Abstract
Hepatitis E virus (HEV) is an important public health concern in many developing countries. HEV is also endemic in some industrialized counties, including the United States. With our recent discovery of swine HEV in pigs that is genetically closely related to human HEV, hepatitis E is now considered a zoonotic disease. Human strains of HEV are genetically heterogenic. So far in the United States, only one strain of swine HEV has been identified and characterized from a pig. To determine the extent of genetic variations and the nature of swine HEV infections in U.S. pigs, we developed a universal reverse transcription-PCR (RT-PCR) assay that is capable of detecting genetically divergent strains of HEV. By using this universal RT-PCR assay, we tested fecal and serum samples of pigs of 2 to 4 months of age from 37 different U.S. swine farms for the presence of swine HEV RNA. Thirty-four of the 96 pigs (35%) and 20 of the 37 swine herds (54%) tested were positive for swine HEV RNA. The sequences of a 348-bp region within the ORF2 gene of 27 swine HEV isolates from different geographic regions were determined. Sequence analyses revealed that the 27 U.S. swine HEV isolates shared 88 to 100% nucleotide sequence identities with each other and 89 to 98% identities with the prototype U.S. strain of swine HEV. These U.S. swine HEV isolates are only distantly related to the Taiwanese strains of swine HEV, with about 74 to 78% nucleotide sequence identities; to most known human strains of HEV worldwide, with <79% sequence identities; and to avian HEV, with 54 to 56% sequence identities. Phylogenetic analysis showed that all the U.S. swine HEV isolates identified in this study clustered in the same genotype with the prototype U.S. swine HEV and the two U.S. strains of human HEV. The data from this study indicated that swine HEV is widespread and enzoonotic in U.S. swine herds and that, as is with human HEV, swine HEV isolates from different geographic regions of the world are also genetically heterogenic. These data further raise potential concerns for zoonosis, xenozoonosis, and food safety.
Hepatitis E is a very important disease in many developing countries and is also endemic in some industrialized countries, including the United States (1, 26, 31, 33). The disease primarily affects young adults and reportedly has a mortality rate of up to 25% in pregnant women (8, 12, 31). Hepatitis E virus (HEV), the causative agent of hepatitis E, is primarily transmitted by the fecal-oral route through contaminated water or water supplies (1, 31). HEV is a positive-sense, single-strand RNA virus without an envelope. The virus remains unclassified, although it was once classified in the family Caliciviridae (13, 30). The genome of HEV is approximately 7.5 kb and contains three open reading frames and a short 5′ and 3′ nontranslated region. ORF1 is the largest of the three open reading frames and encodes nonstructural proteins, ORF2 encodes the putative capsid protein, and ORF3 encodes a small protein of unknown function (1, 11, 17, 20, 31, 38, 44). In the United States, sporadic cases of acute hepatitis E in patients with no known epidemiological exposure have been reported (4, 32). Anti-HEV antibodies have been detected in a significant proportion of healthy individuals in the United States (19, 25, 28, 36). The existence of an animal reservoir for HEV has been proposed (25-27).
Swine HEV, the first animal strain of HEV, was identified and characterized in 1997 from a pig in the United States (21). It has been shown that swine HEV is very closely related to the two U.S. strains of human HEV (US1 and US2) but is genetically distinct from other known strains of HEV worldwide (4, 21, 24, 32). Interspecies transmission of HEV has been documented: swine HEV infects nonhuman primates and the US2 strain of human HEV infects pigs (7, 23). More recently, a strain of avian HEV was identified from chickens with hepatitis-splenomegaly syndrome in the United States (9). Avian HEV is genetically related to but distinct from other known HEV strains (9). The single U.S. swine HEV strain and the several Taiwanese swine HEV strains identified thus far are more closely related to strains of human HEV from the same geographic regions than to those from distant regions (10, 23, 43). Most recently, it was shown that swine veterinarians in the United States (28) and other pig handlers in China, Taiwan, and Thailand (10, 24) are at increased risk of zoonotic HEV infections, suggesting that swine are animal reservoirs for HEV.
The relatively high prevalence of anti-HEV in human populations not at apparent risk of exposure to swine HEV (28) suggests that multiple sources of exposure to HEV may exist in the general U.S. population and that swine are not the only animal reservoir for HEV. Anti-HEV has been detected in wild-caught rats in the United States and other countries (6, 14, 18, 39). In Vietnam, anti-HEV has been detected in 44% of chickens, 36% of pigs, 27% of dogs, and 9% of rats (37). Favorov et al. (5) reported that anti-HEV is detected in about 29 to 62% of cows from three countries where HEV is endemic (Somalia, Tajikistan, and Turkmenistan) and in 12% of cows in a country where it is not (Ukraine). In Turkmenistan, about 42 to 67% of the sheep and goats were also found to be positive for immunoglobulin G anti-HEV. Naturally acquired anti-HEV was also detected in rhesus macaques (2). These serological data strongly suggest that these animal species have been exposed to HEV (or a related agent) and that they could also serve as animal reservoirs for HEV.
Human strains of HEV are genetically very heterogenic with at least eight distinct genotypes worldwide (9, 32-33). Most human strains of HEV identified from the same geographic regions tended to cluster together, although a few strains from the same geographic region differed significantly in their genomic sequences (33, 40-41). Since only one strain of swine HEV has been identified and characterized from a pig in the United States (21), the extent of genetic variations among swine HEV isolates and the nature of swine HEV infections in the U.S. swine herds are not known. The objectives of this study were to develop a universal RT-PCR assay that is capable of detecting genetically divergent strains of HEV and to genetically identify and characterize field isolates of swine HEV from pigs in different geographic regions of the United States.
MATERIALS AND METHODS
Clinical specimens.
Fecal and serum samples used in this study were collected from 95 pigs of 2 to 4 months of age and one 7-month-old pig from 37 different herds in six U.S. states (Arkansas, Iowa, Michigan, Missouri, North Carolina, and Oklahoma). Seventy fecal samples were obtained from pigs submitted to the Iowa State University Veterinary Diagnostic Laboratory. Sixteen serum and 10 fecal samples were obtained from pigs submitted to the University of Missouri Veterinary Medical Diagnostic Laboratory. The pigs were submitted to the Diagnostic Laboratories for a wide variety of health problems that are not related to swine HEV infection. Fecal samples from rectal swabs were resuspended in 10% calcium- and magnesium-free phosphate-buffered saline. Serum and fecal samples were stored at −70°C until analyzed.
Primer design for RT-PCR.
To develop a “universal” reverse transcription-PCR (RT-PCR) assay that is capable of detecting HEV strains with significant sequence variations, a multiple sequence alignment of the ORF2 genes of 18 different known strains of human HEV and the prototype U.S. strain of swine HEV was performed (21). Based upon the multiple sequence alignment, two sets of degenerate HEV primers were designed for the universal nested RT-PCR assay: external primer set 3156N [forward, 5′-AATTATGCC(T)CAGTAC(T)CGG(A)GTTG-3′] and 3157N [reverse, 5′-CCCTTA(G)TCC(T)TGCTGA(C)GCATTCTC-3′] and internal primer set 3158N [forward, 5′-GTT(A)ATGCTT(C)TGCATA(T)CATGGCT-3′] and 3159N [reverse, 5′-AGCCGACGAAATCAATTCTGTC-3′]. The expected product of the universal nested RT-PCR was 348 bp.
In addition, a published nested RT-PCR assay (16, 22, 42) specific for the prototype U.S. strain of swine HEV was also used in this study. The primers for the specific RT-PCR assay were based upon the published sequence of the prototype U.S. strain of swine HEV (21): external primer set 3329 (forward, 5′-AGCTCCTGTACCTGATGTTGACTC-3′) and 3330 (reverse, 5′-CTACAGAGCGCCAGCCTTGATTGC-3′) and internal primer set 3331 (forward, 5′-GCTCACGTCATCTGTCGCTGCTGG-3′) and 3332 (reverse, 5′-GGGCTGAACCAAAATCCTGACATC-3′).
Development and standardization of the universal HEV RT-PCR assay for detection of field isolates of swine HEV.
Since HEV strains (including swine HEV) identified from different geographic regions are genetically heterogenic, a universal HEV RT-PCR assay with degenerate HEV primers was developed in this study to detect genetically divergent strains of HEV. To evaluate if the universal RT-PCR assay with degenerate HEV primers could detect known strains of HEV with significant sequence variations, total RNAs were extracted with TriZol Reagent (GIBCO-BRL) from 100 μl of the US2 strain of human HEV (4, 32), the Pakistani strain (Sar-55) of human HEV (38), and the prototype U.S. strain of swine HEV (21). Total RNAs were resuspended in DNase- and RNase-free water. RT was performed at 42°C for 60 min with Superscript II reverse transcriptase (GIBCO-BRL) using reverse primer 3157N. Five microliters of the resulting cDNA was amplified by the universal RT-PCR assay using Ampli Taq gold DNA polymerase (Perkin-Elmer). The PCR parameters for the first-round PCR with primers 3156N and 3157N included a denaturation step at 95°C for 9 min, followed by 39 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 42°C, extension for 2 min at 72°C, and a final incubation at 72°C for 7 min. The parameters for the second-round PCR were similar, except that primers 3158N and 3159N were used.
To determine the sensitivity of the universal RT-PCR assay, the prototype U.S. strain of swine HEV (21-23) with a known infectious titer of 104.5 50% pig infectious doses (PID50) per ml was serially diluted 10-fold in phosphate-buffered saline. Total RNAs extracted from 100 μl of each dilution were tested using the universal RT-PCR assay, as well as with a published RT-PCR assay (22, 42) specific for the prototype U.S. strain of swine HEV. The PCR parameters for the universal PCR assay are the same as described above. The first- and second-round PCR parameters for the RT-PCR assay specific for the prototype swine HEV were similar, with an initial denaturation step at 95°C for 9 min, followed by 39 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 54°C, extension for 1 min at 72°C, and a final incubation at 72°C for 7 min.
After the universal RT-PCR assay with degenerate HEV primers was standardized, 80 fecal and 16 serum samples from pigs in 37 different herds of different geographic regions (Table 1) were tested by the universal RT-PCR assay. Negative and positive controls were included in each set of PCRs. The negative control was water treated the same way as the fecal suspensions and sera. The positive control was the prototype U.S. strain of swine HEV. The amplified PCR products were examined by agarose gel electrophoresis.
TABLE 1.
Detection of swine HEV RNA from fecal and serum samples of pigs of 2 to 4 months of age from different herds in the United States
| Herd identification | Specimena | No. of pigs positive/tested | Geographic location |
|---|---|---|---|
| 01-23444B | F | 0/1 | Rogers, Ark. |
| 01-15025 | F | 1/3 | Story City, Iowa |
| 01-14185-1 | F | 1/2 | Remsen, Iowa |
| 01-12886 | F | 0/2 | Alden, Iowa |
| 01-12116 | F | 1/2 | Emmetsburg, Iowa |
| 01-15555A | F | 1/1 | Ute, Iowa |
| 01-15675 | F | 0/1 | Stratford, Iowa |
| 01-16137 | F | 1/2 | Bloomfield, Iowa |
| 01-16138 | F | 3/3 | Bloomfield, Iowa |
| 01-16139 | F | 2/2 | Bloomfield, Iowa |
| 01-16140 | F | 1/3 | Bloomfield, Iowa |
| 01-17912 | F | 0/11 | Eldora, Iowa |
| 01-18191 | F | 0/2 | Buffalo Center, Iowa |
| 01-18356 | F | 2/3 | Manson, Iowa |
| 01-18934 | F | 5/6 | Linn Grove, Iowa |
| 01-19248 | F | 3/3 | Alden, Iowa |
| 01-21160 | F | 2/3 | Cherokee, Iowa |
| 01-22171 | F | 1/9 | Everly, Iowa |
| 01-22807 | F | 2/2 | Dayton, Iowa |
| 01-31692 | F | 1/3 | Kamrar, Iowa |
| 01-15552 | F | 0/1 | Holland, Mich. |
| UMC1 (A, B) | S | 0/2 | Marshall, Mo. |
| UMC2 (A, B) | S | 0/2 | Monroe City, Mo. |
| UMC3 (A, B) | S | 0/2 | Tipton, Mo. |
| UMC4 (A, B) | S | 0/2 | Monroe City, Mo. |
| UMC5 (A, B) | S | 0/2 | Columbia, Mo. |
| UMC6 (A, B) | S | 0/2 | Shelbina, Mo. |
| UMC7 (A, B) | S | 2/2 | Columbia, Mo. |
| UMC8 (A, B) | S | 0/2 | Meta, Mo. |
| UMC9 (A, B) | F | 2/2 | Columbia, Mo. |
| UMC11 (A, B, C) | F | 0/3 | Columbia, Mo. |
| UMC12 (A, B, C) | F | 1/3 | Sweet Springs, Mo. |
| UMC13 (A) | F | 1/1 | Pleasant Hill, Mo. |
| UMC14 | F | 0/1 | Columbia, Mo. |
| 01-30609 | F | 0/3 | Rose Hill, N.C. |
| 01-14427 | F | 0/1 | McLeansville, N.C. |
| 01-9913 | F | 1/1b | Holdenville, Okla. |
F, fecal sample; S, serum sample.
A 7-month-old pig.
Nucleotide sequencing.
The expected PCR products amplified from fecal or serum samples of pigs were purified using the glassmilk procedure with a GENECLEAN kit (Bio 101 Inc.). PCR products amplified from 27 selected pigs were directly sequenced at the Virginia Tech DNA Sequencing Facility. Sequences of the PCR products were determined for both DNA strands.
Sequence and phylogenetic analyses.
The primer sequences used to amplify the field isolates of swine HEV were excluded for the final sequence and phylogenetic analyses. The resulting 304-bp sequences in the ORF2 genes of the 27 U.S. isolates of swine HEV were analyzed and compared with the corresponding regions of other known human, swine, and avian HEV strains available in GenBank by the MacVector computer program (Oxford Molecular Inc.) (Table 2). The percentages of nucleotide and amino acid sequence identities among different HEV strains were determined with the MacVector program. Phylogenetic analysis was conducted with the aid of the PAUP program (from David L. Swofford, Smithsonian Institution, Washington, D.C.; distributed by Sinauer Associates Inc., Sunderland, Mass.). A heuristic search with 1,000 replicates was used to produce a phylogenetic tree.
TABLE 2.
Pairwise comparison of the nucleotide sequences of the partial ORF2 gene of 27 swine HEV isolates identified in this study (in boldface) and other selected strains of HEV worldwidea
| Isolate | % Identity
|
||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01-12116-2 | 01-18356-1 | UMC12B | 01-18934D1 | 01-18934-D2 | 01-22807-1 | 01-22807-2 | 01-15555A | 01-15025-3 | 01-19248-3 | UMC7A | UMC7B | 01-31692C1 | 01-16140-3 | 01-16137-1 | MC13A | 01-16139-1 | 01-16139-2 | 01-9913 | 01-14185-1 | 01-22171B1 | UMC9A | 01-19248-1 | 01-18934C1 | 01-16138-3 | 01-21160-1 | Swine HEV | JRA1 | Tw6196e | Tw32sw | Mexico | US1 | US2 | Sar-55 | Ch-T1 | Morocco | Tw74sw | E11 | Avian HEV | |
| 01-16138-2 | 92 b | 91 | 92 | 92 | 92 | 92 | 92 | 93 | 91 | 91 | 93 | 92 | 94 | 99 | 100 | 91 | 99 | 100 | 92 | 89 | 99 | 93 | 99 | 92 | 100 | 91 | 91 | 87 | 75 | 76 | 71 | 91 | 92 | 75 | 77 | 75 | 77 | 82 | 55 |
| 01-12116-2 | 94 | 90 | 92 | 92 | 90 | 90 | 92 | 93 | 91 | 90 | 92 | 92 | 92 | 92 | 91 | 93 | 92 | 96 | 91 | 93 | 90 | 92 | 89 | 92 | 92 | 98 | 87 | 75 | 74 | 73 | 92 | 90 | 76 | 77 | 75 | 76 | 86 | 55 | |
| 01-18356-1 | 91 | 93 | 92 | 90 | 90 | 91 | 93 | 90 | 90 | 92 | 92 | 91 | 91 | 92 | 91 | 91 | 93 | 90 | 91 | 91 | 91 | 89 | 91 | 92 | 93 | 87 | 75 | 74 | 72 | 96 | 91 | 76 | 76 | 76 | 75 | 83 | 54 | ||
| UMC12B | 93 | 91 | 90 | 90 | 92 | 91 | 91 | 95 | 92 | 91 | 91 | 92 | 89 | 93 | 92 | 90 | 89 | 92 | 98 | 93 | 90 | 92 | 91 | 90 | 88 | 77 | 76 | 71 | 91 | 89 | 77 | 79 | 75 | 76 | 83 | 54 | |||
| 01-18934D1 | 92 | 90 | 90 | 93 | 92 | 94 | 92 | 93 | 92 | 91 | 92 | 91 | 92 | 92 | 92 | 90 | 92 | 92 | 92 | 92 | 92 | 94 | 92 | 87 | 78 | 75 | 73 | 93 | 90 | 75 | 78 | 76 | 76 | 83 | 54 | ||||
| 01-18934-D2 | 93 | 93 | 91 | 92 | 91 | 91 | 93 | 92 | 92 | 92 | 95 | 93 | 92 | 91 | 91 | 92 | 90 | 92 | 91 | 92 | 91 | 92 | 86 | 76 | 75 | 72 | 91 | 94 | 76 | 78 | 76 | 76 | 82 | 55 | |||||
| 01-22807-1 | 100 | 92 | 91 | 89 | 91 | 91 | 93 | 92 | 92 | 94 | 93 | 92 | 90 | 91 | 92 | 91 | 92 | 90 | 92 | 90 | 90 | 88 | 75 | 76 | 73 | 90 | 94 | 78 | 77 | 76 | 77 | 81 | 55 | ||||||
| 01-22807-2 | 92 | 91 | 89 | 91 | 91 | 93 | 92 | 92 | 94 | 93 | 92 | 90 | 91 | 92 | 91 | 92 | 90 | 92 | 90 | 90 | 88 | 75 | 76 | 73 | 90 | 94 | 78 | 77 | 76 | 77 | 81 | 55 | |||||||
| 01-15555A | 93 | 91 | 91 | 92 | 94 | 92 | 93 | 93 | 93 | 93 | 92 | 91 | 94 | 93 | 94 | 91 | 93 | 91 | 91 | 86 | 77 | 75 | 72 | 92 | 92 | 75 | 77 | 76 | 77 | 85 | 54 | ||||||||
| 01-15025-3 | 92 | 90 | 92 | 92 | 91 | 91 | 91 | 92 | 91 | 93 | 97 | 92 | 91 | 92 | 90 | 91 | 93 | 93 | 88 | 76 | 77 | 72 | 92 | 92 | 74 | 77 | 76 | 76 | 85 | 56 | |||||||||
| 01-19248-3 | 91 | 91 | 90 | 90 | 91 | 89 | 92 | 91 | 91 | 90 | 91 | 90 | 91 | 90 | 91 | 93 | 91 | 85 | 77 | 75 | 72 | 91 | 89 | 74 | 77 | 76 | 77 | 82 | 54 | ||||||||||
| UMC7A | 93 | 91 | 92 | 93 | 90 | 93 | 93 | 90 | 88 | 93 | 96 | 93 | 89 | 93 | 91 | 89 | 87 | 75 | 75 | 70 | 91 | 90 | 76 | 77 | 74 | 76 | 82 | 53 | |||||||||||
| UMC7B | 92 | 91 | 92 | 92 | 93 | 92 | 92 | 91 | 93 | 92 | 92 | 91 | 92 | 93 | 92 | 87 | 76 | 75 | 72 | 93 | 91 | 76 | 78 | 75 | 76 | 83 | 55 | ||||||||||||
| 01-31692C1 | 93 | 94 | 93 | 94 | 94 | 92 | 92 | 94 | 91 | 94 | 90 | 94 | 93 | 91 | 87 | 76 | 76 | 71 | 92 | 93 | 75 | 77 | 76 | 78 | 83 | 54 | |||||||||||||
| 01-16140-3 | 99 | 91 | 98 | 99 | 92 | 89 | 98 | 92 | 98 | 92 | 99 | 90 | 91 | 86 | 75 | 76 | 72 | 90 | 92 | 76 | 76 | 75 | 76 | 82 | 55 | ||||||||||||||
| 01-16137-1 | 91 | 99 | 100 | 92 | 89 | 99 | 93 | 99 | 92 | 100 | 91 | 91 | 87 | 75 | 76 | 71 | 91 | 92 | 75 | 77 | 75 | 77 | 82 | 55 | |||||||||||||||
| UMC13A | 92 | 91 | 90 | 90 | 91 | 90 | 92 | 91 | 91 | 91 | 90 | 87 | 76 | 75 | 73 | 90 | 96 | 76 | 77 | 75 | 76 | 83 | 54 | ||||||||||||||||
| 01-16139-1 | 99 | 93 | 90 | 99 | 94 | 99 | 92 | 99 | 92 | 92 | 88 | 75 | 76 | 72 | 92 | 93 | 75 | 78 | 75 | 77 | 83 | 55 | |||||||||||||||||
| 01-16139-2 | 92 | 89 | 99 | 93 | 99 | 92 | 100 | 91 | 91 | 87 | 75 | 76 | 71 | 91 | 92 | 75 | 77 | 75 | 77 | 82 | 55 | ||||||||||||||||||
| 01-9913 | 90 | 92 | 90 | 92 | 91 | 92 | 92 | 96 | 86 | 76 | 74 | 74 | 92 | 91 | 75 | 77 | 75 | 76 | 85 | 54 | |||||||||||||||||||
| 01-14185-1 | 90 | 89 | 90 | 89 | 89 | 92 | 90 | 87 | 77 | 78 | 73 | 89 | 91 | 76 | 77 | 78 | 77 | 84 | 56 | ||||||||||||||||||||
| 01-22171B1 | 93 | 99 | 92 | 99 | 91 | 92 | 87 | 75 | 75 | 72 | 91 | 92 | 75 | 77 | 75 | 77 | 83 | 54 | |||||||||||||||||||||
| UMC9A | 93 | 90 | 93 | 91 | 89 | 87 | 77 | 76 | 71 | 91 | 90 | 77 | 78 | 75 | 76 | 82 | 54 | ||||||||||||||||||||||
| 01-19248-1 | 92 | 99 | 92 | 92 | 87 | 75 | 75 | 72 | 92 | 92 | 75 | 77 | 75 | 77 | 83 | 54 | |||||||||||||||||||||||
| 01-18934C1 | 92 | 90 | 89 | 85 | 77 | 76 | 74 | 89 | 90 | 76 | 78 | 75 | 77 | 81 | 54 | ||||||||||||||||||||||||
| 01-16138-3 | 91 | 91 | 87 | 75 | 76 | 71 | 91 | 92 | 75 | 77 | 75 | 77 | 82 | 55 | |||||||||||||||||||||||||
| 01-21160-1 | 92 | 87 | 78 | 75 | 72 | 92 | 93 | 74 | 78 | 75 | 77 | 83 | 55 | ||||||||||||||||||||||||||
| Swine HEV | 86 | 75 | 75 | 73 | 92 | 90 | 75 | 75 | 76 | 77 | 85 | 55 | |||||||||||||||||||||||||||
| JRA1 | 77 | 75 | 73 | 86 | 88 | 75 | 77 | 76 | 77 | 83 | 54 | ||||||||||||||||||||||||||||
| Tw6196e | 84 | 76 | 75 | 76 | 77 | 85 | 77 | 84 | 76 | 54 | |||||||||||||||||||||||||||||
| Tw32sw | 70 | 75 | 75 | 76 | 85 | 74 | 90 | 77 | 56 | ||||||||||||||||||||||||||||||
| Mexico | 71 | 72 | 76 | 76 | 75 | 72 | 71 | 53 | |||||||||||||||||||||||||||||||
| US1 | 90 | 76 | 77 | 76 | 77 | 83 | 54 | ||||||||||||||||||||||||||||||||
| US2 | 75 | 78 | 76 | 77 | 82 | 55 | |||||||||||||||||||||||||||||||||
| Sar-55 | 77 | 89 | 76 | 74 | 55 | ||||||||||||||||||||||||||||||||||
| Ch-T1 | 76 | 86 | 78 | 54 | |||||||||||||||||||||||||||||||||||
| Morocco | 77 | 74 | 56 | ||||||||||||||||||||||||||||||||||||
| Tw74sw | 78 | 54 | |||||||||||||||||||||||||||||||||||||
| E11 | 54 | ||||||||||||||||||||||||||||||||||||||
References for other HEV strains used in the comparisons: the prototype U.S. strain of swine HEV, swine HEV (21); a Japanese strain of human HEV, JRA1 (36); Taiwanese strains of swine HEV (Tw32sw and Tw74sw) and human HEV (Tw6196e) (10, 43); U.S. strains of human HEV (US1 and US2) (4, 32); the Pakistani strain of human HEV, Sar-55 (38); a variant Chinese strain of human HEV, Ch-T1 (40, 41); a Spanish strain of HEV of possible swine origin, E11 (29); and the avian strain of HEV (9).
The geographic origins and the GenBank accession numbers of the nucleotide sequences of the HEV strains used in the phylogenetic and sequence analyses are as follows: TK78/87 (AF020608, Nepal), TK104/91 (AF020603, Nepal), TK15/92 (AF020604, Nepal), Nep4/94 (AF020607, Nepal), TK4/95 (AF020606, Nepal), Hyderabad (AF076239, India), M75 (AF093894, India), AKL-90 (AF124407, India), Y67 (AF093892, India), Madras (X99441, India), Hev037 (X98292, India), Sar-55 (M80581, Pakistan), abb-2B (U40044, Pakistan), Vietnam (AF170450), Burma (M73218), JRA1 (AP003430, Japan), JPHEV (E17109, Japan), Uigh179 (D11093, China), Lanzhou (AF141652, China), K52-87 (L25595, China), Hetian (L08816, China), Ch-T11 (AF151962, China), Ch-T21 (AF151963, China), Ch-T1 (AJ272108, China), Tw6310e (AF117279, Taiwan), Tw8e-2 (AF117275, Taiwan), Tw6196e (AF117278, Taiwan), Tw2494e (AF117276, Taiwan), Tw5483e (AF117277, Taiwan), Tw32sw (AF117280, Taiwan swine HEV), Tw74sw (AF117281, Taiwan swine HEV), Morocco (AF065061, Morocco), Mexico (M74506), VH1 (AF195061, Spain), VH2 (AF195062, Spain), E11 (AF195063, Spain), prototype swine HEV (AF082843, United States), HEV-US1 (AF060668, United States), HEV-US2 (AF060669, United States), and avian HEV (AY043166, United States).
Nucleotide sequence accession number.
The resulting sequences of the 27 swine HEV isolates described in “Nucleotide sequencing” above have been deposited with the GenBank database under accession numbers AF466659 to AF466685.
RESULTS
Sensitivity and specificity of the universal RT-PCR assay.
To evaluate if the universal RT-PCR assay developed in this study is capable of detecting different strains of HEV, we tested three genetically divergent HEV strains available to us: the human Sar-55 strain (38), the US2 human HEV strain (4, 32), and the prototype swine HEV strain (21). These three divergent HEV strains differed in their ORF2 gene sequences by at least 20% (23). Total RNAs extracted from the three HEV strains all tested positive by the universal RT-PCR assay with degenerate HEV primers. We designated this RT-PCR assay with degenerate HEV primers the “universal” RT-PCR assay to distinguish it from the published RT-PCR assay (22, 42) that is specific for the single prototype swine HEV isolate.
To evaluate the sensitivity of the universal RT-PCR assay, we compared it with a published RT-PCR assay specific for the prototype U.S. strain of swine HEV (22, 42) to determine its ability to detect a serially diluted virus stock of the prototype U.S. swine HEV strain. The sensitivity of the published RT-PCR assay specific for the prototype swine HEV was previously determined by using an in vitro-synthesized HEV RNA (42). In this study, the sensitivities of the two assays were compared by using an infectious virus stock of the prototype swine HEV with a known infectious titer. The published RT-PCR assay specific for the prototype swine HEV isolate detected about 3.2 PID50 of swine HEV, while the universal RT-PCR assay detected about 31.6 PID50 of swine HEV. Therefore, the sensitivities of the two assays are comparable, as there is only a 1-log difference in the detection of the prototype swine HEV.
Genetic identification of field isolates of swine HEV from young pigs in different geographic regions of the United States.
Previous seroepidemiological studies indicated that pigs in the United States become infected by swine HEV between the ages of 2 and 4 months (21, 24). Therefore, pigs of 2 to 4 months of age were chosen for this study. Since the universal HEV RT-PCR assay developed in this study is capable of detecting genetically divergent strains of HEV, it was therefore used to detect field isolates of swine HEV from fecal and serum samples randomly collected from pigs in different geographic regions of the United States. The results showed that 34 of the 96 pigs (35%) and 20 of the 37 swine herds (54%) tested in this study were positive for swine HEV RNA. The one 7-month-old pig (no. 01-9913) from a farm in Oklahoma also tested positive for swine HEV RNA (Table 1).
Sequence analyses.
The PCR products from 27 of the 34 positive pigs were sequenced. The PCR products from the other seven positive pigs were not sequenced, since they were from the same herds as those sequenced. The 304-bp sequences within the ORF2 genes of the 27 swine HEV isolates were analyzed and compared to each other as well as to other known human, swine, and avian strains of HEV (Table 2). Sequence analyses revealed that these U.S. swine HEV isolates shared 88 to 100% nucleotide sequence identities with each other, 89 to 98% identities with the prototype U.S. swine HEV strain, 89 to 96% identities with two U.S. human HEV strains (US1 and US2), 85 to 88% identities with a Japanese strain of human HEV (JRA1) thought to be of swine origin (35), and 81 to 86% identities with a Spanish strain of HEV (E11) also thought to be of swine origin (29) (Table 2). However, the U.S. swine HEV isolates were genetically divergent from the Taiwanese strains of swine HEV (Tw32sw and Tw74sw), with about 74 to 78% sequence identities (10, 43), and from other known human strains of HEV (<79% sequence identities) (Table 2). The swine HEV isolates shared 54 to 56% nucleotide sequence identities with the newly identified avian HEV. In general, swine HEV isolates identified from the same geographic region displayed higher percentages of sequence identities than those from different geographic regions (Table 2). Most of these nucleotide changes were found to represent silent mutations and did not result in statistically significant differences at the amino acid level. These U.S. swine HEV isolates displayed 91 to 100% amino acid sequence identities with other swine and human HEV strains but only 52 to 56% amino acid sequence identities with avian HEV.
Phylogenetic analysis.
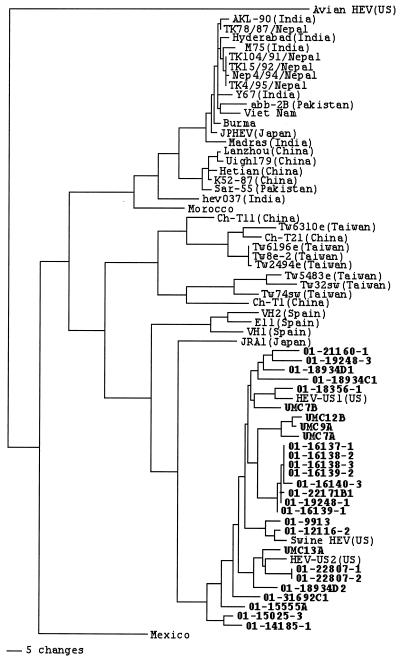
Phylogenetic analysis showed that all the U.S. swine HEV isolates identified in this study clustered in the same genotype with the prototype U.S. swine HEV strain and with the US1 and US2 strains of human HEV (Fig. 1). Swine HEV isolates identified from the same geographic region tended to cluster together, and, thus, minor branches among these U.S. swine and human HEV isolates were observed (Fig. 1). Besides the two U.S. strains of human HEV (US1 and US2), the U.S. swine HEV isolates appeared to be more closely related to the Japanese strain of human HEV (JRA1) and a Spanish strain of HEV (E11) (29, 35) than to other strains of HEV worldwide. The Taiwanese strains of swine HEV were genetically distinct from the U.S. swine HEV strains but were closely related to the Taiwanese strains of human HEV. The U.S. swine HEV isolates were also distinct from the recently discovered avian HEV.
FIG. 1.
A phylogenetic tree based on the nucleotide sequences of a 304-bp region within the ORF2 gene of HEV. The tree was constructed with the aid of the PAUP program. A heuristic search with 1,000 replicates and with a midpoint rooting option was used to construct the tree. A scale bar representing the numbers of character state changes is shown. The swine HEV isolates from this study are indicated in boldface. The GenBank database accession numbers of the sequences of HEV isolates used in the phylogenetic analyses are listed in the text.
DISCUSSION
Recently, numerous novel strains of human HEV have been identified from patients with acute hepatitis in both developing and industrialized countries (4, 10, 15, 29). The intriguing fact is that these novel strains of human HEV are genetically distinct from each other and from other known strains of HEV. The sources of these novel HEV strains are not known; however, it is hypothesized that they may be of animal origin since there exist several potential animal reservoirs for HEV (2-3, 5-6, 14, 25-27, 34, 39). Genetic identifications of swine HEV from pigs (21) and avian HEV from chickens (9) have given credence to this hypothesis. Recently, in a well-controlled, large-scale, seroepidemiological study, it was shown that swine veterinarians in the United States were 1.51 times more likely to be anti-HEV positive than normal U.S. blood donors (28). We found that there was a difference in anti-HEV prevalence in both swine veterinarians and normal blood donors among eight selected states, with subjects from Minnesota (a major swine-producing state) six times more likely to be anti-HEV positive than those from Alabama (a traditionally non-swine-producing state). Age was not a factor for the observed differences from state to state (28). These data provide compelling evidence that swine HEV infects humans.
Since the identification and characterization of the first strain of swine HEV from a U.S. pig, several additional strains of swine HEV have been identified from pigs in Taiwan (10, 43). The Taiwanese swine HEV strains are genetically distinct from the prototype U.S. swine HEV strain. Since only one strain of swine HEV has been identified from a pig in the United States, the extent of genetic variation among swine HEV isolates and the nature of swine HEV infection in pigs in the United States are not known. In order to identify field isolates of swine HEV from pigs in the United States, a sensitive and broadly reactive diagnostic assay is needed, especially swine HEV infection in pigs is subclinical. In this study, we developed a universal RT-PCR assay with degenerate HEV primers that is capable of detecting genetically divergent strains of HEV. We showed that the sensitivity of the universal RT-PCR assay was comparable to that of the published RT-PCR assay (22, 42) specific for the prototype U.S. swine HEV strain. Therefore, this universal HEV RT-PCR assay was used for the detection of genetically divergent strains of HEV from pigs in different geographic regions.
By using this universal RT-PCR assay, we tested fecal and serum samples for swine HEV RNA from pigs of 2 to 4 months of age from 37 herds in different geographic regions of the United States. About 35% of the pigs and 54% of herds tested in this study are positive for swine HEV RNA, indicating that swine HEV infection is enzoonotic and widespread in the United States. It is possible that the negative herds tested by RT-PCR may acquire swine HEV infection at different time points (other than 2 to 4 months of age) or that these negative herds may have better biosecurity measures that could prevent transmission of swine HEV. The swine samples used in this study were submitted to Veterinary Diagnostic Laboratories for diagnoses of diseases unrelated to swine HEV infection. There was no apparent correlation between clinical signs and the presence or absence of swine HEV RNA in the feces. This is consistent with our earlier findings that swine HEV causes only subclinical infection in naturally (21) and experimentally (7, 22) infected pigs.
The U.S. swine HEV isolates identified from pigs in different geographic regions shared significant nucleotide sequence identities with each other (88 to 100%) and with the prototype U.S. strain of swine HEV (89 to 98%). Swine HEV isolates identified from the same pig farm or the same geographic region tended to be more closely related to each other than to those from different farms and geographic regions. Phylogenetic analysis revealed that all the U.S. swine HEV isolates from this study clustered with the US1 and US2 strains of human HEV and the prototype U.S. strain of swine HEV but were distinct from the Taiwanese strains of swine HEV and most strains of human HEV from other countries. These data indicated that, as with human HEV strains (33), swine HEV strains from different geographic regions of the world are also genetically heterogenic.
The enzoonotic nature of swine HEV infection in pigs in the United States and its ability to infect across species raise potential concerns for zoonosis as well as for food and environmental safety (25-26, 34). Since the infected pigs shed virus in feces, swine fouling of irrigation or coastal waters with manure could cause contamination of produce or shellfish (34). Fecal contamination of pork products in meat-packing plants might also serve as a source of HEV infection (34). In addition, since pigs have been considered preferable organ donors for xenotransplantation, the ability of swine HEV to infect across species also poses a concern for xenozoonosis (27). The universal HEV RT-PCR assay developed in this study will be very useful for screening swine HEV infection in xenograft donor pigs.
Acknowledgments
We thank Lee Weigt of Virginia Tech DNA Sequencing Facility for assistance with DNA sequencing and Ray Grover, Pete Thomas, and Jeff Meister for assistance with sample collections. We also thank Suzanne Emerson and Robert Purcell of the National Institutes of Health (Bethesda, Md.) and Isa Mushahwar of Abbott Laboratories (North Chicago, Ill.) for providing the US2 and Sar-55 strains of HEV used in the development of the universal HEV RT-PCR assay.
This study is supported by grants from the National Institutes of Health (AI 01653, AI 46505).
REFERENCES
- 1.Aggarwal, R., and K. Krawczynski. 2000. Hepatitis E: an overview and recent advances in clinical and laboratory research. J. Gastroenterol. Hepatol. 15:9-20. [DOI] [PubMed] [Google Scholar]
- 2.Arankalle, V. A., M. K. Goverdhan, and K. Banerjee. 1994. Antibodies against hepatitis E virus in Old World monkeys. J. Viral Hepat. 1:125-129. [DOI] [PubMed] [Google Scholar]
- 3.Clayson, E. T., B. L. Innis, K. S. Myint, S. Narupiti, D. W. Vaughn, S. Giri, P. Ranabhat, and M. P. Shrestha. 1995. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am. J. Trop. Med. Hyg. 53:228-232. [DOI] [PubMed] [Google Scholar]
- 4.Erker, J. C., S. M. Desai, G. G. Schlauder, G. J. Dawson, and I. K. Mushahwar. 1999. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J. Gen. Virol. 80:681-690. [DOI] [PubMed] [Google Scholar]
- 5.Favorov, M. O., O. Nazarova, and H. S. Margolis. 1998. Is hepatitis E an emerging zoonotic disease? Am. J. Trop. Med. Hyg. 59:242. (Abstract.) [Google Scholar]
- 6.Favorov, M. O., M. Y. Kosoy, S. A. Tsarev, J. E. Childs, and H. S. Margolis. 2000. Prevalence of antibody to hepatitis E virus among rodents in the United States. J. Infect. Dis. 181:449-455. [DOI] [PubMed] [Google Scholar]
- 7.Halbur, P. G., C. Kasorndorkbua, C. Gilbert, D. Guenette, M. B. Potters, R. H. Purcell, S. U. Emerson, T. E. Toth, and X. J. Meng. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamid, S. S., S. M. Jafri, H. Khan, H. Shah, Z. Abbas, and H. A. Fields. 1996. Fulminant hepatic failure in pregnant women: acute fatty liver or acute viral hepatitis? Hepatology 25:20-27. [DOI] [PubMed] [Google Scholar]
- 9.Haqshenas, G., H. L. Shivaprasad, P. R. Woolcock, D. H. Read, and X. J. Meng. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449-2462. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh, S. Y., X. J. Meng, Y. H. Wu, S. T. Liu, A. W. Tam, D. Y. Lin, and Y. F. Liaw. 1999. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 37:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, C. C., D. Nguyen, J. Fernandez, K. Y. Yun, K. E. Fry, D. W. Bradley, A. W. Tam, and G. R. Reyes. 1992. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 191:550-558. [DOI] [PubMed] [Google Scholar]
- 12.Hussaini, S. H., S. J. Skidmore, P. Richardson, L. M. Sherratt, B. T. Cooper, and J. G. O'Grady. 1997. Severe hepatitis E infection during pregnancy. J. Viral Hepat. 4:51-54. [DOI] [PubMed] [Google Scholar]
- 13.Kabrane-Lazizi, Y., X. J. Meng, R. H. Purcell, and S. U. Emerson. 1999. Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 73:8848-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabrane-Lazizi, Y., J. B. Fine, J. Elm, G. E. Glass, H. Higa, A. Diwan, C. J. Gibbs, Jr., X. J. Meng, S. U. Emerson, and R. H. Purcell. 1999. Evidence for wide-spread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61:331-335. [DOI] [PubMed] [Google Scholar]
- 15.Kabrane-Lazizi, Y., M. Zhang, R. H. Purcell, K. D. Miller, R. T. Davey, and S. U. Emerson. 2001. Acute hepatitis caused by a novel strain of hepatitis E virus most closely related to United States strains. J. Gen. Virol. 82:1687-1693. [DOI] [PubMed] [Google Scholar]
- 16.Kasorndorkbua, C., P. G. Halbur, D. K. Guenette, T. E. Toth, and X. J. Meng. 2002. Use of a swine bioassay and a RT-PCR assay to assess the risk of transmission of swine hepatitis E virus in pigs. J. Virol. Methods 101:71-78. [DOI] [PubMed] [Google Scholar]
- 17.Khudyakov, Y. E., E. N. Lopareva, D. L. Jue, T. K. Crews, S. P. Thyagarajan, and H. A. Fields. 1999. Antigenic domains of the open reading frame 2-encoded protein of hepatitis E virus. J. Clin. Microbiol. 37:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maneerat, Y., E. T. Clayson, K. S. A. Myint, G. D. Young, and B. L. Innis. 1996. Experimental infection of the laboratory rat with the hepatitis E virus. J. Med. Virol. 48:121-128. [DOI] [PubMed] [Google Scholar]
- 19.Mast, E. E., I. K. Kuramoto, M. O. Favorov, V. R. Schoening, B. T. Burkholder, C. N. Shapiro, and P. V. Holland. 1997. Prevalence of and risk factors for antibody to hepatitis E virus seroreactivity among blood donors in Northern California. J. Infect. Dis. 176:34-40. [DOI] [PubMed] [Google Scholar]
- 20.Meng, J., J. Pillot, X. Dai, H. A. Field, and Y. E. Khudyakov. 1998. Neutralization of different geographic strains of the hepatitis E virus with anti-hepatitis E virus-positive serum samples obtained from different sources. Virology 249:316-324. [DOI] [PubMed] [Google Scholar]
- 21.Meng, X. J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng, X. J., P. G. Halbur, J. S. Haynes, T. S. Tsareva, J. D. Bruna, R. L. Royer, R. H. Purcell, and S. U. Emerson. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 143:1405-1415. [DOI] [PubMed] [Google Scholar]
- 23.Meng, X. J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by the swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng, X. J., S. Dea, R. E. Engle, R. Friendship, Y. S. Lyoo, T. Sirinarumitr, K. Urairong, D. Wang, D. Wong, D. Yoo, Y. Zhang, R. H. Purcell, and S. U. Emerson. 1999. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. J. Med. Virol. 58:297-302. [PubMed] [Google Scholar]
- 25.Meng, X. J. 2000. Zoonotic and xenozoonotic risks of hepatitis E virus. Infect. Dis. Rev. 2:35-41. [Google Scholar]
- 26.Meng, X. J. 2000. Novel strains of hepatitis E virus identified from humans and other animal species: Is hepatitis E a zoonosis? J. Hepatol. 33:842-845. [DOI] [PubMed] [Google Scholar]
- 27.Meng, X. J. Swine hepatitis E virus: cross-species infection and risk in xenotransplantation. Curr. Topics Microbiol. Immunol., in press. [DOI] [PubMed]
- 28.Meng, X. J., B. Wiseman, F. Elvinger, D. K. Guenette, T. E. Toth, R. E. Engle, S. U. Emerson, and R. H. Purcell. 2002. Prevalence of antibodies to the hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 40:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pina, S., M. Buti, M. Cotrina, J. Piella, and R. Girones. 2000. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J. Hepatol. 33:826-833. [DOI] [PubMed] [Google Scholar]
- 30.Pringle, C. R. 1998. Virus taxonomy—San Diego 1998. Arch. Virol. 143:1449-1459. [DOI] [PubMed] [Google Scholar]
- 31.Purcell, R. H. 1996. Hepatitis E virus, p. 2831-2843. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 32.Schlauder, G. G., G. J. Dawson, J. C. Erker, P. Y. Kwo, M. F. Knigge, D. L. Smalley, J. E. Rosenblatt, S. M. Desai, and I. K. Mushahwar. 1998. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J. Gen. Virol. 79:447-456. [DOI] [PubMed] [Google Scholar]
- 33.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 34.Smith, J. L. 2001. A review of hepatitis E virus. J. Food Prot. 64:572-586. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, K., K. Iwata, N. Watanabe, T. Hatahara, Y. Ohta, K. Baba, and S. Mishiro. 2001. Full-genome nucleotide sequence of a hepatitis E virus strain that may be indigenous to Japan. Virology 287:9-12. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, D. L., P. O. Yarbough, D. Vlahov, S. A. Tsarev, K. E. Nelson, A. J. Saah, and R. H. Purcell. 1997. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J. Clin. Microbiol. 35:1244-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tien, N. T., H. T. Clayson, H. B. Khiem, P. K. Sac, A. L. Corwin, K. S. Myint, and D. W. Vaughn. 1997. Detection of immunoglobulin G to the hepatitis E virus among several animal species in Vietnam. Am. J. Trop. Med. Hyg. 57:211. (Abstract.) [Google Scholar]
- 38.Tsarev, S. A., S. U. Emerson, G. R. Reyes, T. S. Tsareva, L. J. Legters, I. A. Malik, M. Iqbal, and R. H. Purcell. 1992. Characterization of a prototype strain of hepatitis E virus. Proc. Natl. Acad. Sci. USA 89:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsarev, S. A., M. P. Shrestha, J. He, R. M. Scott, D. W. Vaughn, E. T. Clayson, S. Gigliotti, C. F. Longer, and B. L. Innis. 1998. Naturally acquired hepatitis E virus (HEV) infection in Nepalese rodents. Am. J. Trop. Med. Hyg. 59:242. (Abstract.) [Google Scholar]
- 40.Wang, Y., R. Ling, J. C. Erker, H. Zhang, H. Li, S. Desai, I. K. Mushahwar, and T. J. Harrison. 1999. A divergent genotype of hepatitis E virus in Chinese patients with acute hepatitis. J. Gen. Virol. 80:169-177. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., H. Zhang, R. Ling, H. Li, and T. J. Harrison. 2000. The complete sequence of hepatitis E virus genotype 4 reveals an alternative strategy for translation of open reading frames 2 and 3. J. Gen. Virol. 81:1675-1686. [DOI] [PubMed] [Google Scholar]
- 42.Williams, T. P. E., C. Kasorndorkbua, P. G. Halbur, G. Haqshenas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 39:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, J. C., C. M. Chen, T. Y. Chiang, I. J. Sheen, J. Y. Chen, W. H. Tsai, Y. H. Huang, and S. D. Lee. 2000. Clinical and epidemiological implications of swine hepatitis E virus infection. J. Med. Virol. 60:166-171. [PubMed] [Google Scholar]
- 44.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associated with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]