Abstract
Adverse effects of strain persistence and secretion in milk have been encountered with the Brucella melitensis vaccine strain Rev.1. Field isolates obtained from vaccinated animals and from a human resembled the vaccine strain Rev.1 by conventional bacteriological tests. The lack of a specific molecular marker that could specifically characterize the commercial vaccine strain prevented confirmation of the homology of the Rev.1-like field isolates to the vaccine strain. The composition of the omp2 locus from two gene copies with differences in their PstI restriction endonuclease sites was used to establish an epidemiologic fingerprint for the omp2 gene in the Rev.1 vaccine strain. Primers designed to amplify DNA sequences that overlap the PstI site revealed a single 282-bp DNA band common to all Brucella spp. Agarose gel electrophoresis of the PstI digests of the PCR products from strains 16M and the vaccine strain Rev.1 revealed a distinctive profile that included three bands: one band for the intact 282-bp fragment amplified from omp2a and two bands resulting from the digestion of the amplified omp2b gene fragment, 238- and 44-bp DNA fragments, respectively. Amplified fragments of 37 Rev.1-like isolates, including 2 human isolates, also exhibited this pattern. In contrast, DNA digests of all other Israeli field isolates, including atypical B. melitensis biotype 1 and representatives of the biotype 2 and 3 isolates, produced two bands of 238 and 44 bp, respectively, corresponding with the digestion of both omp2a and omp2b genes. This method facilitates identification of the Rev.1 vaccine strain in both animals and humans in Israel.
Brucella melitensis causes a worldwide zoonosis. It is one of the major causes of abortions in sheep and goats,and the organism is secreted in the milk of infected animals. People contract the disease by direct contact with contaminated fetal membranes or, more commonly, as a result of the consumption of contaminated unpasteurized milk and cheese products. The organisms are small, gram-negative coccobacilli that grow in the host as nonobligatory intracellular pathogens of the reticuloendothelial system. Derivatives of tetracycline are often used to treat human infection, while a slaughter policy is recommended for livestock in order to eradicate the disease (11).
In the late 1950s, Elberg developed a live attenuated vaccine, strain Rev.1 (12, 13). It was shown that although the vaccine prevented abortion, it did not provide protection against infection. Bosseray demonstrated that different lots of Rev.1 vaccines showed variable immunogenicity in mice according to their level of virulence (5, 6). This study emphasized the instability of the biological properties of the vaccine strain, stressing the need for stringent control of vaccine production (7).
The problems in the laboratory were reflected by similar results in the field. In South Africa, selection of a few smooth colonies as seed stock led to production of a virulent vaccine strain which infected sheep and caused human disease (22, 27, 28).
Throughout the last decade Israel maintained a conservative vaccination policy in which only young female livestock between the ages of 2 to 6 months were vaccinated, using a full dose by the subcutaneous route. Nevertheless, retrospective data demonstrated that the Rev.1 vaccine led to the adverse effects of strain persistence in the vaccinated animals and was occasionally spread horizontally (4, 30). Moreover, in two cases it was shown that the vaccine strain caused human infection, demonstrating the zoonotic hazards of its virulence. The fact that vaccination did not always protect the animals in the field and the several cases of secretion of the field strain in milk had proven the inefficacy of the whole vaccination program.
International agencies, in their assistance to developing countries, suggested that national control programs should depend on a whole-flock vaccination scheme as a cost-effective method until the prevalence of the disease was reduced. Only then should test and slaughter be implemented to eradicate the disease (10). There was opposition to this proposal (4) due to the adverse effects encountered in the field and the public concerns of possible risks to the human population following secretion of the vaccine strain in milk. The absence of a specific molecular marker that could be associated with the identity of the commercial vaccine strain prevented those opposing the vaccination program from linking Rev.1-like field isolates to the vaccine strain. By the same token, those in favor of the program could ignore the risks posed by the Rev.1 vaccine, using the same rationale.
Data presented below provide evidence supporting the existence of a PstI site polymorphism in the Brucella omp2 gene. The PstI digestion pattern of PCR-amplified fragments from the Israeli isolates was different from that of the prototype strain 16M. Curiously, the PstI digestion profile of the omp2 amplified fragments from the vaccine strain Rev.1 resembled that of strain 16M, allowing the differentiation of Rev.1 isolates from B. melitensis field strains in Israel. This achievement could specifically address the potential misdiagnosis of the atypical B. melitensis biovar 1 strains as Rev.1 isolates due to similarities in their phenotypic susceptibility to penicillin.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study (Table 1) were from the collection maintained in the Israeli reference laboratory. Brucella field strains were isolated by conventional methods (1) from milk samples, aborted fetuses, and placentas. Human isolates were obtained from local medical laboratories. Strain 16M was obtained from J.-M. Verger, Institut National de la Recherche Agronomique, Nouzilly, France. Strain biotyping was performed by standard methods (1). Escherichia coli strain NCTC 9001 was used as a control (for non-Brucella DNA). Rhizobium meliloti strains 158M and 161M were obtained from D. Kishinevsky, and Agrobacterium radiobacter strains At5/r and At96216 were obtained from D. Zutra, The Volcani Center, Beit Dagan, Israel. Bacterial strains other than Brucella were obtained from the Department of Bacteriology, The Kimron Veterinary Institute, Beit Dagan, Israel, or from the Department of Clinical Microbiology, the Hebrew University-Hadassah Medical School, Jerusalem, Israel.
TABLE 1.
Brucella strains used in this study
| Strain no. | Species | Biovar | Strain designation | Strain type, host, or source |
|---|---|---|---|---|
| 1 | B. melitensis | 1 | 16M | Prototype strain |
| 2 | B. melitensis | 1 | 117790 | Human |
| 3 | B. melitensis | 1 | 118762 | Human |
| 4 | B. melitensis | 1 (atypical) | 9413 | Sheep |
| 5 | B. melitensis | 1 (atypical) | 6012 | Human |
| 6 | B. melitensis | 1 (atypical) | 124386 | Human |
| 7 | B. melitensis | 1 | Rev.1 (vaccine) (ocular lot) | Commercial source |
| 8 | B. melitensis | 1 | Rev.1 (vaccine) (subcutaneous lot) | Commercial source |
| 9 | B. melitensis | 1 (Rev.1-like) | 5000 | Human |
| 10 | B. melitensis | 1 (Rev.1-like) | 204216 | Human |
| 11 | B. melitensis | 1 (Rev.1-like) | 134172 | Sheep milk |
| 12 | B. melitensis | 1 (Rev.1-like) | 116375 | Sheep milk |
| 13 | B. melitensis | 1 (Rev.1-like) | 124596 | Sheep milk |
| 14 | B. melitensis | 1 (Rev.1-like) | 225875 | Goat's retropharyngeal gland |
| 15 | B. melitensis | 1 (Rev.1-like) | 226390 | Sheep bulk milk tank |
| 16 | B. melitensis | 1 (Rev.1-like) | 226997 | Sheep milk |
| 17 | B. melitensis | 1 (Rev.1-like) | 223713 | Goat's membranes from aborted fetus |
| 18 | B. melitensis | 2 | 118808 | Human |
| 19 | B. melitensis | 2 | 160621 | Human |
| 20 | B. melitensis | 2 (atypical) | 124906 | Human |
| 21 | B. melitensis | 3 | Ether | Prototype strain |
| 22 | B. melitensis | 3 | 119917 | Human |
| 23 | B. melitensis | 3 | 119919 | Human |
| 24 | B. melitensis | Rough | B115 | Reference strain |
| 25 | B. melitensis | Rough | 119056 | Human |
| 26 | B. abortus | 1 | 544 | Prototype strain |
| 27 | B. abortus | 1 | 2038 | Reference strain |
| 28 | B. abortus | 1 | S19 (vaccine) | |
| 29 | B. abortus | 3 | Tulya | Prototype strain |
| 30 | B. suis | 1 | S2 (vaccine) |
Bacterial DNA.
To prepare chromosomal DNA, bacterial cells were harvested in saline and incubated for 20 min at 4°C with lysozyme (4 mg/ml). Sodium dodecyl sulfate (0.5% [wt/vol]) and proteinase K (200 mg/ml) were then added, and incubation was continued at 37°C for 1 h. The cell lysate was extracted once with phenol-chloroform-isoamyl alcohol (1:1:49) and once with chloroform-isoamyl alcohol (1:24). The purified DNA was alcohol precipitated, resuspended in TE (50 mM Tris-HCl, 1 mM EDTA [pH 8.0]), and stored at 4°C.
PCR and oligonucleotide primers.
The Brucella omp2 gene was used as target DNA. The forward 5′ primer (p1 [TGGAGGTCAGAAATGAAC]) and reverse 3′ primer (p2 [GAGTGCGAAACGAGCGC]) of an omp2 gene segment were obtained from National Biosciences, Inc., Plymouth, Minn.
PCR amplification was performed by the method of Mullis and Faloona (26). A typical reaction mixture contained 50 mM KCl, 1.5 mM MgCl2, 0.1% (wt/vol) Triton X-100, 0.2 mg of bovine serum albumin (fraction IV; Sigma) per ml, and 10 mM Tris-HCl (pH 8.5). Each reaction mixture was supplemented with 100 mM each of the four deoxyribonucleotides, 100 ng of sample DNA, and each oligonucleotide primer. For slide PCR, sample DNA was replaced with brucellae that were laid on a glass slide, air dried, and fixed by being heated. A sample of the dried cells was then collected with a needle, the needle was dipped in 10 μl of double-distilled water, and 2 μl from this solution was put in the PCR mixture. Otherwise, sample DNA (2 μl from a bacterial cell suspension in double-distilled water boiled at 100°C for 20 min) was used. Reactions were initiated by adding 0.5 U of Taq polymerase (Appligene, Illkirch, France). The reaction mixture was covered with 15 μl of mineral oil (Sigma) to prevent evaporation. Following hot start treatment at 95°C for 3 min, PCR was performed with an Eppendorf Thermocycler (Eppendorf, Hamburg, Germany) as follows: 35 cycles of PCR, with 1 cycle consisting of 20 s at 95°C for DNA denaturation, 1 min at 50°C for DNA annealing, and 1 min at 72°C for polymerase-mediated primer extension. The last cycle included incubation of the sample at 72°C for 7 min. Ten microliters of the amplified product was analyzed by electrophoresis in 1.5% agarose gels in TEA buffer (20 mM Tris-acetate, 1 mM EDTA [pH 8.0]).
DNA digestion.
Restriction enzymes were used according to the manufacturer's instructions (Boehringer GmbH, Mannheim, Germany). The digested DNA was separated by electrophoresis on either 1.5% agarose gels (wt/vol in Tris-acetate buffer) or 10% polyacrylamide gels (wt/vol in Tris-borate buffer). DNA fragments were visualized by staining with ethidium bromide (1.5 μg/ml).
RESULTS
Validation of the method with prototype strains.
The PCR was first performed to test specificity by comparing Brucella species DNAs to the DNAs from several other bacteria, including the taxonomically closely related Agrobacterium and Rhizobium strains (29). A single band with the expected size of 282 bp (19) was obtained only when Brucella DNA was used as a template. All other bacterial strains and a water sample failed to produce an amplified fragment (data not shown).
The PCR test was studied with Brucella prototype strains from two of the three important species, namely, B. melitensis and B. abortus (field and vaccine strains). In addition, B. suis vaccine strain S2 was included as a representative of this species. As shown in Fig. 1, the DNAs from all the strains produced a 282-bp band. As shown in Fig. 2, PstI digestions of the amplified fragments from the strains gave different bands on agarose gels. B. abortus (lanes 1, 3, 4, and 5), B. suis S2 (lane 8), and B. melitensis biovar 3 (lane 12) digests revealed a single band, a 238-bp band. Other possible smaller fragments are not shown on the gel. In comparison, PstI digestion of B. melitensis strain 16M (prototype for B. melitensis biovar 1 virulent strain [lane 9]) and strain Rev.1 (a vaccine strain from two different producers administered by a subcutaneous and an ocular route [lanes 10 and 11, respectively]) amplified DNAs, included two visible bands: a large band, which was uncut DNA (lanes 6 and 8), and another band, a 238-bp B. abortus fragment. Other possible smaller bands are not shown on the gel. The PstI digestion pattern of the amplified fragment obtained from B. melitensis strain B115 (a stable rough form obtained from an infected goat in Malta in the early 1970s) (lane 2) was similar to those for strains 16M and Rev.1 (lanes 9, 10, and 11, respectively).
FIG. 1.
Agarose gel electrophoresis of PCR-amplified omp2 gene fragments from Brucella prototype strains. The figure shows a single band, a 282-bp DNA fragment. Lanes: M, molecular size ladder (in base pairs); 1, B. abortus strain 2308; 2, B. melitensis strain B115; 3, B. abortus strain Tulya; 4, B. abortus strain 544; 5, B. abortus vaccine S19; 6, B. melitensis strain 16M; 7, B. suis vaccine S2; 8, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 9, B. melitensis vaccine strain Rev.1 (ocular lot); 10, B. melitensis strain Ether.
FIG. 2.
Agarose gel electrophoresis of PstI digests of amplified omp2 gene fragments from Brucella prototype strains. The figure shows the uncut 282-bp DNA and the larger, PstI-digested DNA fragments. The smaller, 44-bp DNA fragment is not shown. Lanes: 1, B. abortus strain 2308; 2, B. melitensis strain B115; 3, B. abortus strain Tulya; 4, B. abortus strain 544; 5, B. abortus vaccine S19; 6, B. melitensis strain 16M (uncut); M, molecular size ladder (in base pairs); 7, B. abortus strain 544 (uncut); 8, B. suis S2; 9, B. melitensis strain 16M; 10, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 11, B. melitensis vaccine strain Rev.1 (ocular lot); 12, B. melitensis strain Ether.
The digestion profiles of the same DNAs were analyzed by polyacrylamide gel electrophoresis, as shown in Fig. 3. The purpose of this analysis was to identify possible smaller fragments that were not shown by agarose gel electrophoresis. As can be seen in Fig. 3, besides the 282- and 238-bp DNA bands, all samples produced an additional identical smaller fragment which was calculated to be 44 bp. It was calculated that the two smaller bands together were the same size as the uncut DNA, confirming the expected PstI restriction pattern for B. abortus biovar 1 (19).
FIG. 3.
Polyacrylamide gel electrophoresis of PstI digests of amplified omp2 gene fragments from Brucella prototype strains. The figure shows the three DNA fragments, the uncut DNA and the two PstI-digested DNA fragments, with sizes of 282, 238, and 44 bp, respectively. Lanes: M, molecular size ladder (in base pairs); 1, B. melitensis strain 16M (uncut); 2, B. melitensis strain 16M; 3, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 4, B. abortus vaccine S19; 5, B. abortus strain 544; 6, B. abortus strain 2308; 7, B. melitensis strain Ether.
PstI digestion profile of the omp2 gene amplified fragment from atypical B. melitensis biotype 1 strains.
Atypical B. melitensis biotype 1 isolates from a human source (isolate 6012) and sheep source (isolate 9413) were described previously (2). We compared the PstI digestion profiles of the omp2 gene amplified fragment obtained from these strains and those obtained from prototype strains. All Brucella strains produced identical amplified 282-bp fragments (data not shown). Figure 4 depicts a polyacrylamide gel analysis of the digestion profile of these DNAs. As can be seen, PstI digestion of the two B. melitensis atypical strains (lanes 2 and 3) produced a uniform pattern identical to that obtained for B. abortus strains S19 and 544 (lanes 6 and 7) and to that of B. melitensis biotype 3 (lane 8). The PstI digestions of the amplified fragments from the commercial vaccine strain Rev.1 and the prototype strain 16M yielded a different pattern (lanes 4 and 5, respectively). The gel also shows the smaller, 44-bp band common to all digests.
FIG. 4.
Polyacrylamide gel electrophoresis of PstI digests of amplified omp2 gene fragments from atypical B. melitensis biotype 1 isolates from humans and animals compared to prototype strains. The figure shows the three DNA fragments, the uncut DNA and the two PstI-digested DNA fragments, with sizes of 282, 238, and 44 bp, respectively. Lanes: M, molecular size ladder (in base pairs); 1, B. melitensis strain 6012 (human isolate) (uncut); 2, B. melitensis strain 6012 (human isolate); 3, B. melitensis strain 9413 (sheep isolate); 4, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 5, B. melitensis strain 16M; 6, B. abortus vaccine S19; 7, B. abortus strain 544; 8, B. melitensis strain Ether.
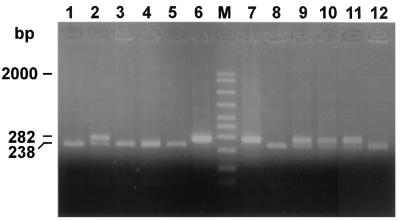
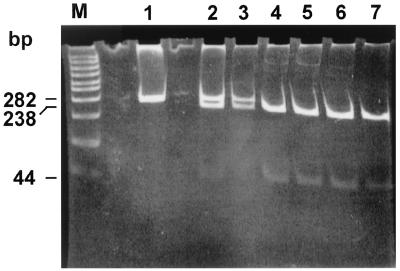
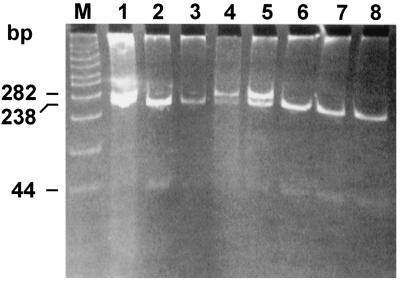
We then analyzed the digestion profile of the omp2 DNA fragment of selected human isolates as shown in Fig. 5. The samples included biovar 1 strains (lanes 1, 2, 3, 4, and 14), a rough isolate (lane 7), atypical biotype 1 (lane 15) and atypical biotype 2 (lane 16) strains, and two Rev.1 human isolates (lanes 10 and 12). In addition, we included the prototype strain 16M (lane 11) and two uncut DNAs from strain Rev.1 (lane 8) and strain 16M (lane 9). Besides strain Rev.1 (lanes 5 and 6), Rev.1-like isolates (lanes 10 and 12), and B. melitensis strain 16M (lane 11), all other isolates had similar digestion profiles (as shown in Fig. 2 for B. abortus strains) and B. melitensis biotype 3 strain Ether.
FIG. 5.
Agarose gel electrophoresis of PstI digests of amplified omp2 gene fragments from Brucella field strains. The figure shows the uncut 282-bp DNA and the larger, PstI-digested DNA fragment. The smaller, 44-bp DNA fragment is not shown. Lanes: 1, B. melitensis biotype 1 strain 117790; 2, B. melitensis biotype 1 strain 118762; 3, B. melitensis biotype 2 strain 118808; 4, B. melitensis biotype 2 strain 160621; 5, B. melitensis vaccine strain Rev.1 (ocular lot); 6, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 7, B. melitensis rough strain 119056; 8, B. melitensis vaccine strain Rev.1 (subcutaneous lot) (uncut); M, molecular size ladder (in base pairs); 9, B. melitensis strain 16M (uncut); 10, B. melitensis biotype 1 strain 5000 (human Rev.1-like isolate); 11, B. melitensis strain 16M; 12, B. melitensis biotype 1 strain 204215 (sheep Rev.1-like isolate); 13, B. melitensis biotype 3 strain 119917; 14, B. melitensis biotype 3 strain 119919; 15, B. melitensis biotype 1 atypical strain 124386; 16, B. melitensis biotype 2 atypical strain 124906.
PstI digestion profile of the omp2 gene amplified fragment from B. melitensis Rev.1-like isolates.
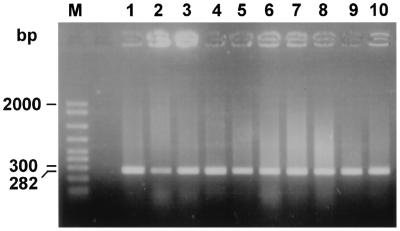
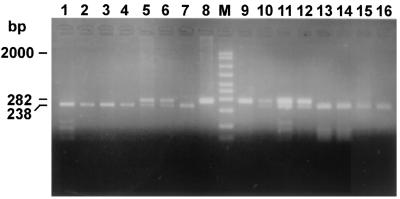
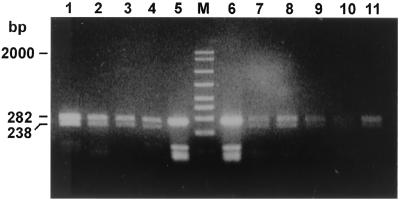
A total of 23 B. melitensis biotype 1 field isolates, 5 B. melitensis atypical biovar 1 field isolates (2), 13 B. melitensis biotype 2 field isolates including 3 atypical strains, and 13 B. melitensis biotype 3 field isolates were studied. All exhibited the pattern shown for B. abortus strains (data not shown). In contrast, 37 field isolates with Rev.1-like bacteriological characteristics produced an identical PstI digestion pattern (as did the commercial vaccine, strain Rev.1). A representative selection is shown in Fig. 6, lanes 1, 2, 3, 8, 9, 10, and 11, compared to lanes 4 and 7, respectively.
FIG. 6.
Agarose gel electrophoresis of PstI digests of amplified omp2 gene fragments from Brucella field strains that resemble Rev.1 according to biochemical biotyping. The figure shows the uncut 282-bp DNA and the larger, PstI-digested DNA fragment. The smaller, 44-bp DNA fragment is not shown. Lanes: 1, B. melitensis strain 134172; 2, B. melitensis strain 116375; 3, B. melitensis strain 124596; 4, B. melitensis vaccine strain Rev.1 (subcutaneous lot); 5, B. melitensis vaccine strain Rev.1 (subcutaneous lot) (uncut); M, molecular size ladder (in base pairs); 6, B. melitensis vaccine strain (ocular lot) (uncut); 7, B. melitensis vaccine strain Rev.1 (ocular lot); 8, B. melitensis strain 225877; 9, B. melitensis strain 226390; 10, B. melitensis strain 226997; 11, B. melitensis strain 223713.
DISCUSSION
The PCR technique has increasingly been used as a supplementary method in Brucella diagnosis (8, 14, 15, 21, 23, 25). Recently, a molecular biotyping approach has been proposed on the basis of restriction endonuclease polymorphism in the genes encoding the major 25- and 36-kDa outer membrane proteins of Brucella (9, 17, 20). The omp2 gene exists as a locus of two nearly homologous repeated copies that differ slightly among Brucella spp. and biotypes (18). We used this information to design specific primers that amplify a 282-bp fragment (Fig. 1), flanking upstream sequences of the 5′ terminus of the two genes and expanding downstream of the PstI and KpnI sites (17). We assumed that the sensitivity of the test would be doubled by selecting duplicated DNA sequences of the two genes. Moreover, we assumed that because of the existing PstI site polymorphism between B. melitensis and B. abortus, the test would distinguish between the two species. According to the working hypothesis, DNA fragments obtained from B. melitensis strain 16M should produce three bands, an intact 282-bp fragment from the amplified omp2a gene that lacks the PstI site and two smaller fragments of 238 and 44 bp, the products obtained from digestion of the omp2b amplified fragment (17). In contrast, B. abortus DNA should produce only the two smaller fragments from both genes, a 238-bp fragment and a 44-bp fragment, respectively (Fig. 2 and 3).
We used this method to study the stability of the omp2 gene among local B. melitensis isolates derived from small ruminants, cattle, and humans and representing the current Brucella population in Israel. Our data confirmed the expected paradigm for B. melitensis strains B115, 16M, and the vaccine strain Rev.1 (Fig. 2, lanes 2, 9, 10, and 11), as well as for B. abortus (Fig. 2, lanes 3, 4, and 5) and B. suis strain 2 (Fig. 2, lane 8). The Israeli B. melitensis field isolates from the three biotypes, including the atypical biotype 1 strains, unexpectedly exhibited the PstI digestion profile which occurs in B. abortus, i.e., two bands of 238 and 44 bp, respectively (Fig. 4, lane 3, and Fig. 5, lanes 1, 2, 3, 4, 7, 13, 14, 15, and 16; also data not shown).
In a comprehensive study, Meyer has shown that unlike B. abortus, B. melitensis lacked plasticity in the features characterized by the conventional biotyping methods (24). Our data indicated that in contrast to these findings, B. melitensis has undergone genetic diversions in a pattern similar to that previously shown to occur in other Brucella spp. From the data, one could infer that the prevailing Israeli biotype 1 strains have acquired a new PstI site in the omp2a gene (compared to the sequences established for strain 16M). On the other hand, strains belonging to biotypes 2 and 3 acquired this change earlier, since all isolates demonstrated the same pattern, which was similar to the B. melitensis biovar 3 prototype strain Ether (Fig. 2, lane 12).
Results obtained by Cloeckaert et al. (9) confirmed these data, showing that B. melitensis isolates were split between those with a single PstI site located in the omp2b gene and those with two PstI sites, one in omp2a and one in omp2b.
Interestingly, from the list presented by Cloeckaert et al., it can be seen that even Israeli isolates from the 19702 all had two PstI sites, one in omp2a and the second in omp2b, similar to the results presented above for isolates from the later period. This suggests that the Israeli B. melitensis biotype 1 strains emerged separately from 16M, a strain that originated in the United States.
It is interesting that recent field isolates in Mexico produced a digestion profile of the omp2 amplified gene fragments similar to that of strain 16M and strain Rev.1 (T. A. Ficht, personal communication). Our data and those obtained by Cloeckaert et al. have further shown that the described phenomenon applied not only to strain 16M and Rev.1 but also to the rough strain B115 and H38 as well (Fig. 2, lane 2) (9). We could propose, therefore, that at least two separate B. melitensis biotype 1 lines have evolved, one represented by strain 16M and the other represented by the Israeli isolates. A similar conclusion was drawn by Cloeckaert et al. regarding the absence of a BglII restriction site in the omp2b genes of the Israeli isolates (9).
This study included 37 isolates that according to conventional bacteriological methods were characterized as vaccine strain Rev.1. The PstI digestion pattern of the omp2 amplified gene fragments resembled that of strain 16M, the prototype strain for virulent B. melitensis biovar 1, and that of the vaccine strain Rev.1 (Fig. 2, lanes 9, 10, and 11). In contrast, the PstI digestion profile of the omp2a gene amplified fragments from all other Israeli isolates, representing the three biotypes, depicted a reproducible and conserved pattern that was different from that shown for strains 16M and Rev.1 (Fig. 5, lanes 1, 2, 3, 4, 7, 13, 14, 15, and 16). This suggests that a genetic link might be established between the prototype strain 16M and the vaccine strain Rev.1. A few other geographically remote isolates may have shared the same ancestral strain.
Human infection with the vaccine strain Rev.1 in South Africa has been reported, following horizontal infection among sheep. Clonal selection of virulent colonies during the preparation of a working seed stock probably led to production of a vaccine lot with undesirable characteristics (22, 27, 28). It is interesting that, in Israel, we also identified a human case of infection with the vaccine strain Rev.1. The owner of an intensively managed sheep farm was infected with the Rev.1 vaccine strain 6 months after a series of abortions in ewes and isolation of Rev.1 from the fetal membranes (3). In a report from South Africa, the researchers biotyped the isolates by conventional Brucella biotyping methods, and no direct molecular linkage was shown between the field isolates and the commercial Rev.1 vaccine strain. To the best of our knowledge, our report is the first to associate animal and human infection with the vaccine strain Rev.1 based on molecular identification of the strain.
The resemblance of the phenotypic properties of the vaccine strain Rev.1 and the atypical strain characterized in Israel (2) regarding susceptibility to penicillin and dyes has raised the possibility that the atypical strain had originated from a mutation of the vaccine strain. The unique PstI pattern described for strain 16M and the vaccine strain Rev.1 has allowed us to elaborate on this subject by comparing their omp2 gene PstI digestion patterns. If the atypical strain had originated from a Rev.1 mutant, its omp2 gene PstI digestion pattern should have matched that of strain Rev.1 and strain 16M. The contrary would be true if it had originated from a field strain mutant. The similarity between the PstI omp2 gene digestion profiles of the atypical strains and the virulent field isolates (Fig. 4) clearly implied that the latter was the case, lending a mutation in a virulent strain to render it susceptible to penicillin and dyes. The zoonotic competence of the atypical isolates (2) that caused human infection further supported the idea that these strains had originated from a virulent strain.
The results presented in this study have highlighted some of the potential hazards associated with use of the Rev.1 vaccine in national control programs. It has been argued that vaccine quality could be impaired if its production did not adhere to stringent standards (5, 7). Having encountered the adverse effects of the subcutaneous vaccine, we assumed that the commercial Rev.1 vaccine supplied to Israel originated from a defective seed stock, similar to the events described in South Africa. To overcome these problems, Israel changed the vaccine source in November 1997, purchasing it from a company that had sustained its seed stock on a true Elberg strain (passage 101, 1970; M. Banai, personal communication). Other expected advantages of the new vaccine were the safety to adult animals, attributed to the lower dose (108 CFU instead of 109 CFU), and the method of inoculation as an ocular preparation (16). Implementation of the new vaccine in whole-flock vaccination, including vaccination of pregnant animals, led to outbreaks of abortions in several intensively managed flocks and isolation of the strain from the milk of the aborting animals. In this study, besides conventional biotyping, we also applied the new PCR method to confirm the Rev.1 identity of the isolates according to the omp2a PstI digestion profile (Fig. 6). This new technique made it possible to associate a second human case of strain Rev.1 infection in a 15-year-old girl (Fig. 5, lane 12). Since then, the Israeli veterinary services changed the vaccination policy back to the consensus method of vaccination of only young females, and no additional problems have been encountered.
Acknowledgments
This research was supported in part by a grant from the Israeli Ministry of Health in 1993 (“Rapid diagnosis of Human Brucellosis by the PCR”); grant no. TA MOU CA 13-007; U.S.-Israel Cooperative Developmental Research Program, Office of the Science Advisor, U.S. Agency for International Development. The research was completed under the support by research grant award no. 2781-96 from BARD, The United States-Israel Binational Agricultural Research and Development Fund.
The omp2 gene primers were designed by Gad Frenkel, National Institute for Medical Research, London, United Kingdom.
REFERENCES
- 1.Alton, G. G., L. M. Jones, R. D. Angus, and J.-M. Verger (ed.). 1988. Techniques for the brucellosis laboratory, p. 24-61. Institut National de la Recherche Agronomique, Paris, France.
- 2.Banai, M., I. Mayer, and A. Cohen. 1990. Isolation, identification, and characterization in Israel of Brucella melitensis biovar 1 atypical strains susceptible to dyes and penicillin, indicating the evolution of a new variant. J. Clin. Microbiol. 28:1057-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banai, M., G. Hoida, E. Rapoport, O. Zamir, M. Haimovich, E. Katz, I. Mayer, A. Cohen, and M. Davidson. 1991. Is Brucella melitensis undergoing evolutionary development or a change in the vaccinal Rev.1 strain?, p. 152. In A. Tumbay, S. Hilmi, and O. Ang (ed.), Brucella and brucellosis in man and animals. Turkish Microbiological Society publication no. 16. Ege University Press, Izmir, Turkey.
- 4.Banai, M., M. Abramson, I. Mayer, K. Chechik, G. Hoida, O. Zamir, S. Bardenstein, A. Cohen, and M. Davidson. 1996. Problems associated with the persistence and possible horizontal transfer of Brucella melitensis Rev.1 vaccine in connection with serological surveillance in Israel, p. 69-76. In B. Garin-Bastugi and A. Benkirane (ed.), FAO/WHO/OIE Round Table on the Use of Rev.1 Vaccine in Small Ruminants and Cattle. Centre National d’Etudes Vétérinaires et Alimentaires, Maison-Alfort, France.
- 5.Bosseray, N. 1985. Quality control of four Rev.1 anti-Brucella vaccines, p. 229-236. In J. M. Verger and M. Plommet (ed.), Brucella melitensis. Martinus Nijhoff Publishers, Dordrecht, The Netherlands.
- 6.Bosseray, N. 1991. Brucella melitensis Rev.1 living attenuated vaccine: stability of markers, residual virulence and immunogenicity in mice. Biologicals 19:355-363. [DOI] [PubMed] [Google Scholar]
- 7.Bosseray, N. 1992. Control methods and thresholds of acceptability for anti-Brucella vaccines. Dev. Biol. Stand. 79:121-128. [PubMed] [Google Scholar]
- 8.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 32:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., J.-M. Verger, M. Grayon, and O. Grepinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 141:2111-2121. [DOI] [PubMed] [Google Scholar]
- 10.Elberg, S. 1996. Rev.1 Brucella melitensis vaccine. Part III, 1981-1995. Vet. Bull. 66:1193-1200. [Google Scholar]
- 11.Elberg, S. S. (ed.). 1981. A guide to the diagnosis, treatment and prevention of human brucellosis. World Health Organization publication VPH/81.31. World Health Organization, Geneva, Switzerland.
- 12.Elberg, S. S., and K. Faunce, Jr. 1957. Immunization against Brucella infection. VI. Immunity conferred on goats by a nondependent mutant from a streptomycin-dependent mutant strain of Brucella melitensis. J. Bacteriol. 73:211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elberg, S. S., and K. F. Meyer. 1958. Caprine immunization against brucellosis. A summary of experiments on the isolation, properties and behaviour of a vaccine strain. Bull. W. H. O. 19:711-724. [PMC free article] [PubMed] [Google Scholar]
- 14.Fekete, A., J. A. Bantle, and S. M. Halling. 1992. Detection of Brucella by polymerase chain reaction in bovine fetal and maternal tissues. J. Vet. Diagn. Investig. 4:79-83. [DOI] [PubMed] [Google Scholar]
- 15.Fekete, A., J. A. Bantle, S. M. Halling, and R. W. Stich. 1992. Amplification fragment length polymorphism in Brucella strains by use of polymerase chain reaction with arbitrary primers. J. Bacteriol. 174:7778-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fensterbank, R., P. Pardon, and J. Marly. 1982. Comparison between subcutaneous and conjunctival route of vaccination with Rev.1 strain against B. melitensis infection in ewes. Ann. Rech. Vet. 13:185-190. [PubMed] [Google Scholar]
- 17.Ficht, T. A., S. W. Bearden, and H. Marquis. 1990. Genetic variation at the omp2 porin locus of the Brucellae: species-specific markers. Mol. Microbiol. 4:1135-1142. [DOI] [PubMed] [Google Scholar]
- 18.Ficht, T. A., S. W. Bearden, B. A. Sowa, and L. G. Adams. 1988. A 36-kilodalton Brucella abortus cell envelope protein is encoded by repeated sequences closely linked in the genomic DNA. Infect. Immun. 56:2036-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ficht, T. A., S. W. Bearden, B. A. Sowa, and L. G. Adams. 1989. DNA sequence and expression of the 36-kilodalton outer membrane protein gene of Brucella abortus. Infect. Immun. 57:3281-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ficht, T. A., H. S. Husseinen, and S. W. Bearden. 1996. Species-specific sequences at the omp2 locus of Brucella type strains. Int. J. Sys. Bacteriol. 46:329-331. [DOI] [PubMed] [Google Scholar]
- 21.Guarino, A., L. Serpe, G. Fusco, A. Scaramuzzo, and P. Gallo. 2000. Detection of Brucella species in buffalo whole blood by gene-specific PCR. Vet. Rec. 147:632-634. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, P., S. M. Pefanis, C. C. Williamson, W. J. S. Botha, and M. S. Schalkwyk. 1989. Horizontal transmission in sheep and delayed clearance in guinea pigs and mice of a Brucella melitensis Rev.1 mutant. J. S. Afr. Vet. Assoc. 60:92-94. [PubMed] [Google Scholar]
- 23.Leal-Klevezas, D. S., I. O. Martínez-Vázquez, A. López-Merino, and J. P. Martínez-Soriano. 1995. Single-step PCR for detection of Brucella spp. from blood and milk of infected animals. J. Clin. Microbiol. 33:3087-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, M. E. 1984. Inter- and intra-strain variants in the genus Brucella. Dev. Biol. Stand. 56:73-83. [PubMed] [Google Scholar]
- 25.Morata, P., M. I. Queipo-Ortuño, J. M. Reguera, M. A. García-Ordoñez, C. Pichardo, and J. de Dios Colmenero. 1999. Posttreatment follow-up of brucellosis by PCR assay. J. Clin. Microbiol. 37:4163-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullis, K. B., and F. A. Faloona. 1987. Specific synthesis of DNA in vitro via a polymerase catalysed chain reaction. Methods Enzymol. 155:335-350. [DOI] [PubMed] [Google Scholar]
- 27.Pefanis, S. M., B. Gummow, P. M. Pieterson, C. Williamson, C. G. Venter, and S. Herr. 1988. The isolation and serology of the “FSA” Brucella melitensis Rev.1 mutant in a flock of sheep. Onderstepoort J. Vet. Res. 55:181-183. [PubMed] [Google Scholar]
- 28.Pieterson, P. M., B. Gummow, S. Pefanis, C. G. Venter, and S. Herr. 1988. The characteristics of a variant strain of Brucella melitensis Rev.1. Onderstepoort J. Vet. Res. 55:15-17. [PubMed] [Google Scholar]
- 29.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. 1998. The Israeli experience of field application of S19 and Rev.1, p. 15-17. In The development of new/improved brucellosis vaccines. Report of a WHO Meeting. World Health Organization publication WHO/EMC/ZD1/98.14. Department of Communicable Disease Surveillance and Response, World Health Organization, Geneva, Switzerland.