Scholar]
- 36.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39:430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renesto, P., K. Lorvellec-Guillon, M. Drancourt, and D. Raoult. 2000. rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema, and Leptospira. J. Clin. Microbiol. 38:2200-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Med. Biol. E 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Stein, A., and D. Raoult. 1992. A simple method for amplification of DNA from paraffin-embedded tissues. Nucleic Acids Res. 20:5237-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, T., M. Kaneto, Y. Mori, M. Tsuji, N. Kikuchi, and T. Hiramune. 1997. Phylogenetic analyses of Staphylococcus based on the 16S rDNA sequence and assignment of clinical isolates from animals. J. Vet. Med. Sci. 59:775-783. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomson-Carter, F. M., P. E. Carter, and T. H. Pennington. 1989. Differentiation of staphylococcal species and strains by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 135:2093-2097. [DOI] [PubMed] [Google Scholar]
- 44.Vannuffel, P., M. Heusterspreute, M. Bouyer, B. Vandercam, M. Philippe, and J.-L. Gala. 1999. Molecular characterization of femA from Staphylococcus hominis and Staphylococcus saprophyticus, and femA-based discrimination of staphylococcal species. Res. Microbiol. 150:129-141. [DOI] [PubMed] [Google Scholar]
- 45.Webster, J. A., T. L. Bannerman, R. J. Hubner, D. N. Ballard, E. M. Cole, J. L. Bruce, F. Fiedler, K. Schubert, and W. E. Kloos. 1994. Identification of the Staphylococcus sciuri species group with EcoRI fragments containing rRNA sequences and description of Staphylococcus vitulus sp. nov. Int. J. Syst. Bacteriol. 44:454-460. [DOI] [PubMed] [Google Scholar]
- 46.Wieser, M., and H. J. Busse. 2000. Rapid identification of Staphylococcus epidermidis. Int. J. Syst. E vol. Microbiol. 50:1087-1093. [DOI] [PubMed] [Google Scholar]
- 47.Yugueros, J., A. Temprano, B. Berzal, M. Sanchez, C. Hernanz, and J. M. Luengo. 2000. Glyceraldehyde-3-phosphate dehydrogenase-encoding gene, a new taxonomic tool for Staphylococcus spp. J. Clin. Microbiol. 38:4351-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]