Abstract
A sensitive, specific, and high-throughput oligonucleotide ligation assay (OLA) for the detection of genotypic human immunodeficiency virus type 1 (HIV-1) resistance to Food and Drug Administration-approved protease inhibitors was developed and evaluated. This ligation-based assay uses differentially modified oligonucleotides specific for wild-type or mutant sequences, allowing sensitive and simple detection of both genotypes in a single well of a microtiter plate. Oligonucleotides were designed to detect primary mutations associated with high-level resistance to amprenavir, nelfinavir, indinavir, ritonavir, saquinavir, and lopinavir, including amino acid substitutions D30N, I50V, V82A/S/T, I84V, N88D, and L90M. Plasma HIV-1 RNA from 54 infected patients was amplified by reverse transcription-PCR and sequenced by using dideoxynucleotide chain terminators for evaluation of mutations associated with drug resistance. These same amplicons were genotyped by the OLA at positions 30, 50, 82, 88, 84, and 90 for a total of 312 codons. The sensitivity of detection of drug-resistant genotypes was 96.7% (87 of 90 mutant codons) in the OLA compared to 92.2% (83 of 90) in consensus sequencing, presumably due to the increased sensitivity of the OLA. The OLA detected genetic subpopulations more often than sequencing, detecting 30 mixtures of mutant and wild-type sequences and two mixtures of drug-resistant sequences compared to 15 detected by DNA sequencing. Reproducible and semiquantitative detection of the mutant and the wild-type genomes by the OLA was observed by analysis of wild-type and mutant plasmid mixtures containing as little as 5% of either genotype in a background of the opposite genome. This rapid, simple, economical, and highly sensitive assay provides a practical alternative to dideoxy sequencing for genotypic evaluation of HIV-1 resistance to antiretrovirals.
Viral resistance to the antiretroviral drugs used for treatment of human immunodeficiency virus type 1 (HIV-1) infection has been an important cause of treatment failure and limits options for alternative antiretroviral regimens. The utility of monitoring HIV-1 for drug resistance in the clinical setting has not been fully defined. Several studies have suggested that evaluating HIV-1 drug resistance increased the likelihood of suppressing viral replication with subsequent treatment (2, 7, 11; C. Cohen, S. Hunt, M. Sension, C. Farthing, M. Conant, S. Jacobson, J. Nadler, W. Verbiest, K. Hertogs, M. Ames, and A. Rinehart, Abstr. 7th Conf. Retrovir. Opportunistic Infect., abstr. 237, 2000), whereas other studies found that testing for drug resistance did not improve outcome (R. Haubrich, P. Keiser, C. Kemper, M. Witt, J. Leedom, D. Forthal, M. Leibowitz, J. Hwang, E. Seefried, J. A. McCutchan, N. Hellmann, D. Richman, et al., Antivir. Ther., abstr. 80, 2001; J. L. Meynard, M. Vray, L. Morand-Joubert, et al., Antivir. Ther., abstr. 85, 2000). In addition, transmission of HIV-1 drug-resistant variants has been reported (1, 16, 30, 31, 33; S. J. Little, S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Coup, B. Conway, E. Connick, M. S. Saag, A. Mwatha, L. Corey, P. H. Keiser, M. Kilby, K. Dawson, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman, Antivir. Ther., abstr. 25, 2001) with an increasing incidence, which is apparently related to the widespread use of antiretrovirals. Early reports suggest that highly active antiretroviral therapy was less effective in suppressing viral replication in individuals infected with drug-resistant virus, and when suppression of replication occurred, it was less durable (Little et al., Antivir. Ther.; K. Van Vaerenbergh, L. Debaisieux, I. Derdelinckx, N. De Cabooter, K. De Smet, K. Fransen, D. Marissens, K. Miller, G. Muyldermans, S. Sprecher, D. Vaira, C. Verhofstede, G. Zissis, M. Van Ranst, E. De Clercq, and A. M. Vandamme, Antivir. Ther., abstr. 130, 2001). In this context, HIV-1 drug resistance screening may prove useful in guiding the choice of initial therapeutic regimens by identifying drugs that are unlikely to suppress viral replication.
Oligonucleotide ligation assays (OLAs) are rapid, specific, and sensitive reactions for the detection of known point mutations (15, 27). Ligation assays are based on the covalent joining of two adjacent oligonucleotide probes by a DNA ligase when they are hybridized to a cDNA template, usually a PCR product. The specificity of the ligation is regulated by three factors: (i) the hybridization of the oligonucleotides to complementary sequences within the template, (ii) the need for these primers to anneal directly adjacent to one another in a 5′ to 3′ orientation on the target, and (iii) the requirement that the oligonucleotides have two bases complementary with the target in each direction at the site of the junction (15). These characteristics allow nonstringent ligation conditions, which can be used to type multiple nucleotide substitutions in a single assay yet require specificity at the site of interest.
Our laboratory has developed an OLA for the detection of mutations in the HIV-1 pol gene associated with resistance to zidovudine, dideoxyinosine, and lamivudine (9) to nonnucleoside reverse transcriptase inhibitors (unpublished data) and, in collaboration with others, to mutations associated with multinucleoside drug resistance (29). This high-throughput ligation-based system uses differentially modified oligonucleotides specific for wild-type or mutant sequences, allowing sensitive detection of both genotypes in a single well of a microtiter plate. In this report, we describe an OLA for the detection of primary mutations in HIV-1 pol associated with high-level resistance to protease inhibitors currently in clinical use: nelfinavir, saquinavir, ritonavir, amprenavir, indinavir, and lopinavir.
While sequencing HIV-1 nucleic acids is commonly used to evaluate the presence of drug resistance mutations and provides the opportunity to comprehensively evaluate multiple mutations associated with HIV-1 antiretroviral resistance and novel mutations, the OLA has several advantages over consensus sequencing. First, the assay has a high throughput that makes it ideal for epidemiologic studies or clinical trials that evaluate a large number of specimens for specific mutations. Second, the OLA is highly sensitive in the detection of small populations of mutant genotypes among wild-type viral sequences (9). Third, the results of the OLA are simple to interpret either visually or by a spectrophotometer. These attributes, together with its low cost, make this assay suitable for genotypic evaluation of HIV-1 drug resistance in laboratories where the costly equipment, software, and technical expertise needed for sequencing analysis may not be available.
MATERIALS AND METHODS
Patients’ specimens.
PCR products, derived from the plasma of clinical specimens submitted to the University of Washington Clinical Virology Laboratory for genotypic evaluation of HIV-1, were selected for OLA testing based on the detection of mutations by direct dideoxynucleotide sequencing that have been associated with high-level resistance to protease inhibitors. The PCR products that remained following direct dideoxy sequencing were evaluated, in a blinded fashion, by the OLA, and the genotypes derived by the two methods were later compared.
RT-PCR and PCR.
HIV-1 RNA obtained by silica extraction (3) from 1 or 2 ml of plasma was used for reverse transcription (RT) with the GeneAmp RNA PCR Core kit (Applied Biosystems, Foster City, Calif.). The first round of PCR was carried out in a 50-μl reaction volume containing 10 μl of cDNA, 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM concentrations of deoxynucleoside triphosphates, 20 pmol of each primer (PRL [GGGACCAGCGGCTACACTAGAAGAAATGATGACAGCATGTCAGG] and RT2 [9]), and 2.5 U of Taq DNA polymerase. Cycling conditions consisted of an initial denaturation step of 94°C for 5 min, followed by 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 2 min, and a final extension step of 72°C for 7 min. The second round of PCR contained 2 μl of first round product and 20 pmol of primers PRC (CTCCCCCTCAGAAGCAGGAGCCGATAGACAAGGAAACTGTATCC) and RT3 (9). Cycling conditions were as follows: 94°C for 5 min; 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min; and final extension of 72°C for 7 min. The product amplified, a 1,033-bp DNA fragment extending from HIV-1 gag to amino acid 237 of the reverse transcriptase gene, was visualized in a 1.5% agarose gel following electrophoresis and ethidium bromide staining.
Sequencing.
Residual PCR primers and deoxynucleoside triphosphates were removed from the amplified DNA products by treatment with shrimp alkaline phosphatase and exonuclease I (PCR Product Pre-Sequencing kit; Amersham Laboratories, Arlington Heights, Ill.). Cleaned PCR products were then directly bidirectionally sequenced by using forward primers PRC and RT4 (9) and reverse primers PR2 (GGAGTATTGTATGGATTTTCAGGCC) and RT3 with fluorescence-labeled dideoxy chain terminators (ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems) and an ABI PRISM 310 automated sequencer (Applied Biosystems). Sequences encoding HIV-1 protease and reverse transcriptase were assembled and edited with Sequencher 3.0 (GenCodes, Ann Arbor, Mich.).
Molecular cloning of controls for OLA.
HIV-1 mutant and wild-type controls for the OLA were obtained, respectively, by cloning (TOPO TA Cloning kit with TOP10 cells; Invitrogen Corporation, Carlsbad, Calif.) PCR-amplified nucleic acid from clinical specimens, containing the drug resistance mutations of interest, and DNA from 8E5 cells (10). Plasmid DNA extraction and purification were performed with the QIAprep Spin Miniprep kit (Qiagen Inc., Valencia, Calif.). The cloned HIV-1 sequences were amplified from the plasmid constructs by using the second round PCR primers and conditions described above. The plasmid DNA products were sequenced to confirm the presence of the mutations of interest.
Sensitivity of the OLA in the detection of drug-resistant genotypes mixed into wild-type virus.
The plasmid constructs with drug resistance mutations and wild-type sequences were used as controls for the OLA and to evaluate the sensitivity of the assay in detecting small populations of drug-resistant and wild-type genotypes. Drug-resistant plasmid DNA was mixed with the wild-type DNA at 5, 10, 25, 50, 75, 90, and 95% of the total. One nanogram of DNA from each of the mixtures was PCR amplified with the second round primers as described above. The amplicons were analyzed by the OLA in duplicate in two or three different assays.
Oligonucleotides for ligation detection.
Ligation oligonucleotides specific for wild-type or mutant codon sequences (Table 1) were modified at the 5′ end by addition of digoxigenin or fluorescein, respectively. The oligonucleotide complementary to each type common sequence adjacent to the wild-type or mutant codon was biotinylated at the 3′ end and phosphorylated at the 5′ end (9). Modified, highly purified, salt-free oligonucleotides were obtained from MWG-Biotech Inc. (High Point, N.C.).
TABLE 1.
Oligonucleotides used in a ligation assay to detect mutations in HIV-1 pol associated with resistance to protease inhibitors
| Drug(s) | Codon | Amino acid substitution | Genotype detected | Sequence (5′→3′)a |
|---|---|---|---|---|
| Nelfinavir | 30 | WTb | dig-TATTAGATACAGGAGCAGATG | |
| D30N | Mutant | f-TATTAGATACAGGAGCAGATA | ||
| Common | p-ATACAGTATTAGAAGAAATGAAT-bio | |||
| 88 | WT | dig-CCTGTCAACATAATTGGAAGAA | ||
| N88D | Mutant | f-CCTGTCAACATAATTGGAAGAG | ||
| Common | p-ATCTGTTGACTCAGATTGGTTG-bio | |||
| Amprenavir | 50 | WT | dig-AAACCAAAAATGATAGGGGGAA | |
| I50V | Mutant | f-AAACCAAAAATGATAGGGGGAG | ||
| Common | p-TTGGAGGTTTTATCAAAGTAAGA-bio | |||
| Ritonavir, indinavir, lopinavirc | 82 | WT | dig-TATTAGTAGGACCTACACCTGT | |
| V82A | Mutant A | f-TATTAGTAGGACCTACACCTGC | ||
| V82S | Mutant S | f-TATTAGTAGGACCTACACCTAG | ||
| V82T | Mutant T | f-TATTAGTAGGACCTACACCTAC | ||
| Common | p-CAACATAATTGGAAGAAATCTGT-bio | |||
| Amprenavir, indinavir | 84 | WT | dig-AGGACCTACACCTGTCAACA | |
| Nelfinavir, ritonavir | I84V | Mutant | f-AGGACCTACACCTGTCAACG | |
| Saquinavir, lopinavirc | Common | p-TAATTGGAAGAAATCTGTTGACT-bio | ||
| Saquinavir, nelfinavir | 90 | WT | dig-CAACATAATTGGAAGAAATCTGT | |
| L90M | Mutant | f-CAACATAATTGGAAGAAATCTGA | ||
| Common | p-TGACTCAGATTGGTTGCACTTT-bio |
Bases comprising the codons of interest are in boldface type. dig, digoxigenin; f, fluorescein; p, phosphate; bio, biotin.
WT, wild-type.
Mutations associated with HIV-1 resistance to lopinavir in phenotypic susceptibility assays and with virologic failure in clinical trials (13; Harrigan et al., Antivir. Ther.; Kempf et al., Antivir. Ther.; Molla et al., Antivir. Ther.).
OLA.
The procedure and reaction conditions were as described previously (9), except each reaction mixture contained 0.167 U of Ampligase DNA ligase (Epicentre Technologies, Madison, Wis.) and 0.333 pmol of each of the ligation oligonucleotides (the two genotype-specific oligonucleotides and the adjacent type common primer for the specific codon tested). Three different mutant genotypes at amino acid 82 were assayed in separate reactions. All patient samples and controls were analyzed in duplicate. Cutoff values for absorbance readings of oligonucleotides designed to detect mutant genotypes were determined for each codon by analysis of multiple reactions of these oligonucleotides with HIV-1 DNA from 8E5 cells or plasmid constructs that had 100% of the wild-type genome. Optical densities at 490 nm (OD490) were measured in a Dynatech MR 5000 plate reader (Dynatech Laboratories, Chantilly, Va.), and the cutoff values were established as the mean OD490 + 2.5 standard deviations (SD). Samples were defined as positive for the mutant genotype at an OD490 of ≥0.060 for codon 30, an OD490 of ≥0.040 for codon 50, an OD490 of ≥0.030 for codon 82A, an OD490 of ≥0.025 for codons 82S, 82T, and 90, an OD490 of ≥0.100 for codon 84, and an OD490 of ≥0.080 for codon 88. For reactions using oligonucleotides designed to detect wild-type genomes, the absorbance threshold for each codon was determined by analysis of reactions containing 100% of the appropriate drug-resistant DNA from plasmid constructs. Using the mean OD450 + 2.5 SD, samples were called positive for the wild-type genotype at an OD450 of ≥0.160 for codon 30, an OD450 of ≥0.060 for codons 50 and 82A, an OD450 of ≥0.033 for codon 82S, an OD450 of ≥0.025 for codon 82T, an OD450 of ≥0.040 for codon 84, an OD450 of ≥0.140 for codon 88, and an OD450 of ≥0.026 for codon 90. To control for high background and assay conditions, HIV-1 mutant and wild-type controls for each codon tested were tested along with specimens in every assay.
RESULTS
Selection of oligonucleotides for the assay.
Oligonucleotides were designed to detect mutations at amino acids 30, 50, 82, 84, 88, and 90 of the HIV-1 protease gene, as they are regarded as the primary mutations responsible for resistance to the currently Food and Drug Administration-approved protease inhibitors: amprenavir, indinavir, nelfinavir, ritonavir, saquinavir, and lopinavir (5, 6, 8, 12, 13, 17, 18, 19, 21, 22, 32; P. R. Harrigan, C. Van Den Eynde, and B. A. Larder, Antivir. Ther., abstr. 49, 2001; D. Kempf, M. King, J. Isaacson, R. Rode, S. Brun, and E. Sun, Antivir. Ther., abstr. 61, 2001; A. Molla, S. Brun, K. Garren, H. Mo, B. Richards, T. Marsh, J. Sylte, M. King, L. Han, E. Sun, and D. Kempf, Antivir. Ther., abstr. 64, 2001; J. G. Prado, T. Wrin, J. Beauchaine, L. Ruiz, C. J. Petropoulos, B. Clotet, R. D'Aquila, and J. Martinez-Picado, Antivir. Ther., abstr. 67, 2001). Table 1 shows the oligonucleotide sequences, the amino acid substitutions detected, and the drug(s) to which these mutations appear to confer resistance. The G48V substitution is also considered a primary mutation in the development of resistance to saquinavir; however, L90M has been more commonly associated with the use of the drug in vivo (12, 24). Our analysis of 256 sequences from HIV-1-infected patients on different antiretroviral regimens showed that only 1 of 9 patients with the G48V substitution did not have any of the other mutations detected by our assay. An analysis of the 256 sequences in our clinical laboratory sequence database in 1999 indicated that screening sequences for the mutations included in our OLA would detect 99.2% (121 of 122) of the specimens with primary mutations associated with HIV-1 resistance to protease inhibitors (data not shown).
Comparison of OLA and dideoxy consensus sequencing.
Specimens from 54 HIV-infected patients with unknown antiretroviral histories were chosen for resistance analysis by the OLA. These specimens were selected from a database of genotyping results obtained by dideoxynucleotide sequencing that listed all nonsynonymous mutations based on the presence of any of the protease primary mutations screened by the OLA and, in 11 cases, additional mutations in the vicinity of the ligation site to evaluate their effect on the sensitivity of the assay. These 11 specimens had mutations at codon 85 or 89, the only mutations found in the sequence database situated immediately adjacent to a codon screened by the OLA. Plasma HIV-1 RNA from these specimens was amplified by RT-PCR, and a consensus sequence was obtained by using dideoxynucleotide chain terminators. The amplified HIV-1 pol genome that remained after dideoxynucleotide sequencing was genotyped by the OLA at codons 30, 82, 84, 88, and 90 in all 54 amplicons and at codon 50 in 42 amplicons, for a total of 312 codons analyzed. A comparison of the genotypes obtained by OLA and dideoxy sequencing is summarized in Table 2.
TABLE 2.
Summary of subjects' genotypes for codons associated with HIV-1 drug resistance to protease inhibitors as determined by OLA and dideoxy sequencing
| Genotyping result | No. of codons detected by OLA and sequencing at protease amino acid:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D30N
|
I50V
|
V82A/S/T
|
I84V
|
N88D
|
L90M
|
|||||||
| OLA | Seqa | OLA | Seq | OLA | Seq | OLA | Seq | OLA | Seq | OLA | Seq | |
| No. of WTb | 44 | 43 | 35 | 37 | 30 | 30 | 38 | 43 | 40 | 47 | 27 | 28 |
| Total no. of MTc | 10 | 11 | 5 | 5 | 22 | 24e | 14 | 11 | 10 | 7 | 26 | 26 |
| No. of MT only | 4 | 7 | 2 | 3 | 18 | 23 | 8 | 10 | 5 | 6 | 20 | 20 |
| No. of mixtures (MT and WT) | 6 | 4 | 3 | 2 | 4 | 1 | 6 | 1 | 5 | 1 | 6 | 6 |
| No. of indeterminantd | 0 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 4 | 0 | 1 | 0 |
| No. of falsely negative | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 1 | 1 |
Seq, dideoxy sequencing.
WT, wild-type.
MT, mutant. Total number of MT equals the number of samples with MT genotype plus the samples with mixtures of wild-type and MT genotype.
Indeterminant refers to an OLA negative for both wild-type and mutant genomes.
Includes one specimen with a V82I mutation, a mutation not associated with HIV-1 drug resistance (24).
Of the 312 codons analyzed, 80 were genotyped as mutant both by OLA and direct consensus sequencing of the same PCR product. The OLA detected mutant viral sequences in an additional seven that had been genotyped as wild type by dideoxynucleotide sequencing. Each of these seven had mixtures of mutant and wild-type sequences in the OLA, three at HIV-1 protease amino acid 84, three at amino acid 88, and one at amino acid 90. These specimens were assayed by the OLA on two separate occasions, and both indicated the presence of the mutant genotype. A reevaluation of the sequencing electrophoretograms at these sites revealed the presence of small peaks corresponding to the mutant base detected by OLA, confirming the mixture of genotypes, for four of the seven codons. There was no indication of the mutant genome in the other three by dideoxynucleotide sequencing. On the other hand, dideoxynucleotide sequencing detected a total of 83 drug-resistant codons, 3 of which were not detected by the OLA. One of these had an indeterminate genotype by OLA, i.e., neither the mutant nor the wild-type reactions were positive, and the other two genotyped as wild-type sequence (Table 3 and see below).
TABLE 3.
Nucleotide sequences of HIV-1 amplicons that failed to be genotyped by OLA, including the region complementary to the ligation oligonucleotides on either side of the ligation site
| Patient no. | Direct sequencing (5′→3′)a | Protease amino acid | OLA resultb |
|---|---|---|---|
| azh32 | TA TTA GAT ACA GGA GCA GACA ∗ AT ACA GTA TTA GAA GAA GTG GAT | D30N | WT |
| ays52 | AAA CCA AAA ATA ATA GTG GGA A ∗ TT GGA GGT TTT ACC AAA GTA AGA | I50 | INDTR |
| aqu27 | AAA CCA AAA CTG ATA GGG GGTA ∗ TT GGA GGT TTT GTC AAA GTA AGA | I50 | INDTR |
| awz34 | TA TTA GTA GGA CCT ACA CCT GC ∗ T AAC ATA ATT GGA AGA AAT GTG T | V82A | INDTR |
| bmy37 | TA TTA ATA GGA CCT ACA CCT AT ∗ C AAC ATA GTT GGA AGA AAT CTG T | V82I | INDTR |
| ajz26 | A GGA CCT ACA CCT AGC AAC A ∗ TAGTT GGA AGA AAT CTG TTG ACT | I84 | INDTR |
| bhl34 | A GGA CCT ACA CCG AGC AAC A ∗ TA ATT GGA AGA AAT CTG TTG ACT | I84 | INDTR |
| awm29 | CCT GTC AAC ATA ATT GGA AGGA ∗ AT CTG ATG ACT CAG ATT GGT TG | N88 | INDTR |
| axh51 | CCT ACC AAC GTA GTT GGA AGA A ∗ AT CTG ATG ACT CAA ATT GGC TG | N88 | INDTR |
| awz34 | CCT GCC AAC ATA ATT GGA AGA A ∗ ATGTG TTG ACT CAG ATT GGT TG | N88 | INDTR |
| bwo55 | CCT ACC AAC GTA ATT GGA AGA A ∗ ATATT TTG ACT CAA ATT GGC TG | N88 | INDTR |
| bwo55 | C AAC GTA ATT GGA AGA AAT ATTT ∗ TG ACT CAA ATT GGC TGC ACT TT | L90 | INDTR |
| ays52 | C AAC ATA ATT GGA AGA AAT CTG/AT/A ∗ TG ACT CAG CTT GGC TGC AC | L90L/M | WT |
Boldface type indicates base changes no longer complementary to the ligation oligonucleotides included in our assay. The drug resistance codon tested in the assay is underlined. An asterisk indicates the ligation site.
WT, wild type; INDTR, indeterminate genotype with negative reaction for both wild-type and mutant genomes.
Of the 312 protease codons analyzed, 11 (3.5%) had indeterminate results by the OLA due to mutations not included in the assay at or near the ligation site. Table 3 shows the nucleotide sequence surrounding the ligation site of these specimens. Eight of the 11 sequences had mutations within three nucleotides of the ligation site. One sequence (patient bhl34) had three consecutive base changes located six nucleotides from the ligation site. No mutations were found near the ligation site for the remaining two sequences (patient ays52 and patient axh51); however, they had multiple base changes along the region corresponding to the ligation oligonucleotides, which apparently interfered with annealing of the oligonucleotides. Table 3 also shows the nucleotide sequences of the two specimens with mutant codons that were genotyped as wild type by the OLA (patient azh32 and patient ays52). Patient azh32, in addition to the D30N mutation, had a nucleotide substitution two bases upstream of the ligation site, which should have inhibited the ligation reaction and produced an indeterminate result. However, the specimen appeared to contain a small population of virus with the wild-type sequence, and this was apparently detected in the OLA, whereas the larger mutant population detected by dideoxynucleotide sequencing could not be detected in the OLA due to the additional mutation interfering with the ligation. Review of the electrophoretogram from dideoxy sequencing of patient ays52 supported this assessment, as it clearly showed dual peaks, indicating a mixture of viruses containing either a double mutation at codons 89 and 90 or the wild-type sequence at these sites.
Sensitivity of OLA in detecting minority genotypes.
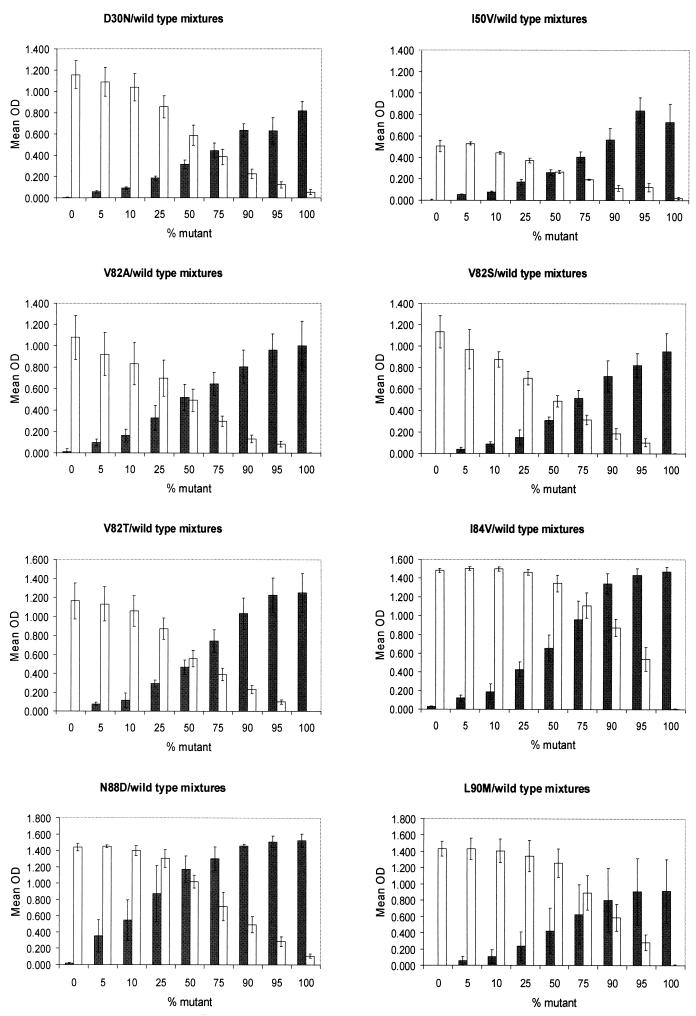
Thirty of the 312 codons were genotyped as mixtures of wild-type and mutant sequences by the OLA compared to only 15 mixtures detected by sequencing. The OLA also detected two mixtures containing the mutation V82A in addition to the V82T mutation detected by sequencing, suggesting a greater sensitivity of the OLA for the detection of mixed genotypes. Assays of mixtures of plasmid DNA with drug-resistant sequences and wild-type sequences at ratios ranging from 5 to 95% confirmed the high sensitivity of the OLA. Mutant virus was detected by the OLA at the lowest concentration tested, 5%, at codons 50, 82 (A, S and T), 84, 88, and 90 (Fig. 1) and at 10% at codon 30. The corresponding wild-type codons were also detected at the 5% level when mixed with the mutant genome, except for codon 30, which was detected at 10%. Mutant and wild-type viruses at codon 30 were visually positive at the 5% concentration compared to their respective negative controls; however, the absorbance readings fell below the cutoff value established for the assay. These assays had high reproducibility, as evidenced by the relatively low SD (Fig. 1) when the mixtures were assayed in four to six reactions.
FIG. 1.
The sensitivity of the OLA was evaluated for the detection of HIV-1 genotypes of low prevalence. Mixtures of wild-type and drug-resistant genomes derived from plasmid DNA were prepared. One nanogram of DNA from each mixture was PCR amplified and evaluated by the OLA. The mean absorbance readings (± SD) for four or six reactions are shown for mutant genome (shaded bars, OD490) and wild-type genome (white bars, OD450). Low concentrations (5%) of mutant and wild-type genomes were detected both by OD readings and direct observation of color in the microtiter plate wells.
DISCUSSION
We have developed a rapid, highly sensitive, and specific assay for the detection of mutations encoding amino acids 30, 50, 82, 84, 88, and 90 of the HIV-1 protease that are associated with high-level drug resistance to currently available protease inhibitors.
The overall sensitivity for the detection of drug-resistant genotypes was 96.7% (87 of 90 mutant codons tested). Of the three mutant codons not detected by the OLA, one had an indeterminate genotype due to the absence of ligation to the wild-type and mutant oligonucleotide detectors, and two erroneously typed as wild type. The codon with indeterminate results corresponded to a specimen that had the V82A mutation, but it also had an additional base change within the ligation site (GTC→GCT) that precluded ligation. The two mutant codons erroneously genotyped as wild type had mutations in addition to those associated with drug resistance that precluded ligation to the mutant detector. The wild-type result obtained by the OLA for these two codons suggested that these samples contained a small population of wild-type virus, which appeared to be the case for one of the two upon reexamination of the sequence electrophoretogram.
The presence of alternative mutations, mutations no longer complementary to the OLA oligonucleotides, was the most common cause of the assay failing or giving indeterminate results. Of the 312 codons analyzed by OLA, 11 (3.5%) had an indeterminate assay. This is not surprising, considering that polymorphisms and accessory mutations in the HIV-1 protease gene are common (4, 14, 25), even among therapy-naive HIV-1-infected patients. All but 1 of the 54 specimens evaluated included drug-resistant mutants, and 20% of the specimens (11 of 54) were selected for evaluation due to the presence of genetic polymorphisms in the vicinity of the codons analyzed by the OLA (codons 85 and 89). Thus, the 3.5% failure rate would be typical of highly drug-experienced populations. Most of the indeterminate OLA reactions were due to mutations located within three bases of the ligation site or to the presence of two or more base changes in the patient's specimen in the region complementary to one of the ligation oligonucleotides. Mutations within two bases of the ligation site have been shown to preclude ligation (9, 15). The third base from the ligation site appeared in this study to occasionally interfere with the ligation. For example, polymorphisms at codon 89, including the L89I mutation, which contained two base changes (CTG→ATT), interfered with the ligation reaction both at codons 88 (third base) and 90 (second base), and the L89V mutation (CTG→GTG) inhibited the ligation at codon 88 (third base) in one of two specimens. The L89M substitution (CTG→ATG), on the other hand, did not affect the ligation at codon 88 in either of two specimens with this mutation. I85V (ATT→GTT), a polymorphism present in six of the specimens analyzed, precluded ligation at codon 84 in only one of the specimens. Mutations at codons 82, 84, 88, and 90, despite their proximity, did not interfere with the OLA at the neighboring codons, except when present in combination with additional mutations in the region of the ligation oligonucleotides. Of note, a failed ligation did not lead to a miscall of the genotype, as dually negative, or indeterminate, results in the OLA are indicative of a genotype not evaluated by the assay and dideoxynucleotide sequence analysis is required.
False-negative results were infrequent (2.2% of instances) with the OLA, occurring when a mutant virus not evaluated or detected by the OLA coexisted in a specimen with wild-type virus. A higher rate of false negatives (7.8%) occurred with direct dideoxynucleotide sequencing, all presumably due to the insensitivity of consensus fluorescence-based sequencing to detect minor (<25%) genetic subpopulations (20, 23; R. Shuurman, D. Brambilla, T. de Groot, et al., Antivir. Ther., abstr. 58, 1999). The greater sensitivity of the OLA to detect minor HIV-1 species was further demonstrated by our experiments testing various proportions of mutant and wild-type plasmid-derived sequences, where as little as 5% of either the mutant or the wild-type genotype was detected in a background of the opposite genome.
The hybridization line probe assay (Murex Diagnostics Corp., Norcross, Ga.) appears to have a sensitivity similar to that of the OLA in the detection of mutations encoding HIV-1 mutants resistant to reverse transcriptase inhibitors (26, 28). The sensitivity and specificity of the line probe assay for detection of mutations encoding drug-resistant protease have not been evaluated, and the multiple polymorphisms in the protease gene could compromise this type of assay that relies on detection by DNA probes.
Thus, the high sensitivity of the OLA reduces the potential for false-negative results when mutant virus populations are present at very low levels, such as when resistant variants are first emerging during antiretroviral treatment or when selective pressure by a drug has been removed. Detection of these minor populations of mutant viruses may be important in the design of effective antiretroviral therapeutic drug regimens, especially among therapy naive individuals when a drug history would not be available to guide the choice of drugs (Little et al., Antivir. Ther.).
In summary, the OLA is a rapid, sensitive, and specific assay for monitoring HIV-1 primary mutations associated with resistance to nucleosides (9, 29), nonnucleosides (M. Mahalanabis, I. A. Beck, G. Pepper, A. Wright, S. Hamilton, W. E. Naugler, G. Ellis, and L. M. Frenkel, unpublished data), and protease inhibitors. The assay is highly adaptable and can easily incorporate additional oligonucleotides to detect new mutations as they are identified. Finally, because the OLA is simple to perform and interpret and does not require expensive equipment or technical expertise, it offers a practical alternative to dideoxynucleotide sequencing for the detection of HIV-1 mutations associated with high-level resistance to antiretrovirals where resources are limited.
Acknowledgments
This project was supported by National Institutes of Health grant no. RO1 HD36184 and UNAIDS/WHO grant no. HQ/98/468435 MOLECU 01.
REFERENCES
- 1.Balotta, C., A. Berlusconi, A. Pan, M. Violin, C. Riva, M. C. Colombo, A. Gori, L. Papagno, S. Corvasce, R. Mazzucchelli, G. Facchi, R. Velleca, G. Saporetti, M. Galli, S. Rusconi, and M. Moroni. 2000. Prevalence of transmitted nucleoside analogue-resistant HIV-1 strains and pre-existing mutations in pol reverse transcriptase and protease region: outcome after treatment in recently infected individuals. Antivir. Ther. 5:7-14. [PubMed] [Google Scholar]
- 2.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, T. C. Merigan, et al. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossi, P., M. Mouroux, A. Yvon, F. Bricaire, H. Agut, J.-M. Huraux, C. Katlama, and V. Calvez. 1999. Polymorphism of the human immunodeficiency virus type 1 (HIV-1) protease gene and response of HIV-1-infected patients to a protease inhibitor. J. Clin. Microbiol. 37:2910-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 7.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomized controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 8.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelstein, R., D. Nickerson, V. Tobe, L. Manns-Arcuino, and L. M. Frenkel. 1998. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J. Clin. Microbiol. 36:569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folks, T., D. Powell, M. Lightfoote, S. Koenig, A. Fauci, S. Benn, A. Rabson, D. Daugherty, H. E. Gendelman, M. Hoggan, S. Venkatesan, and M. A. Martin. 1986. Biological and biochemical characterization of a cloned Leu-3− cell surviving infection with the acquired immune deficiency syndrome retrovirus. J. Exp. Med. 164:280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haubrich, R., and L. Demeter. 2001. International perspectives on antiretroviral resistance. Clinical utility of resistance testing: retrospective and prospective data supporting use and current recommendations. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S51-S59. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen, H., M. Hänggi, M. Ott, I. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 13.Kempf, D. J., J. Isaacson, M. King, S. Brun, Y. Xu, K. Real, B. Bernstein, A. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 15.Landegren, U., R. Kaiser, J. Sanders, and L. Hood. 1988. A ligase-mediated gene detection technique. Science 241:1077-1080. [DOI] [PubMed] [Google Scholar]
- 16.Little, S. J. 2000. Transmission and prevalence of HIV resistance among treatment-naive subjects. Antivir. Ther. 5:33-40. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz, M., H. Mo, D. J. Kempf, D. W. Norbeck, T. N. Bhat, J. W. Erickson, and D. D. Ho. 1995. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J. Virol. 69:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz, M., M. Conant, A. Hurley, R. Schluger, M. Duran, J. Peterkin, S. Chapman, A. Patick, A. Hendricks, G. J. Yuen, W. Hoskins, N. Clendeninn, and D. D. Ho. 1998. A preliminary evaluation of nelfinavir mesylate, an inhibitor of human immunodeficiency virus (HIV)-1 protease, to treat HIV infection. J. Infect. Dis. 177:1533-1540. [DOI] [PubMed] [Google Scholar]
- 19.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 20.Parker, L. T., H. Zakeri, Q. Deng, S. Spurgeon, P.-Y. Kwok, and D. A. Nickerson. 1996. AmpliTaq® DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. BioTechniques 21:694-699. [DOI] [PubMed] [Google Scholar]
- 21.Partadelis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. J. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patick, A. K., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuurman, R., L. Demeter, P. Reichelderfer, J. Tijnagel, T. de Groot, and C. Boucher. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafer, R. W., D. R. Jung, B. J. Betts, Y. Xi, and M. J. Gonzales. 2000. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 28:346-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafer, R. W., P. Hsu, A. K. Patick, C. Craig, and V. Brendel. 1999. Identification of biased amino acid substitution patterns in human immunodeficiency virus type 1 isolates from patients treated with protease inhibitors. J. Virol. 73:6197-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuyver, L., A. Wyseur, A. Rombout, J. Louwagie, T. Scarcez, C. Verhofstede, D. Rimland, R. F. Schinazi, and R. Rossau. 1997. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob. Agents Chemother. 41:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobe, V., S. Taylor, and D. A. Nickerson. 1996. Single-well genotyping of diallelic sequence variations by a two-color ELISA-based oligonucleotide ligation assay. Nucleic Acids Res. 24:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Laethem, K., K. Van Vaerenbergh, J.-C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clercq, and A.-M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 29.Villahermosa, M. L., I. Beck, L. Perez-Alvarez, et al. J. Hum. Virol., in press. [PubMed]
- 30.Wainberg, M. A., and G. Friedland. 1998. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 279:1977-1983. [DOI] [PubMed] [Google Scholar]
- 31.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 32.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:697-698. [DOI] [PubMed] [Google Scholar]