Abstract
Between 1998 and 2001, tissues from four captive white-tailed deer were observed to have histologic lesions of systemic lymphocytic vasculitis. These lesions suggested malignant catarrhal fever, although epizootic hemorrhagic disease and bluetongue were included in the differential diagnosis. Initial diagnostic efforts, including virus isolation and reverse transcription-PCR for epizootic hemorrhagic disease virus and bluetongue virus, failed to identify an etiologic agent. However, consensus primer PCR targeted to the herpesvirus DNA polymerase gene detected viral genomic DNA in each of these four cases. Nucleotide sequence analysis of the amplified product demonstrated that the detected virus was identical over the compared region to the recently described malignant catarrhal fever virus of white-tailed deer (H. Li, N. Dyer, J. Keller, and T. B. Crawford, J. Clin. Microbiol. 38:1313-1318, 2000). Additional nucleotide sequencing of both the DNA polymerase gene and DNA packaging gene followed by phylogenetic analysis solidified this newly recognized herpesvirus as a member of the Gammaherpesvirinae and suggests that this virus, along with ovine herpesvirus 2, alcelaphine herpesvirus 1, alcelaphine herpesvirus 2 and caprine herpesvirus 2, may be part of a separate clade within this subfamily.
Malignant catarrhal fever (MCF) affects a broad range of domestic and exotic ruminants (12, 20), including at least 13 species of deer (22). The distribution of MCF is worldwide, and the clinical course is typically severe and frequently fatal (20), although inapparent infections do occur in susceptible hosts (13) and cattle have been reported to be chronically infected with and to recover from MCF (18). MCF is usually sporadic, with limited morbidity in a population, although severe epizootics with high morbidity and mortality have been reported (3, 10, 27). Two gammaherpesviruses, alcelaphine herpesvirus 1 (AHV1) and ovine herpesvirus 2 (OHV2) (previously referred to as sheep-associated MCF virus), are considered to be the principal etiologic agents of MCF. Both AHV1 and OHV2 are apathogenic in their natural hosts, i.e., wildebeest and sheep, respectively. Transmission to susceptible ruminants can usually be associated with close contact between carrier sheep or wildebeest calves. However, spread by contact between infected cattle, and presumably other susceptible ruminants as well, does not readily occur (5, 20). While the reason for the lack of transmission between susceptible animals has not been proven, it is likely that the clinically susceptible hosts do not shed sufficient levels of cell-free, infectious virus to infect herdmates.
Recently, a third gammaherpesvirus was detected from the tissues of white-tailed deer with lesions that were characteristic of MCF (14). A combination of molecular and virological testing demonstrated that the affected deer were negative for OHV2 and AHV1 as well as a number of other potential etiologic agents. Genetically, this newly identified herpesvirus was found to be closely related to the other viruses known to cause MCF, with 82 and 71% identity to OHV2 and AHV1, respectively, in the amplified region of the DNA polymerase gene. In this epizootic five of six white-tailed deer on the premises died, and the sixth was euthanized. Tissues from all six deer were found to be positive for this newly recognized virus. These deer were part of a small zoo collection that contained approximately 200 animals, including several nonruminant species plus a variety of exotic and domestic ruminants.
The present study describes detection of the newly recognized MCF-associated virus of deer (deer herpesvirus [DHV]) in necropsy samples from four captive white-tailed deer with lesions characteristic of MCF. A majority of the cases were examined retrospectively, using DNA extracted from formalin-fixed, paraffin-embedded tissues as the template for PCR analysis (4, 5, 8, 9, 16, 25). Affected deer originated from three unrelated premises, and the clinical history did not include a high morbidity or mortality rate within the source herds. Furthermore, contact with exotic ruminants was reported in only one of the cases examined, suggesting a domestic rather than an exotic reservoir host. A survey of multiple tissues from captive and wild white-tailed deer without lesions characteristic of MCF demonstrated that DHV was not detected in any of these deer and thus is probably not widespread within the deer population of Missouri. Additional nucleotide sequence and phylogenetic analysis of both the DNA polymerase and the DNA packaging genes solidify identification of this virus as a member of the Gammaherpesvirinae and its relationship to AHV1 and OHV2.
MATERIALS AND METHODS
Cases.
Between 1998 and 2001, four white-tailed deer from three separate, unrelated captive herds in Missouri were necropsied by veterinary practitioners and various tissues were submitted to the University of Missouri's Veterinary Medical Diagnostic Laboratory. Two of the cases were submitted in the month of December, and two were submitted in February. Each of these deer came from a captive herd, and contact with other ruminants was generally limited to cattle, sheep, elk, and/or goats. None of the farms had exotic ruminants on the premises when the deer died or became clinically ill, although contact with exotic species likely occurred in the past for one of the deer.
Thirty-nine control cases were selected from the archives of the University of Missouri's Veterinary Medical Diagnostic Laboratory. The control cases were submitted between 1998 and 2001, and none had lesions characteristic of MCF. A combination of tissues from both captive and wild white-tailed deer were examined.
Nucleic acid extraction and PCR amplification.
For one case with lesions characteristic of MCF, spleen and lymph nodes collected at necropsy were homogenized in phosphate-buffered saline (pH 7.4) (2 to 4 g of tissue per 5 ml). One milliliter of this homogenate was centrifuged at 12,000 × g for 1 min, and the cell pellet was used for DNA extraction. For all other cases (and a majority of control cases), formalin-fixed, paraffin-embedded tissue sections of spleen, kidney, heart, lung, and/or skeletal muscles (two sections per paraffin block; each section 20 μm thick) were deparaffinized by two extractions with xylene, followed by centrifugation. Tissue pellets were then washed twice with 100% ethanol and dried for 10 min at room temperature. For DNA extraction, both fresh and fixed tissue samples were digested with proteinase K (Qiagen, Inc., Valencia, Calif.) in buffer ATL (Qiagen, Inc.) for a minimum of 12 h at 55°C. DNA extractions were performed using the QIAamp DNA minikit (Qiagen, Inc.) according to the manufacturer's instructions. For each of the DNA extraction steps, strict protocols were followed to avoid cross-contamination of samples. Samples were stored at −80°C until used as templates for amplification.
Amplification of a region of the herpesvirus DNA polymerase gene was performed with nested, degenerate primers targeted to highly conserved regions (26). First-round amplification was performed using two upstream primers (DFA and ILK) and one downstream primer (KG1), each at a final concentration of 0.6 μM with 1.0 U of HotStarTaq (Qiagen, Inc.) in the manufacturer's buffer containing 1.5 mM MgCl2 and 0.2 mM (each) deoxynucleoside triphosphates in a final reaction volume of 25 μl. Thermocycling conditions for the first-round amplification were as follows: 95°C (12 min), followed by 10 cycles of denaturation (94°C, 30 s), annealing (70°C, 30 s), and extension (72°C, 90 s) with the annealing temperature in these cycles reduced by 2°C each cycle. An additional 40 cycles of denaturation (94°C, 30 s), annealing (48°C, 30 s), and extension (72°C, 90 s) were performed, followed by a final extension at 72°C for 7 min. Second-round amplification was performed using 2 μl of the first-round reaction product with one upstream primer (TGV) and one downstream primer (IYG). Reaction and thermocycling conditions were as described above. Following second-round amplification, two amplification products (approximately 225 and 440 bp) were consistently observed. For each positive sample, the 440-bp product was consistently the dominant product. Nucleotide sequence analysis demonstrated that the 225-bp fragment was an amplification product of the TGV and IYG primers and the 440-bp fragment was a product of the TGV and KG1 primers. Given these findings, all subsequent second-round PCRs were performed as heminested reactions with the TGV and KG1 primers. A 388-bp product (excluding the primer sequences) was used for sequence analyses.
Amplification of a region of the herpesvirus DNA packaging protein (terminase) gene was performed with nested, degenerate primers targeted to highly conserved regions (11). First-round amplification was performed with primers A2 and B1, and second-round amplification was performed with primers A3 and B2. Reaction and thermocycling conditions were as described above for amplification of the DNA polymerase gene fragment. A single amplification product of approximately 415 bp was observed after the second-round reaction. A 371-bp product (excluding the primer sequences) was used for subsequent analyses.
To allow screening of additional white-tailed deer samples for the presence of DHV DNA, specific primers were designed to amplify an internal region of the DNA polymerase gene. The forward primer (5′-GACATTCAGTACCTGCAGC-3′) and reverse primer (5′-GGATCTGAGAATAGGAGGC-3′) corresponded to bases 98 to 116 and 276 to 294, respectively, of the DHV DNA polymerase sequence reported here. The reaction mixture contained a 0.6 μM concentration of each primer, 1.0 U of HotStarTaq (Qiagen, Inc.) in the manufacturer's buffer containing 2.0 mM MgCl2 and 0.2 mM (each) deoxynucleoside triphosphates in a final reaction volume of 25 μl. Thermocycling conditions were as follows: 95°C (12 min), followed by 10 cycles of denaturation (95°C, 30 s), annealing (70°C, 30 s), and extension (72°C, 90 s) with the annealing temperature in these cycles reduced by 1°C each cycle. An additional 30 cycles of denaturation (95°C, 30 s), annealing (60°C, 30 s), and extension (72°C, 90 s) were performed, followed by a final extension at 72°C for 7 min. Amplification with the DHV-specific primers yielded a product of 197 bp.
Amplification products were visualized in a 2% agarose-1× Tris-acetate-EDTA gel by ethidium bromide staining and UV transillumination (24). Products of the appropriate size were excised from the gel and purified with a Qiaex II gel extraction kit (Qiagen, Inc.). Purified products were either sequenced directly using the herpesvirus PCR primers or cloned using the pCR-Blunt TOPO II system (Invitrogen, Inc., Carlsbad, Calif.). When amplified fragments were sequenced directly, a minimum of three sequencing reactions were performed for each product. When amplified fragments were cloned, a minimum of three clones were sequenced. Nucleotide sequencing was performed by the DNA Core Facility, University of Missouri, Columbia. Nucleotide sequence analysis was performed with GeneTool, version 1.0 (BioTools, Inc.).
Alignments and phylogenetic analysis.
DNA and deduced amino acid sequences of amplified herpesvirus PCR products, excluding the primer sequences, were aligned with BLAST (1) and CLUSTAL W (version 1.74) software. Sequences for comparison were obtained from GenBank and corresponded to AHV1 (GenBank accession no. NC_002531) nucleotide positions 21557 to 21944 for the DNA polymerase protein gene and 52324 to 52694 for the DNA packaging protein gene. Phylogenetic analyses of DNA and amino acid alignments were performed using maximum-parsimony methods (DNAPARS or PROTPARS) and distance matrix methods (DNADIST or PROTDIST followed by NEIGHBOR) within the PHYLIP software package (7). Data sets were subjected to bootstrap analysis (6), based on 100 resamplings of the original data set, using the SEQBOOT program to produce a majority-rule consensus tree. Completed tree files were visualized using TreeView 1.5 (19).
Nucleotide sequence accession numbers.
The herpesviral sequences obtained in this study have been deposited in the National Center for Biotechnology Information database and assigned GenBank accession numbers AF387516 (DNA polymerase gene fragment) and AY055731 (DNA packaging gene fragment).
RESULTS
Gross lesions and histopathology.
Macroscopic lesions were observed in tissue samples from two of the four deer. These lesions were gray-white nodules up to 5 mm in diameter in heart, kidney, or liver. In the heart, nodules were mainly in subendocardial ventricular myocardium; in the kidney, they were concentrated around arcuate vessels at the corticomedullary junction. Histologically, aggregates of small and large lymphocytes with fewer histiocytes and plasma cells comprised these nodules. Small to medium-sized muscular arteries were usually at the center of lymphoid nodules. Lymphocytes infiltrated arterial adventitia with focal extension into the tunica media, where inflammation was associated with segmental fibrinoid necrosis. Lymphocytic arteritis and perivasculitis were observed microscopically in the heart, lung, kidney, adrenal gland, spleen, brain, meninges, skeletal muscle, abomasum, and/or intestine in each of the four cases. Lymph nodes had inconspicuous follicles, but paracortical regions were expanded by lymphocytes. A morphological diagnosis of systemic lymphocytic vasculitis prompted consideration of MCF in the differential diagnosis (16, 17).
PCR amplification and nucleotide sequence analysis.
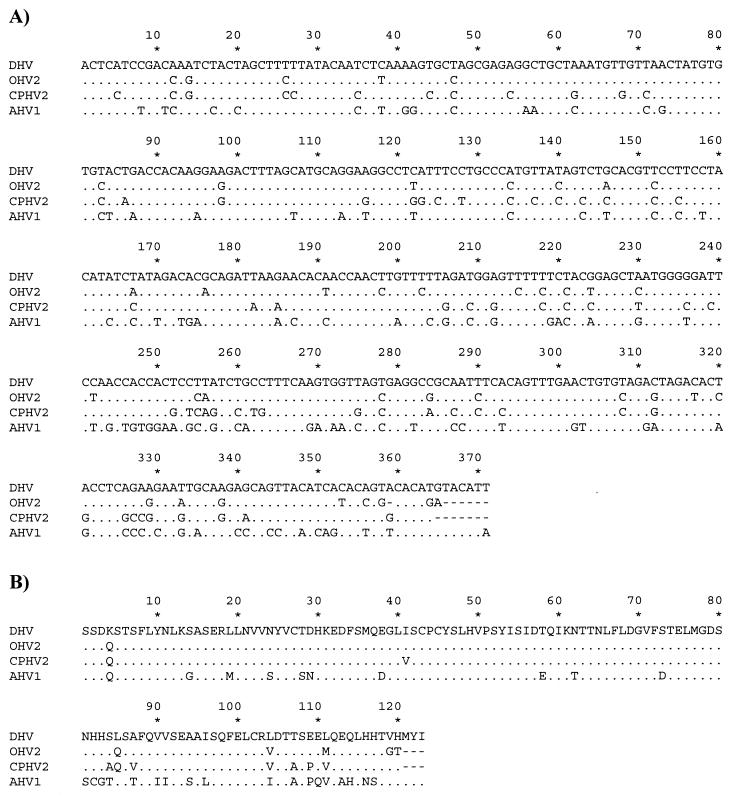
Consensus primer PCR targeted to the herpesvirus DNA polymerase gene amplified specific products from the tissues of each of the four deer with lesions characteristic of MCF. A second round of amplification of the DNA polymerase gene produced two fragments of approximately 225 and 440 bp, with the larger product consistently present in higher concentrations. Nucleotide sequencing demonstrated that the 440-bp fragment was a product of amplification with the TGV and KG1 primers and that the 225-bp fragment was a product of amplification with the IYG and TGV primers and thus was an internal fragment of the larger amplification product. The nucleotide sequence of the smaller fragment has previously been reported (14). For the larger fragment, exclusion of the primer sequences yielded a 388-bp fragment. The sequence of this fragment was determined for amplification products from each of the four PCR-positive deer cases and was found to be 100% identical for all samples from these cases. Pairwise alignment of the nucleotide sequence and deduced amino acid sequence (Fig. 1) demonstrated that the 388-base DNA polymerase gene fragment was 83.2 and 87.5% identical, respectively, to OHV2 (Table 1). Somewhat lower levels of identity were found when this sequence was compared to AHV1, as well as the recently identified AHV2 (15), caprine herpesvirus 2 (CPHV2) (2, 15), and bovine lymphotrophic herpesvirus (BLHV) (23). Phylogenetic analysis of the deduced amino acid sequence from the DNA polymerase gene sequence revealed that the herpesvirus detected from these deer, along with OHV2, AHV1, AHV2, and CPHV2, form a distinct clade within the Gammaherpesvirinae subfamily (Fig. 2).
FIG. 1.
Alignment of nucleotide sequences (A) and deduced amino acid sequences (B) from herpesvirus DNA polymerase genes. Only nucleotides or amino acids which differ from the sequence of DHV are shown. Identical bases or amino acids are shown by dots, and gaps are shown by dashes.
TABLE 1.
Nucleotide and deduced amino acid sequence identities of the DNA polymerase gene fragment
| Virus | % Identitya with:
|
|||||
|---|---|---|---|---|---|---|
| DHV | OHV2 | AHV1 | AHV2b | CPHV2 | BLHV | |
| DHV | 83.2 | 70.8 | 66.2 | 70.1 | 64.9 | |
| OHV2 | 87.5 | 72.1 | 70.2 | 70.8 | 64.4 | |
| AHV1 | 77.5 | 78.2 | 80.0 | 64.1 | 62.2 | |
| AHV2 | 72.8 | 69.4 | 84.4 | 60.6 | 57.3 | |
| CPHV2 | 72.8 | 71.3 | 65.1 | 61.0 | 61.0 | |
| BLHV | 68.2 | 67.4 | 65.6 | 49.1 | 61.2 | |
Values for deduced amino acid sequence identities are in boldface.
Similarity values for AHV2 are based on 174 nucleotides (or 58 amino acids).
FIG. 2.
Phylogenetic tree for the DNA polymerase gene proteins of representative alpha-, beta-, and gammaherpesviruses plus the MCF virus of white-tailed deer (DHV). The unrooted tree was generated using the distance matrix program NEIGHBOR with the jumble option evoked. Distance values were calculated using the PROTDIST program with the Dayhoff percent accepted mutation matrix. Bootstrap values of greater than 50 are shown, and the branch lengths represent relative genetic distances. The herpesviruses used for comparison and their accession numbers are as follows: ateline herpesvirus 2 (AtHV2), U63457; AHV1, NC_002531; AHV2, AF275942; asinine herpesvirus 4 (AsHV4), AY054990; AsHV5, AY054993; bovine herpesvirus 1 (BHV1), Z78205; BHV2, AF031810; BHV4, AF031811; BLHV, AF031808; canine herpesvirus 1 (CHV1), X89500; CPHV2, AF275941; DHV, AF181468; Epstein-Barr virus (EBV) (human herpesvirus 4 [HHV4]), V01555; equine herpesvirus 1 (EHV1), M86664; EHV2, NC_001650; EHV5, AF141886; gallid herpesvirus 2 (GHV2) (Marek's disease virus), L40431; human cytomegalovirus (HCMV) (HHV8), M14709; HHV6, X83413; HHV7, U43400; HHV8 (Kaposi's sarcoma virus), U93872; herpes simplex virus type 1 (HSV1), X04771; HSV2, M16321; murine gammaherpesvirus 68 (HV68), U97553; saimiriine herpesvirus 2 (HVS), M31122; OHV2, AF031812; pseudorabies virus (PRV), L24487; retroperitoneal fibromatosis herpesvirus (RFHVMn), AF005478; varicella-zoster virus (VZV) (HHV3), X04370; wildass herpesvirus (WAHV), AF141888; and zebra herpesvirus (ZHV), AF141889.
Consensus primer PCR targeted to the herpesvirus DNA packaging gene amplified a 415-bp product from samples of each of the four cases with lesions characteristic of MCF. Nucleotide sequence analysis demonstrated that a novel gene fragment corresponding to the DNA packaging genes of other herpesviruses had been detected. The sequence of this fragment was determined for amplification products from each of the four deer with MCF and was found to be 100% identical for all samples tested. Exclusion of the primer sequences yielded a 371-bp fragment. Pairwise alignment demonstrated that the nucleotide and deduced amino acid sequences of this fragment were 86.1 and 91.9% identical, respectively, to the corresponding region of the OHV2 genome (Table 2). Slightly lower levels of identity were found for both the nucleotide and deduced amino acid sequences when the putative DHV DNA packaging gene fragment was compared to those of CPHV2 and AHV1 (Fig. 3; Table 2). The results of phylogenetic analysis for the deduced amino acid sequence of the DNA packaging gene fragment (Fig. 4) were very similar to those of analysis of the DNA polymerase gene, although sequences for comparison were not available for AHV2 and BLHV.
TABLE 2.
Nucleotide and deduced amino acid sequence identities of the DNA packaging gene fragment
| Virus | % Identitya with:
|
|||
|---|---|---|---|---|
| DHV | OHV2 | CPHV2 | AHV1 | |
| DHV | 86.1 | 79.7 | 74.2 | |
| OHV2 | 91.9 | 84.1 | 69.4 | |
| CPHV2 | 89.5 | 93.3 | 71.2 | |
| AHV1 | 77.4 | 72.5 | 75.8 | |
Values for deduced amino acid sequence identities are in boldface.
FIG. 3.
Alignment of the nucleotide sequences (A) and deduced amino acid sequences (B) from herpesvirus DNA packaging genes. Only nucleotides or amino acids which differ from the sequence of DHV are shown. Identical bases are shown by dots, and gaps are shown by dashes.
FIG. 4.
Phylogenetic tree for the DNA packaging gene proteins of representative alpha-, beta-, and gammaherpesviruses plus the MCF virus of white-tailed deer (DHV). The unrooted tree was generated using the distance matrix program NEIGHBOR with the jumble option evoked. Distance values were calculated using the PROTDIST program with the Dayhoff percent accepted mutation matrix. Bootstrap values of greater than 50 are shown, and the branch lengths represent relative genetic distances. The herpesviruses used for comparison and their accession numbers are as follows: AHV1, NC_002531; ateline herpesvirus 3 (AtHV3), NP_001987; bovine herpesvirus 1 (BHV1), Z48053; BHV4, AF139096; callitrichine herpesvirus 3 (CaHV3), AF091062; CPHV2, AF327834; cercopithecine herpesvirus 15 (CeHV15), AF091054; elephant endotheliotropic herpesvirus (EETV), AAD24548; Epstein-Barr virus (EBV) (human herpesvirus 4 [HHV4]), P03219; equine herpesvirus 1 (EHV1), NP_041054; EHV2, U20824; EHV4, NP_045262; feline herpesvirus 1 (FHV1), AF079124; gallid herpesvirus 1 (GHV1), AJ131832; GHV2, AF147806; human cytomegalovirus (HCMV) (HHV5), P16732; HHV6, P24443; HHV7, NP_043815; HHV8 (Kaposi's sarcoma virus), U93872; herpes simplex virus type 1 (HSV1), P04295; saimiriine herpesvirus 2 (HVS), Q01020; murine gammaherpesvirus 68 (HV68), U97553; leporid herpesvirus 2 (LHV2), AF091069; macaca mulatta rhadinovirus 17577 (MMRV), AF083501; OHV2, AF327835; and varicella-zoster virus (VZV) (HHV3), P09294.
A DHV-specific PCR assay was designed to allow rapid screening of a larger number of samples without the requirement of a nested amplification strategy. The analytical sensitivity of a single round of amplification with the specific primers was demonstrated to be approximately equivalent to that of both the DNA polymerase and DNA packaging gene nested consensus primer PCR assays (Fig. 5). The use of a cloned, quantitated DNA template indicated that the specific (nonnested) reaction was capable of detecting 0.05 fg of DNA, which corresponds to approximately 10 genomic copies (Fig. 5B). As an initial effort to determine the prevalence of DHV in the deer population of Missouri, DNA extracted from either fresh tissue or formalin-fixed, paraffin-embedded tissues of 39 control white-tailed deer was tested for the presence of this virus by using a DHV-specific PCR assay. None of the control cases had lesions characteristic of MCF, and all were negative by PCR for DHV. However, all four of the previously identified cases which had histologic lesions of MCF were positive by the DHV-specific PCR assay.
FIG. 5.
Detection of the DHV DNA polymerase and packaging genes by PCR. (A) Consensus primer PCR for the DNA polymerase gene (lanes 1 to 6) and DNA packaging gene (lanes 7 to 12) was used to detect serial 10-fold dilutions of DNA purified from formalin-fixed, paraffin-embedded tissue. Amplification products of the second-round (nested) reaction are shown. For the DNA polymerase gene, primers TGV and KG1 were used. For the DNA packaging gene, primers A3 and B2 were used. DNA dilutions used were as follows: undiluted (lanes 1 and 7), 1:10 (lanes 2 and 8), 1:100 (lanes 3 and 9), 1:1,000 (lanes 4 and 10), and 1:10,000 (lanes 5 and 11). Results of the extraction (negative) control reaction are shown in lanes 6 and 12. (B) Specific primers targeting the DHV DNA polymerase region were used to amplify serial 10-fold dilutions of DNA purified from formalin-fixed, paraffin-embedded tissue (lanes 1 to 6) or dilutions of a plasmid clone containing the 440-bp fragment of the DHV DNA polymerase gene. DNA dilutions used were as follows: undiluted (lane 1), 1:10 (lane 2), 1:100 (lane 3), 1:1,000 (lane 4), and 1:10,000 (lane 5). Results of the extraction (negative) control reaction are shown in lanes 6 and 12. For reactions in which cloned DHV DNA was the template, the quantities of DNA added were as follows: 50 fg (lane 7), 5 fg (lane 8), 0.5 fg (lane 9), 0.05 fg (lane 10), and 0.005 fg (lane 11).
DISCUSSION
Over a 4-year period, tissues from four captive white-tailed deer were observed to have histologic lesions of severe and systemic lymphocytic vasculitis (arteritis). MCF, epizootic hemorrhagic disease, and bluetongue were included in the differential diagnosis. Initial virological and molecular testing eliminated epizootic hemorrhagic disease virus and bluetongue virus as potential etiologic agents, and other diagnostic testing (including bacteriologic culture) failed to identify plausible causes of disease for these cases. However, consensus primer PCR targeted to conserved regions of the herpesvirus DNA polymerase gene (26) detected viral genomic DNA in each of the four cases with lesions characteristic of MCF, suggesting, though not proving, that this virus was associated with the lesions observed. Nucleotide sequence analysis of the 225-bp amplification product demonstrated that the virus detected was identical to that from white-tailed deer with lesions characteristic of MCF reported recently by Li et al. (14). Additional analysis of a larger amplification product of the DNA polymerase gene, as well as consensus primer PCR to the herpesviral DNA packaging gene (11), provided further evidence that this newly recognized herpesvirus of white-tailed deer (DHV) was genetically distinct but most closely related to the two viruses known to cause MCF in ruminants, AHV1 and OHV2, as well as two newly recognized gammaherpesviruses of ruminants, AHV2 and CPHV2 (2, 15). Significant but lower levels of similarity were observed when the DNA polymerase gene nucleotide and deduced amino acid sequences were compared to those of BLHV (23). Phylogenetic analysis of the deduced amino acid sequences for these two gene fragments places DHV within the Gammaherpesvirinae subfamily and suggests that AHV1, AHV2, OHV2, CPHV2, and DHV may form a distinct clade within this subfamily.
In this second study to describe detection of the newly recognized gammaherpesvirus of white-tailed deer, both similarities to and differences from the first report were noted. First, in the initial report of this virus causing disease in white-tailed deer (14), the deer that were positive had concurrent contact with a broad range of species, including exotic ruminants. In contrast, contact species for the deer studied in the present report were generally limited to other domestic ruminants and most often included cattle, sheep, elk, and/or goats. Only one of the four deer had contact with exotic species, and this was not concurrent with the onset of clinical disease. Similar to the report of Li et al. (14), the cases described here occurred during the late fall or early winter. Both the extent of contact with other species (especially ruminants) and the potential seasonal nature of clinical MCF associated with DHV may offer a starting point for epizootiological investigations. For MCF caused by OHV2, an association of contact by susceptible species with lambing ewes suggests that perinatal lambs play an important role in transmission. Similarly, the peak incidence of AHV1 causing MCF in cattle occurs when young wildebeests are in maximum numbers (21). Infected wildebeests up to the age of 3 months have a continuous viremia, and AHV1 can be detected in a cell-free form in both nasal and ocular secretions. Thus, for investigation into the epizootiology of the disease caused by DHV, efforts directed toward increased awareness and/or surveillance would logically be focused during the late fall and early winter months and possibly on contact animals that have recently given birth.
A second major distinction between the present report and that of Li et al. (14) is the level of morbidity and subsequent mortality. From the herd histories gathered for this report, it was noted that while some deer were fatally afflicted, other white-tailed deer on the same premises remained clinically normal. In the epizootic investigated by Li et al. (14), five of six deer in the zoological collection died of MCF and the sixth was euthanized and found to be PCR positive for DHV. The sporadic nature of the cases described here is similar to the typical epizootiologic patterns for MCF of cattle in which the morbidity and mortality is generally low, presumably due to the fact that transmission of AHV1 and OHV2 does not readily occur following contact between infected and uninfected cattle. This similarity suggests that deer-to-deer transmission of DHV may not readily occur. Furthermore, if the biology of the two other MCF viruses, AHV1 and OHV2, can be used as a guide, DHV probably does not endemically infect deer but rather is carried by a different ruminant species and transmitted to the deer by direct contact.
To facilitate screening of additional samples from tissue archives and future epizootiologic investigations, a specific PCR assay was developed to amplify a region of the DHV DNA polymerase gene. A single (nonnested) round of amplification with the DHV-specific primers was shown to be equally sensitive to the nested, consensus primer reactions that were initially used to detect both the DNA polymerase and DNA packaging genes of DHV. This DHV-specific assay failed to detect virus in any tissues from deer without lesions characteristic of MCF, suggesting that DHV was not widespread in the white-tailed deer population of Missouri. However, it is important to recognize that the presence of this virus may be difficult to detect if the host is latently infected. It is anticipated that this assay will be useful in future efforts to identify the reservoir host for DHV, which will likely require screening of a large number of antemortem samples such as nasal and/or ocular swabs.
In summary, additional cases of fatal MCF have been associated with a newly recognized herpesvirus of white-tailed deer (14). These cases were sporadic, spanned a period of 4 years, and involved three separate and unrelated captive herds of white-tailed deer located within the state of Missouri. Phylogenetic analysis based on the deduced amino acid sequences of both the DNA polymerase and DNA packaging gene fragments clearly demonstrated that this herpesvirus detected in deer was similar to AHV1 and OHV2, two gammaherpesviruses that are known etiologic agents of MCF in cattle and other ruminants.
Acknowledgments
We thank John M. Kreeger and Sunny J. Troxell for invaluable discussions during the preparation of the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chmielewicz, B., M. Goltz, and B. Ehlers. 2001. Detection and multigenic characterization of a novel gammaherpesvirus. Virus Res. 75:87-93. [DOI] [PubMed] [Google Scholar]
- 3.Collery, R., and A. Foley. 1996. An outbreak of malignant catarrhal fever in cattle in the Republic of Ireland. Vet. Rec. 139:16-17. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, T. B., H. Li, and D. O'Toole. 1999. Diagnosis of malignant catarrhal fever by PCR using formalin-fixed, paraffin-embedded tissues. J. Vet. Diagn. Investig. 11:111-116. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, T. B., D. O'Toole, and H. Li. 1999. Malignant catarrhal fever, p. 306-309. In J. L. Howard (ed.), Current veterinary therapy IV: food animal practice. The W. B. Saunders Co., Philadelphia, Pa.
- 6.Felsenstein, J. 1985. Confidence intervals on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- 8.Greer, C. E., J. K. Lund, and M. M. Manos. 1991. PCR amplification from paraffin-embedded tissues: recommendations on fixatives for long-term storage and prospective studies. PCR Methods Appl. 1:46-50. [DOI] [PubMed] [Google Scholar]
- 9.Greer, C. E., S. L. Peterson, N. B. Kiviat, and M. M. Manos. 1991. PCR amplification from paraffin-embedded tissues. Am. J. Clin. Pathol. 95:117-124. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton, A. F. 1990. Account of three outbreaks of malignant catarrhal fever in cattle in the Republic of Ireland. Vet. Rec. 127:231-232. [PubMed] [Google Scholar]
- 11.Hargis, A. M., P. E. Ginn, J. E. K. L. Mansell, and R. L. Garber. 1999. Ulcerative facial and nasal dermatitis and stomatitis in cats associated with feline herpesvirus 1. Vet. Dermatol. 10:267-274. [DOI] [PubMed] [Google Scholar]
- 12.Heuschele, W. P. 1988. Malignant catarrhal fever: a review of a serious disease hazard for exotic and domestic ruminants. Zool. Garten. N. F. 58:123-133. [Google Scholar]
- 13.Li, H., D. T. Shen, D. A. Jessup, D. P. Knowles, J. R. Gorham, T. Thorne, D. O'Toole, and T. B. Crawford. 1996. Prevalence of antibody to malignant catarrhal fever virus in wild and domestic ruminants by competitive-inhibition ELISA. J. Wildl. Dis. 32:437-443. [DOI] [PubMed] [Google Scholar]
- 14.Li, H., N. Dyer, J. Keller, and T. B. Crawford. 2000. Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 38:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, H., J. Keller, D. P. Knowles, and T. B. Crawford. 2001. Recognition of another member of the malignant catarrhal fever virus group: an endemic gammaherpesvirus in domestic goats. J. Gen. Virol. 82:227-232. [DOI] [PubMed] [Google Scholar]
- 16.Liggitt, H. D., and J. C. DeMartini. 1980. The pathomorphology of malignant catarrhal fever. I. Generalized lymphoid vasculitis. Vet. Pathol. 17:59-73. [DOI] [PubMed] [Google Scholar]
- 17.Liggitt, H. D., and J. C. DeMartini. 1980. The pathomorphology of malignant catarrhal fever. II. Multisystemic epithelial lesions. Vet. Pathol. 17:74-84. [DOI] [PubMed] [Google Scholar]
- 18.O'Toole, D., H. Li, D. Miller, W. R. Williams, and T. B. Crawford. 1997. Chronic and recovered cases of sheep-associated malignant catarrhal fever in cattle. Vet. Rec. 140:519-524. [DOI] [PubMed] [Google Scholar]
- 19.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 20.Plowright, W. 1990. Malignant catarrhal fever virus, p. 123-150. In Z. Dinter and B. Morein (ed.), Virus infections of ruminants, 1st ed. Elsevier Science Publishing Co., Inc., New York, N.Y.
- 21.Radostits, O. M., C. C. Gay, D. C. Blood, and K. W. Hinchcliff. 2000. Veterinary medicine: a textbook of the diseases of cattle, sheep, pigs, goats and horses, p. 1081-1085. Harcourt Publishers Limited, London, United Kingdom.
- 22.Reid, H. W. 1992. The biology of a fatal herpesvirus infection of deer (malignant catarrhal fever), p. 93-100. In R. D. Brown (ed.), The biology of deer. Springer-Verlag, New York, N.Y.
- 23.Rovnak, J., S. L. Quackenbush, R. A. Reyes, J. D. Baines, C. R. Parrish, and J. W. Casey. 1998. Detection of a novel bovine lymphotropic herpesvirus. J. Virol. 72:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Tham, K. M. 1997. Molecular and clinicopathological diagnosis of malignant catarrhal fever in cattle, deer, and buffalo in New Zealand. Vet. Rec. 141:303-306. [DOI] [PubMed] [Google Scholar]
- 26.VanDevanter, D. R., P. Warrener, L. Bennet, E. R. Schultz, S. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver, L. D. 1979. Malignant catarrhal fever in two California dairy herds. Bov. Pract. 14:121-124. [Google Scholar]