Abstract
G12 rotavirus has not been detected anywhere in the world since the first detection of a human strain, L26 (G12, P1B[4]), in the Philippines in 1990. In this study, we isolated a human rotavirus (strain T152) with a VP7 of G12 specificity from the stool of an 11-month-old diarrheic patient in Thailand. The strain T152 exhibited a long RNA pattern and subgroup I specificity. In the comparison of the nucleotide and amino acid sequences of the VP7 gene of strain T152 with those of rotaviruses with different G type specificities, strain T152 showed the highest identity, 90.9 and 93.9%, respectively, to G12 prototype strain L26. In contrast, the VP4 gene of strain T152 showed the highest identity with P[9] specificity of human strains K8 and AU-1 and feline strains Cat2 and FRV-1, with homologies of 89.3 to 90.6% at the nucleotide level and 93.9 to 95.6% at the amino acid level. Thus, strain T152 was found to be a natural reassortant strain with G12 and P[9] specificities.
Rotavirus is the major cause of acute gastroenteritis in infants of animals and humans. In developing countries, rotavirus infection results in high mortality, and an annual death rate of 800,000 persons has been estimated (6). Furthermore, in developed countries, rotavirus infection is a cause of high morbidity. However, to date no vaccine has been successful. Rotavirus VP7 and VP4 have independent serotype specificities of the G serotype and P serotype, respectively. A total of 14 G serotypes have been reported. Among them, 10 G serotypes have been detected in humans. G1 to G4 are the major G serotypes, and G5, G6, G8 to G10, and G12 are minor or unusual ones (2, 6). In contrast, 21 P genotypes have been recognized, and at least 10 P genotypes have been detected in humans. Recently, a number of strains with an unusual G or P type and a rare combination of G and P types have been detected in human rotaviruses worldwide (1, 3, 10-12, 16, 17, 25-27).
G12 was first detected in stool specimens collected from diarrheic children under 2 years of age between December 1987 and February 1988 in the Philippines (20, 27). In 40 rotavirus-positive stool specimens, 20 samples showed subgroup I and long RNA profile (7). Four samples were adapted to cell culture, and at least two (L26 and L27) of them were found to have G12 and P1B[4] specificities by serological and sequence analyses of their VP4 and VP7 (20, 27). Since then, however, no further report on the detection of G12 in humans or animals has been presented, although extensive surveys on the G serotype distribution worldwide have been conducted. In this study, we isolated a human rotavirus with G12 and P[9] specificities in Thailand.
MATERIALS AND METHODS
Stool specimens.
A total of 405 stool specimens were collected from diarrheic children in a hospital of the Queen Sirikit National Institute of Child Health, Thailand, between 1998 and 1999. An approximately 10% (wt/vol) stool suspension was prepared in phosphate-buffered saline. For virus isolation in MA-104 cells in roller tube culture, each stool extract was pretreated with 10 μg of trypsin (type IX, obtained from porcine pancreas and crystallized; Sigma) per ml, inoculated onto MA-104 cells in the presence of trypsin (1 μg/ml), and then harvested 5 to 7 days after infection. At least three cycles of passage in roller tube cultures were performed.
ELISA.
An enzyme-linked immunosorbent assay (ELISA) with monoclonal antibodies (MAbs) was carried out as described previously (21). The following MAbs were used: group A-common YO-156 (directed to VP6), subgroup I-specific S2-37 (VP6), subgroup II-specific YO-5 (VP6), G1-specific KU-4 (VP7), G2-specific S2-2G10 (VP7), G3-specific YO-1E2 (VP7), G4-specific ST-2G7 (VP7), and a group A-common YO-2C2 (VP4).
Polyacrylamide gel electrophoresis (PAGE).
Rotavirus double-stranded RNA was extracted from stool and culture fluid with a disruption solution containing 1% sodium dodecyl sulfate, 0.1% 2-mercaptoethanol, and 50 mM EDTA and then with phenol-chloroform. The RNA was electrophoresed in 10% acrylamide gels (2-mm thick) for 16 h at 20 mA at room temperature. RNA segments were visualized by silver staining.
RT-PCR.
Reverse transcriptase PCRs (RT-PCRs) for G typing and P typing were carried out as described previously (23, 28). For G typing, a full-length VP7 gene was amplified with a pair of primers, 5′ GGCTTTAAAAGAGAGAATTTCCGTCTGG 3′ (T31) and 5′ GGTCACATCATACAATTCTAATCTAAG 3′ (T32), corresponding to the common 5′ and 3′ ends of the gene, respectively. In the second PCR, T32 and G1, G2, G3, G4, G8, and G9 serotype-specific primers were used to identify G types. For P typing, a pair of primers, 5′ TGGCTTCGTTCATTTATGACA 3′ and 3′ CTAAATGCTTTT GAATCATCCCA, corresponding to the common sequences of nucleotides 11 to 32 and 1,072 to 1,094, was used for the first amplification, and a mixture of primers specific to each of the variable regions of P1A[8], P1B[4], P2[6], and P3[9] and a primer corresponding to nucleotides 11 to 32 were employed for the second amplification. PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and then visualized with a UV transilluminator.
Sequence determinations.
Full-length cDNA of the VP7 and VP4 genes of culture-adapted strain T152 was prepared by RT-PCR. Direct sequencing was carried out using ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kits (PE Biosystems, Chiba, Japan) with an automated sequencer, the ABI Prism 310 Genetic Analyzer (PE Applied Biosystems, Foster City, Calif.). Nucleotide sequences (VP8*, encoding part of the VP4 gene) were analyzed for construction of a phylogenetic tree using the unweighted pair group method with arithmetic means.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper for the VP7 and VP4 genes of strain T152 were submitted to the GenBank database and have been assigned the accession numbers AB071404 (VP7 gene) and AB077766 (VP4 gene).
RESULTS
Detection of the G12 strain.
We first screened 405 stool specimens for rotavirus by RNA-PAGE analysis, and 194 specimens were found to be positive for rotavirus. RT-PCR showed that the G type of some rotaviruses remained unassigned (unpublished data), since no DNA bands were detected in the second PCR, although cDNA product was obtained in the first PCR. We detected a G12 rotavirus (specimen no. T152) by sequencing a part of the VP7 gene cDNA product obtained by RT-PCR. Symptoms of the patient with strain T152, a female that was 11 months old, were cough, nasal discharge, anorexia, high fever, vomiting, and watery diarrhea (10 times a day). The subgroup specificity of T152 was found to be subgroup I by ELISA subgrouping using subgroup-specific MAbs, and the G serotype specificity could not be determined by ELISA serotyping using MAbs specific to G1, G2, G3, or G4. The T152 isolate was then culture adapted to MA-104 cells.
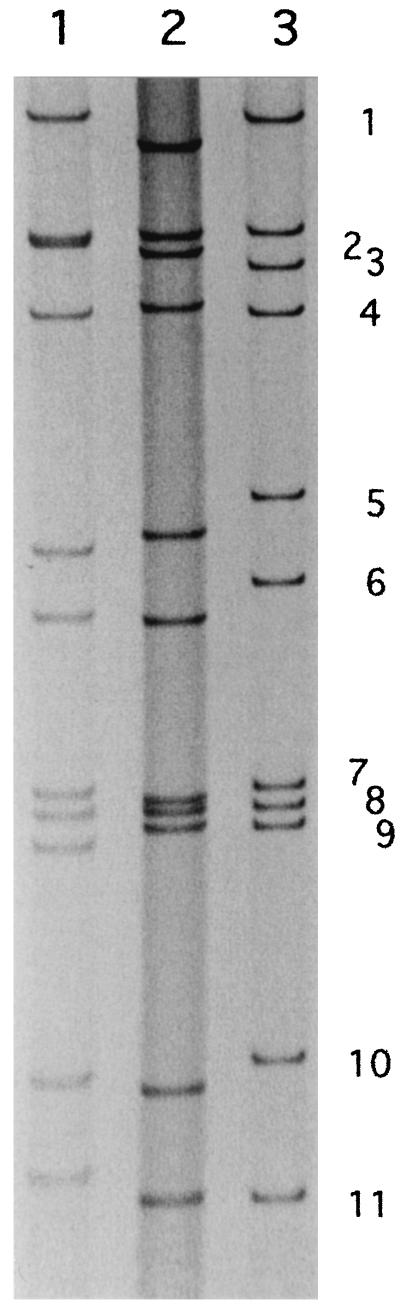
Figure 1 shows the RNA profile of strain T152, which is quite distinct from that of strain L26. In T152 RNA, the mobilities of genes 5 and 6 are slower and the distance between genes 10 and 11 is wider than those in strain L26 RNA. The latter profile has been generally observed in animal rotaviruses and P[9] human rotaviruses. No difference was observed in the RNA-PAGE profiles from stool suspension or culture fluid of T152. Culture-adapted strain T152 was subjected to sequence analysis.
FIG. 1.
RNA pattern of strain T152 in PAGE. Lane 1, KU; lane 2, L26; lane 3, T152.
Sequence analysis.
The VP7 gene of strain T152 was found to be 1,062 bp in length and encodes 326 amino acids. The sequence of the VP7 gene of strain T152 was compared with those of other representative human and animal rotaviruses of different G serotype specificities (Table 1). The VP7 gene of T152 showed the highest identity to that of strain L26 with G12 specificity, with 90.9% homology at the nucleotide level and 93.9% homology at the amino acid level. In contrast, low identities of nucleotide and amino acid sequences were observed with strains other than strain L26, with homologies of 65.3 to 77.6% and 59.9 to 81.9%, respectively. Figure 2 shows the alignment of the VP7 of strains T152 and L26. Amino acid sequences in three major antigenic regions (B, amino acids 87 to 101; D, amino acids 143 to 152; and E, amino acids 208 to 221) were compared between T152 and strains with G serotypes 1 to 14. The amino acid sequence homologies in the three regions of T152 were only 35.9 to 53.8% for those of G1 to 11, G13, and G14 specificities, but a high identity was found for L26 (92.3%). Thus, strain T152 is suggested to have G12 serotype specificity.
TABLE 1.
Nucleotide and amino acid sequence homology of strain T152 VP7 with VP7 from other rotavirus strains with different G serotype specificities
| G serotype | Strain | Homology (%)
|
|
|---|---|---|---|
| Nucleotide | Amino acid | ||
| G1 | KU | 76.2 | 79.5 |
| G2 | S2 | 73.5 | 76.4 |
| G3 | YO | 76.3 | 81.9 |
| G4 | VA70 | 73.7 | 74.8 |
| G5 | OSU | 76.9 | 80.7 |
| G6 | NCDV | 75.5 | 79.5 |
| G7 | Ty1 | 65.3 | 59.9 |
| G8 | 69M | 73.8 | 79.5 |
| G9 | WI61 | 77.6 | 81.9 |
| G10 | A44 | 74.4 | 77.9 |
| G11 | YM | 75.2 | 81.3 |
| G12 | L26 | 91.0 | 94.2 |
| G13 | L338a | 74.4 | 78.8 |
| G14 | CH3 | 75.4 | 77.9 |
The nucleotide sequence of the VP7 gene from strain L338 has not been completed, and so the sequence of nucleotides 3 to 1038 was compared.
FIG. 2.
Comparison of VP7 amino acid sequences from strains T152 and L26. Only the amino acids of strain L26 that are different from those of strain T152 are shown below the T152 amino acid sequence. Three major antigenic regions are boxed.
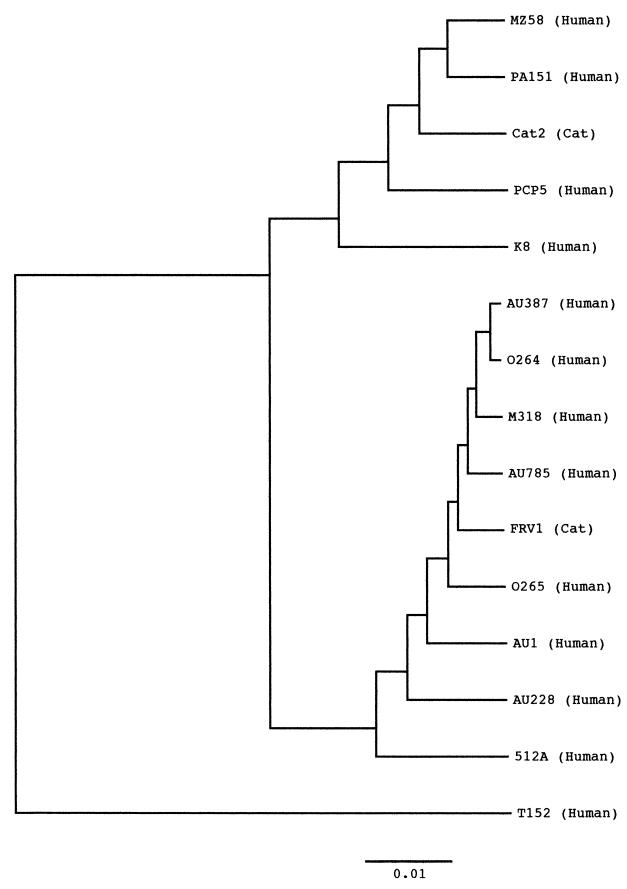
The VP4 nucleotide sequence of strain T152 was 2,359 bases long and contained a single open reading frame encoding 775 amino acids. The VP4 gene of strain T152 was found to have an insertion of one amino acid after residue 135 and it lacked one amino acid at residue 575, compared to those of the representative human rotaviruses with P[8], P[4], or P[6] specificity. The genomic organization of the VP4 gene of strain T152 is commonly found in P[9] rotavirus strains. The nucleotide and amino acid sequences of the T152 VP4 are indeed highly homologous to those of P[9] rotaviruses, with 89.3 to 90.6% identity at the nucleotide sequence level and 93.6 to 95.6% identity at the amino acid sequence level, but it showed more than 13.3% divergence at the amino acid level from the sequences of other rotaviruses (Table 2). Phylogenetic analysis of the VP8* region (nucleotides 1 to 750 of the VP4 gene) of human and feline rotaviruses with P[9] was carried out since the region has been found to be P-type specific. As shown in Fig. 3, VP8* of strain T152 was most distantly related to other P[9] rotaviruses.
TABLE 2.
Nucleotide and amino acid sequence homology of strain T152 VP4 with VP4 from other rotavirus strains with different VP4 genotypes
| VP4 genotype | Strain | Homology (%)
|
|
|---|---|---|---|
| Nucleotide | Amino acid | ||
| 1 | NCDV | 69.4 | 70.7 |
| 2 | SA11 | 69.3 | 70.6 |
| 3 | CU1 | 69.2 | 69.5 |
| 4 | DS1 | 66.9 | 65.0 |
| 5 | UK | 48.1 | 67.1 |
| 6 | M37 | 67.6 | 65.9 |
| 7 | OSU | 48.4 | 69.4 |
| 8 | KU | 67.4 | 63.8 |
| 9 | K8 | 90.0 | 95.5 |
| AU1 | 90.3 | 95.6 | |
| 02/92 | 89.4 | 93.9 | |
| Cat2 | 89.3 | 94.3 | |
| FRV-1 | 90.6 | 95.4 | |
| 10 | 69M | 69.8 | 69.7 |
| 11 | B223 | 62.9 | 55.1 |
| 12 | FI14 | 69.5 | 68.7 |
| 13 | MDR13 | 68.3 | 66.2 |
| 14 | Mc35 | 77.7 | 86.7 |
| 15 | Lp14 | 68.7 | 67.9 |
| 16 | EB | 65.1 | 63.4 |
| 17 | 993/83 | 64.5 | 58.9 |
| 18 | L338 | 68.5 | 68.3 |
| 19 | Mc323 | 67.4 | 66.5 |
| 20 | EHP | 35.0 | 67.4 |
| 21 | Hg18 | 37.7 | 66.0 |
FIG. 3.
Phylogenetic tree for the nucleotide sequences of the VP8*-encoding region of the VP4 gene of human and feline rotaviruses. The bar indicates the variation scale.
DISCUSSION
Global surveillances of rotavirus G and P types have been extensively performed and indicate that types G1 to G4 and P[8] and P[6] are most frequently distributed worldwide (2, 6). In addition, four frequent combinations of VP7 and VP4, G1P[8], G2P[4], G3P[8], and G4P[8], are common worldwide (4). In contrast, unusual G types, P types, or G-P combinations have also been detected. These epidemiological surveys on VP7 and VP4 types are important for developing efficient rotavirus vaccines and for elucidating rotavirus ecology and evolution.
G12 strains have been detected only in the Philippines, and there have been no reports on the prevalence of strains with a G12 specificity since the first detection in 1990 (20, 27). The G12 prototype strain L26 has long RNA pattern and subgroup I specificity. In this study, interestingly, we detected a strain T152 with G12, long RNA profile, and subgroup I specificity in Thailand.
The nucleotide and amino acid sequences of the VP7 genes of strains T152 and L26 showed the highest identity to those of strain L26 with G12 specificity, with 90.9 and 93.9% identities, respectively. However, the homology percentages are not necessarily high compared to those found with strains of the same G type. For example, the nucleotide and amino acid sequence homology percentages for the VP7 gene are 97.4 and 98.8% between the Japanese G1 strain KU and the American G1 strain Wa. It is conceivable that strain T152 was not directly transferred from the Philippines to Thailand and that it evolved gradually from the prototype L26 or a relevant strain. Thus, the strains with G12 such as T152 might be more prevalent than expected from the only two reports on the detection, including this one. We are preparing G12-specific MAbs for long-term and large-scale surveys on the distribution of G12 strains in other countries, including Southeast Asia.
The P specificity of strain L26 is P1B[4], which is commonly linked with the G2 type. In contrast, strain T152 was found to have P[9] specificity. P[9] specificity has been detected only in humans and cats. Furthermore, the prevalence of human rotaviruses with P[9] is low; in the surveys which detected P[9] strains, 0.2% of 1,316 rotaviruses between 1996 and 1999 in the United States (4) and 3.8% of 282 rotaviruses between 1991 and 1994 in Israel (18) were of this P[9] specificity. All the strains with P[9] specificity are associated with G3 or G1 specificity (5, 18). By hybridization experiments, it has been shown that G3P[9] is the prototype strain and that G1P[9] strains such as strain K8 appear to be a reassortant between the AU-1 and Wa genogroups (9). It would be interesting to know how strain T152 with G12P[9] specificity has occurred. Hybridization experiments or Northern blotting experiments might provide a clue to this question.
The relatedness of G12 strain L26 to human and animal rotaviruses has been examined. In our previous RNA-RNA hybridization assays (8), the L26 probe hybridized with only two or three RNA segments from various animal strains and subgroup II human strains in the Wa genogroup. In contrast, five or six RNA hybrids were observed between the L26 probe and genomic RNA from subgroup I human rotavirus strains in the DS-1 genogroup. In reciprocal hybridizations using a DS-1 probe, the probe formed six hybrids with genomic RNA from strains L26 and 69M. Thus, L26 is more similar to human rotaviruses in the DS-1 genogroup. Indeed, sequences of the VP4 gene and NSP1 gene of L26 are highly homologous to those of strains DS-1 and S2 (8). Since the related RNA segments could not be identified exactly in the liquid RNA-RNA hybridization experiments, we should carry out Northern blotting analysis or sequence determination of other genes in order to elucidate the segment-to-segment relatedness of L26 and T152 with other animal and human rotaviruses.
We have conducted surveys on the properties of rotaviruses in Thailand and have found several interesting features (13-15, 17, 19, 22, 24, 25): distinct yearly change of G serotype in human rotaviruses, detection of G8 and G10 bovine rotaviruses in a high frequency, detection of a G10 porcine rotavirus, detection of a G10 human rotavirus, detection of a G8 bovine rotavirus with rearrangement in the NSP1 gene, detection of human rotavirus with rearrangement in the NSP5 gene, and detection of human rotaviruses with a new P serotype. Thus, there seem to exist numerous rotaviruses with unusual properties in Thailand. It will be valuable to continue the surveillance of rotaviruses prevalent in Thailand.
Acknowledgments
This study was supported in part by a grant-in-aid from the Ministry of Education and Science, Japan.
REFERENCES
- 1.Armah, G. E., C. T. Pager, R. H. Asma, F. R. Anto, A. B. Oduro, F. Binka, and D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 2.Estes, M. K. 1996. Rotaviruses and their replication, p. 1625-1656. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 3.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 4.Griffin, D. D., C. D. Kirkwood, U. D. Parasgar, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and The National Rotavirus Strain Surveillance System Collaborating Laboratories. 2000. Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol. 38:2784-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaga, E., M. Iizuka, T. Nakagomi, and O. Nakagomi. 1994. The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol. Immunol. 38:317-320. [DOI] [PubMed] [Google Scholar]
- 6.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 7.Kobayashi, N., I. C. Lintag, T. Urasawa, K. Taniguchi, M. C. Saniel, and S. Urasawa. 1989. Unusual human rotavirus strains having subgroup I specificity and “long” RNA electropherotype. Arch. Virol. 109:11-23. [DOI] [PubMed] [Google Scholar]
- 8.Kojima, K., K. Taniguchi, T. Urasawa, and S. Urasawa. 1996. Sequence analysis of normal and rearranged NSP5 genes from human rotavirus strains isolated in nature: implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology 224:446-452. [DOI] [PubMed] [Google Scholar]
- 9.Nakagomi, O., E. Kaga, and T. Nakagomi. 1992. Human rotavirus strain with unique VP4 neutralization epitopes as a result of natural reassortment between members of the AU-1 and Wa genogroups. Arch. Virol. 127:365-371. [DOI] [PubMed] [Google Scholar]
- 10.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada, J., T. Urasawa, N. Kobayashi, K. Taniguchi, A. Hasegawa, K. Mise, and S. Urasawa. 2000. New P serotype of group A human rotavirus closely related to that of a porcine rotavirus. J. Med. Virol. 60:63-69. [PubMed] [Google Scholar]
- 12.Palombo, E. A., P. J. Masendycz, H. C. Bugg, N. Bogdanovic-Sakran, G. L. Branes, and R. F. Bishop. 2000. Emergence of G9 in Australia. J. Clin. Microbiol. 38:1305-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pongsuwanna, Y., K. Taniguchi, M. Chiwakul, T. Urasawa, F. Wakasugi, C. Jayavasu, and S. Urasawa. 1996. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J. Clin. Microbiol. 34:1050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pongsuwanna, Y., K. Taniguchi, M. Choonthanom, M. Chiwakul, T. Susansook, S. Saguanwongse, C. Jayavasu, and S. Urasawa. 1989. Subgroup and serotype distribution of human, bovine, and porcine rotavirus in Thailand. J. Clin. Microbiol. 27:1956-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pongsuwanna, Y., K. Taniguchi, F. Wakasugi, Y. Sutivijit, M. Chiwakul, P. Warachit, C. Jayavasu, and S. Urasawa. 1993. Distinct yearly change of serotype distribution of human rotavirus in Thailand as determined by ELISA and PCR. Epidemiol. Infect. 111:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, R. I. Glass, and The National Rotavirus Strain Surveillance System (NRSSS) Collaborating Laboratories. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos, N., R. C. C. Lima, C. F. A. Pereiira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silberstein, I., L. M. Shulman, E. Mendelson, and I. Shif. 1995. Distribution of both VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J. Clin. Microbiol. 33:1421-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi, K., K. Kojima, and S. Urasawa. 1996. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153 to 155. J. Virol. 70:4125-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi, K., T. Urasawa, N. Kobayashi, M. Gorziglia, and S. Urasawa. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J. Virol. 64:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi, K., T. Urasawa, Y. Morita, H. B. Greenberg, and S. Urasawa. 1987. Direct serotyping of human rotaviruses in stools by enzyme-linked immunosorbent assay using serotype 1-, 2-, 3-, and 4-specific monoclonal antibodies to VP7. J. Infect. Dis. 155:1159-1166. [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi, K., T. Urasawa, Y. Pongsuwanna, M. Choonthanom, C. Jayavasu, and S. Urasawa. 1991. Molecular and antigenic analyses of serotype 8 and 10 of bovine rotaviruses in Thailand. J. Gen. Virol. 72:2929-2937. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi, K., F. Wakasugi, Y. Pongsuwanna, T. Urasawa, S. Ukae, S. Chiba, and S. Urasawa. 1992. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol. Infect. 109:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urasawa, S., A. Hasegawa, T. Urasawa, K. Taniguchi, F. Wakasugi, H. Suzuki, S. Inouye, B. Pongprot, J. Supawadee, S. Suprasert, P. Rangsiyanond, S. Tonusin, and Y. Yazaki. 1992. Antigenic and genetic analysis of human rotaviruses prevailing in Chiang Mai, Thailand: evidence for a close relationship between human and animal rotaviruses. J. Infect. Dis. 166:227-234. [DOI] [PubMed] [Google Scholar]
- 25.Urasawa, T., K. Taniguchi, N. Kobayashi, K. Mise, A. Hasegawa, Y. Yamazi, and S. Urasawa. 1993. Nucleotide sequence of VP4 and VP7 genes of a unique human rotavirus strain Mc35 with subgroup I and serotype 10 specificity. Virology 195:766-771. [DOI] [PubMed] [Google Scholar]
- 26.Urasawa, T., K. Taniguchi, N. Kobayashi, F. Wakasugi, I. Oishi, Y. Minekawa, M. Oseto, M. U. Ahmed, and S. Urasawa. 1990. Antigenic and genetic analyses of human rotavirus with dual subgroup specificity. J. Clin. Microbiol. 28:2837-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urasawa, S., T. Urasawa, F. Wakasugi, N. Kobayashi, K. Taniguchi, I. C. Lintag, M. C. Saniel, and H. Goto. 1990. Presumptive seventh serotype of human rotavirus. Arch. Virol. 113:279-282. [DOI] [PubMed] [Google Scholar]
- 28.Wu, H., K. Taniguchi, F. Wakasugi, S. Ukae, S. Chiba, M. Ohseto, A. Hasegawa, T. Urasawa, and S. Urasawa. 1994. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol. Infect. 112:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]