Abstract
Infections caused by methicillin-resistant Staphylococcus aureus (MRSA) are increasingly a main health concern worldwide for hospitalized patients. In addition, the prevalence of community-acquired infection has risen continuously during the last few years. Some MRSA clones spread easier than others within the hospital environment and therefore are frequently implicated in outbreaks. Thus, the spread of a unique epidemic multiresistant clone, the so-called South American clone, is the main cause of nosocomial infections produced by this bacterium in Brazil and in some regions of Argentina, Chile, and Uruguay. In the present work we describe the identification of a novel clone of MRSA that is involved in nosocomial infections and that shows a prevalence as high as that for the South American clone. A total of 53 consecutive single-patient MRSA isolates were recovered during a 3-month period (May to July 1999) from six different hospitals (955 beds) in Córdoba. The isolates were initially typed according the antibiotic resistance and phage susceptibility patterns, followed by genotyping using pulsed-field gel electrophoresis (PFGE). PFGE analysis of the 53 MRSA isolates revealed six major types (A to F) and 25 subtypes. The B-type DNA pattern was indistinguishable from that of the South American epidemic clone observed in 34% of the isolates. A novel highly prevalent clone, showing the A-type DNA pattern and representing 38% of the isolates, was also identified. Moreover, the most frequent subtype of the A clonal family triggered an outbreak in a hospital 2 months later, further confirming its epidemic feature.
The prevalence of antimicrobial agent-resistant bacteria has dramatically increased in hospitals worldwide during the last few years (41).
Staphylococcus aureus remains a major human pathogen, both in nosocomial and community-acquired infections. The prevalence of methicillin-resistant S. aureus (MRSA) among the S. aureus isolates differs widely among different countries as well as from one hospital to another in the same country (8, 9, 30).
In Argentina, the percentage of MRSA strains causing nosocomial infections during the periods April to May and October to November of 1999 was 52% (n = 919) according to a report from the Antibiotic Resistance Informatic System, a national health network composed of 22 hospitals (4). An extended survey showed that 41% of the 896 S. aureus isolates recovered from hospitalized patients from April to May of 1998 were methicillin resistant (56).
It is well documented that several MRSA strains became endemic within the hospital environment and that, as a general rule, specific epidemic clones have emerged occasionally on the endemic background (6).
Genotype and phenotype analyses of MRSA strains confirmed the existence of epidemic clones with enhanced ability to spread within and among hospitals and to cross national boundaries (6, 9, 22, 30). In this context, it has been shown that long-term geographic dissemination of this bacterium is associated with genetic diversification within a common clonal background of an epidemic MRSA strain (6). Considering all of the MRSA epidemic clones described so far, the two that have spread the most extensively are the so-called Iberian MRSA and the Brazilian MRSA, renamed as the South American clone. The former is widely dispersed in hospitals in Spain, Italy, Belgium, Germany, and more recently the United States (14, 26, 33, 36, 51, 54). The South American clone is broadly disseminated in hospitals in Brazil (38, 46, 47), Uruguay (9), Chile (2), and several cities of Argentina such as Buenos Aires, Tucuman, and Posadas (9), as well as in Portugal (1) and Italy (13).
The existence of a MRSA strain mainly infecting pediatric hosts (children and newborn), named the pediatric MRSA clone, in Portugal was recently reported; this strain was also identified in international samples from Poland, Argentina, Colombia, and the United States (New York). Distinctive features of this clone are its low-level and heterogeneous methicillin resistance and a resistance profile limited to β-lactams. In addition, the pediatric clone was found in areas of low antimicrobial pressure and low patient turnover (39).
As an essential step in the epidemiological surveillance of MRSA infections in Córdoba, Argentina, the analysis of endemic strains of this bacteria was initiated in 1998. Twenty-three MRSA isolates collected over the first 3 months of 1998 from two hospitals were examined to determine their phenotypic features and genetic pattern. Forty-three percent (9) of these isolates were identified as belonging to subtypes of the South American clone, which showed the main single clonal pattern according to the analysis of the DNA restriction pattern by pulsed-field gel electrophoresis (PFGE) (C. Sola and J. L. Bocco, unpublished data). As expected, these results confirmed that the South American clone was the most prevalent MRSA clone in Córdoba as it was in other regions of the country until 1998 (2, 9, 10).
The analysis of MRSA infections carried out during the period of May to July of the following year (1999) was further extended to six hospitals in Córdoba, and this allowed us to identify a new clone of MRSA genetically unrelated to the major South American and Iberian epidemic clones known worldwide. The MRSA clone identified here was able to provoke infectious outbreaks within the hospital environment, confirming its epidemic nature.
MATERIALS AND METHODS
Participating institutions.
Six hospitals of Córdoba, designated H1 to H6 (to preserve confidentiality), participated in this collaborative study for 3 months (May to July) of 1999. The features of the hospitals where the strains were isolated are summarized in Table 1. As indicated, the total number of beds of the six hospitals together was 955.
TABLE 1.
MRSA isolates from six hospitals of Córdoba, Argentina, from May to July 1999
| Hospital | Specialty | Type | No. of beds | No. of specimens/patients positive for:
|
% of specimens/patients positive for MRSA | |
|---|---|---|---|---|---|---|
| S. aureus | MRSA | |||||
| H1 | Community/university | Public | 270 | 57/55 | 20/18 | 35/33 |
| H2 | Community/university | Public | 180 | 26/23 | 11/8 | 42/35 |
| H3 | Community/university | Private | 160 | 27/27 | 12/12 | 44/44 |
| H4 | Community/university | Private | 179 | 26/22 | 12/8 | 54/36 |
| H5 | Community/university | Public military | 100 | 8/8 | 4/4 | 50/50 |
| H6 | Community | Private | 66 | 8/8 | 3/3 | 38/38 |
| Total | 955 | 152/143 | 62/53 | 41/37 | ||
The collaborating health personnel from each hospital included both a member of the Clinical Infectious Disease Service and a member of the Clinical Microbiology Laboratory. Four hospitals were private, and two were public. The six collaborating hospitals were widely dispersed throughout Córdoba, which is the second-most-populous city of Argentina, with more than 1 million people. Córdoba is situated at the center of the country, 750 km northwest of Buenos Aires.
Patient features.
Medical records were reviewed for the following clinical data: age (categories were as follows: newborn, <1 year; child, 1 to 14 years; adult, 14 to 65 years; elderly, >65 years), location (inpatient or outpatient), clinical inpatient service, underlying diseases, associated infection, time of hospitalization, prior surgery or antibiotic therapy, prior stay in an intensive care unit (ICU), transfer from another hospital or from another ward service, infection focus, origin of the infection (community acquired or nosocomial), and whether the isolates were epidemiologically related to a suspected outbreak.
The Centers for Disease Control and Prevention definitions for nosocomial infections were used for suspected MRSA infections (16). Infections were considered nosocomial if they appeared during the 48-h period after hospital admission. An infection was considered community acquired if a specimen met the following criteria: (i) it was obtained from nonhospitalized patients, (ii) it was obtained from patients more than 1 month after their last discharge from the hospital, or (iii) it was recovered within 48 h of patient admission to the hospital.
Microbiological features and clinical isolates.
Consecutive MRSA isolates were identified at each of the six hospitals involved in the present study from May to July 1999. After the initial phenotypic identification as S. aureus and determination of antibiotic susceptibility by classical microbiological procedures the isolates were received at the Departamento de Bioquíica Clínica, Universidad Nacional de Córdoba, for further test analysis by phage typing, PCR for the mecA gene, and DNA restriction pattern determination by PFGE.
Antibiotic susceptibility test.
The susceptibility of these isolates to different antibiotics was determined by the disk diffusion method according the recommendations of the National Committee for Clinical Laboratory Standards (27).
The following antibiotics were tested: penicillin (10 U), oxacillin (1 μg), minocycline(30 μg), erythromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), gentamicin (10 μg), trimethoprim-sulfamethoxazole (1.25 [trimethoprim] and 23.75 μg [sulfamethoxazole]), ciprofloxacin (5 μg), rifampin (5 μg), vancomycin (30 μg), teicoplanin (30 μg), and nitrofurantoin (300 μg; isolates collected in urine).
The phenotypic resistance to oxacillin of each isolate was measured. In addition, multiplex PCR to detect the mecA gene was performed as described elsewhere (17).
To classify the strains as hetero- or homoresistant to methicillin, the phenotypic evaluation was carried out by the plating efficiency (EOP) method as described by Hartman and Tomasz (19). Briefly, petri dishes containing tryptic soy agar supplemented or not with oxacillin (32 μg/ml) were inoculated in parallel with roughly 109 CFU of individual isolates/ml or with a control strain (indicated below) and incubated at 35°C for 96 h. EOP was determined by the ratio of CFU of isolates with oxacillin/CFU of isolates without oxacillin. The strains were considered as homo- or heteroresistant when ≥1 and <1% of cells showed high-level resistance, respectively (29).
S. aureus ATCC 25912 and S. aureus ATCC 43300 were used as control strains that were susceptible and resistant to methicillin, respectively.
Phage typing.
Phage typing was carried out by the Blair and Williams method with the group of 23 phages from the Basic International Set of Typing Phages for S. aureus (group I, 29, 52, 52A, 79, and 80; group II, 3A, 3C, 55, 71; group III, 6, 42E, 47, 53, 54, 75, 77, 83A, 84, and 85; group V, 94 and 96; group I-III, phages from groups I and III and miscellaneous 81 and 95) (5).
All phages were used at routine test dilution (RTD) and 100 times the RTD. Isolates not susceptible to RTD or 100 times the RTD were tested at 1,000 times the RTD and were subjected to heat treatment, as described previously (12, 55).
Individual isolates were considered to belong to different specific phage types if they were susceptible to lysis produced by at least two different phages (57).
Analysis of DNA restriction pattern by PFGE.
Chromosomal DNA was prepared as previously described (11), digested with SmaI restriction endonuclease (Promega), and analyzed by gel electrophoresis in a CHEF-DR II (contour-clamped homogeneous electric field) apparatus (Bio-Rad) for 22 h in 0.5× TBE buffer (1× TBE buffer is 44.5 mM Tris-HCl, 44.5 mM boric acid, and 1 mM EDTA, pH 8). The running parameters were as follows: initial pulse, 5 s; final pulse, 50 s; voltage, 6 V/cm; temperature, 12°C.
Gels were stained with ethidium bromide, visualized under UV illumination, and photographed.
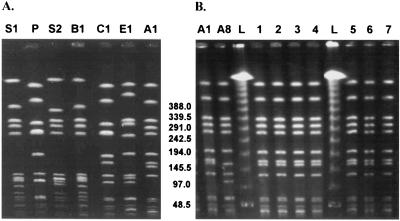
MRSA isolates that have already been described and that are representative of the two international major clonal families, the South American clone (ARG288 and ARG114 [9]) and the pediatric clone (ARG164 [9]), were also included as controls in PFGE analysis (see Fig. 3A, lanes S1, S2, and P, respectively).
FIG. 3.
(A) PFGE of the SmaI-digested genomic DNA of the Cordobes clone in comparison with the South American and pediatric MRSA clones. MRSA epidemic clones isolated in Cordoba, Argentina (A1 and B1) were analyzed by PFGE to compare their DNA patterns with those of the South American (S1 and S2) and the pediatric (P) MRSA clones already described (9). C1, DNA pattern for local MRSA isolates that is closely related to that for the pediatric clone (P); E1, sporadic MRSA local isolate included for comparative analysis. (B) PFGE of SmaI-digested genomic DNA of A1 and A8 MRSA subtypes isolated in hospital H5 from May to July 1999. Lanes 1 to 7, seven strains that produced an outbreak 2 months later in the same hospital; lane L, DNA molecular size markers (lambda DNA ladder; Promega).
The analysis of the SmaI high-molecular-weight DNA restriction pattern was carried out by visual inspection of the bands using the criteria described by Tenover et al. (48, 49). A dendrogram was created to estimate the similarity among strains on the basis of the relative genetic distances. To this end, the Dice similarity coefficient was used and strains were clustered by the minimum-linkage method (43).
Analysis of reproducibility and similarity.
To control the reproducibility of the restriction patterns obtained, each new pattern that was observed frequently was included in subsequent PFGE analysis for comparison. High reproducibility in the DNA patterns obtained from one experiment to another was observed.
Statistical analysis.
Data from patients infected with MRSA strains were analyzed with version 6.04 of the Epi Info 6 software.
RESULTS
Clinical features of the patients.
A total of 62 MRSA isolates were recovered from 53 patients; all the isolates were available for DNA typing, and the medical records from all patients were reviewed.
Ninety-eight percent (52) of the patients were either adults (58%) or elderly (40%). Moreover 47 patients (89%) were hospitalized and 6 (11%) were from an outpatient clinic service. For all inpatients the MRSA isolates were from nosocomial infections.
The majority of the inpatients were assisted either at the surgical service (SS; 32%, 15 of 47 patients) or the ICU (26%, 12 of 47 patients). The percentage of inpatients from SS showed a variation among different hospitals of between 50 and 75% (mean, 60%); for those from the ICU the variation was 22 to 57% (mean, 38%).
A significant increase in MRSA infections at hospital H5 in the 2 months following the time period of this study was detected. The patients suffering MRSA infections were hospitalized in ICU (four patients) and in a surgical ward (three patients).
In summary, the patients infected by MRSA strains showed a broad array of underlying clinical conditions. These included (i) vascular infections or sepsis (25%, 13 of 52), (ii) surgical interventions (19%, 10 of 52), of which 70% (7 of 10) resulted in surgical wound infection, (iii) respiratory disorders (15%, 8 of 52), and (iv) endocrine disorders such as diabetes (11%, 6 of 52). In addition, three of six patients (50%) with MRSA community-acquired infections had diabetes.
The average time of hospitalization for inpatients from admission until the first positive culture for MRSA was 17 ± 14 days, with a maximum of 60 days.
Microbiological features.
The total number of single-patient S. aureus isolates from the six hospitals was 143, and 53 (37%) of them were resistant to methicillin. As shown in Table 1, the percentage of patients with MRSA infection at each hospital was between 33 and 50%. More than one isolate per patient was obtained in three hospitals (H1, H2, and H4); however, only one isolate (the most clinically relevant) per patient was considered for epidemiological analysis during the present work. The isolates were recovered from the following sources: blood and intravascular device (18, 34%); mucocutaneous sites such as surgical and skin wounds (17, 32%); respiratory sources such as tracheal aspirate, sputum, and bronchoalveolar lavage (8, 15%); bone prosthesis (5, 9%); abscess (3, 6%); urine (2, 4%).
The significant increase in the incidence of MRSA infections detected in H5 during August and September 1999 was confirmed as an outbreak (see below). Of the isolates recovered during this outbreak, 71% (n = 5) were obtained from clinically relevant samples such as blood and/or an intravascular device and 29% (n = 2) were isolated from surgical wounds.
Phenotypic characterization of the isolates. (i) Antibiotic resistance patterns.
All the strains were resistant to methicillin according to a standard antibiogram assay. This was confirmed by the detection of the mecA gene (data not shown). Most strains (75%) were multiresistant (MR+), meaning that, in addition to being resistant to β-lactam antibiotics, they were resistant to ciprofloxacin, clindamycin, erythromycin, and gentamicin and variably resistant to minocycline (35%), chloramphenicol (65%), trimethoprim-sulfamethoxazole (45%), and rifampin (60%). For these strains more than 1% of cells had high-level methicillin resistance on the basis of EOP, and therefore the strains were considered homoresistant (Fig. 1). It is important that the seven strains that were recovered from the outbreak that occurred in H5 during August and September had the same MR+ pattern and were also homoresistant.
FIG. 1.
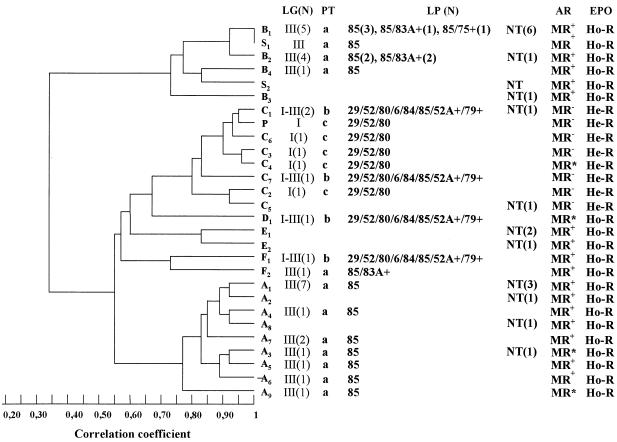
(Left) Dendrogram of the similarity between SmaI-digested DNA patterns obtained by PFGE from representative MRSA strains of each pulsotype and subtype isolated in Córdoba, Argentina, and representative South American (S1 and S2) and pediatric (P) clones described in Buenos Aires, Argentina. (Right) Distribution of the lytic groups (LG), phage types (PT), lytic phages (LP), antibiotic resistance pattern (AR), and phenotypic evaluation of methicillin resistance (EPO) of each subtype of MRSA clinical isolates from Córdoba, Argentina. NT, nontypeable; N, number of isolates; MR+, strains resistant to β-lactam antibiotics, ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), and gentamicin (GEN) and variably resistant to minocycline, chloramphenicol (CHL), trimethoprim-sulfamethoxazole, and rifampin (RIF); MR−, strains resistant only to β-lactam antibiotics and GEN; MR*, strains resistant to β-lactam antibiotics and GEN and variably resistant to RIF, ERY, CLI, CIP, and CHL; Ho-R, homoresistant strains; He-R, heteroresistant strains.
The second group of MRSA isolates (eight strains) did not shown multiresistance (MR−) to antibiotics: they were resistant only to β-lactam antibiotics and gentamicin except for two strains that were also resistant to rifampin. These strains were classified as heteroresistant because <1% of cells displayed high-level methicillin resistance on the basis of EOP frequencies of 10−5and 10−7 (Fig. 1).
The five remaining MRSA isolates indicated as MR* in Fig. 1 had small variations of the two main antibiogram patterns. These strains had variable resistance to clindamycin (two of five, 40%), rifampin (one of five, 20%), ciprofloxacin (two of five, 40%), and chloramphenicol (two of five, 40%) and were susceptible to trimethoprim-sulfamethoxazole and minocycline. Four isolates were homoresistant, with >1% of cells having high-level methicillin resistance in the EOP assay.
Finally, all MRSA isolates analyzed were susceptible to vancomycin and teicoplanin, at least in the disk-based antibiogram assay.
(ii) Phage typing.
Thirty-six percent (n = 19) of all MRSA isolates could not be typed by phage susceptibility. Among the phage-susceptible strains 12 (35%) and 22 (65%) could be typed at 100 times the RTD or at 1,000 times the RTD combined with heat treatment, respectively. Of these 34 strains, 25 were susceptible to lysis mediated by the lytic group III phages. The same strains were lysed by phage 85 alone (20 isolates) as well as by phages 85 and 83A+ (4 isolates) and 85 and 75+ (1 isolate). All these isolates had the same phage susceptibility pattern, which was named phage type a (Fig. 1).
Four MRSA strains were lysed by phages 29, 52, and 80, which belong to lytic group I; this clearly distinct susceptibility pattern was designated phage type c. On the other hand, five isolates showed a mixed lytic pattern (phage type b); they were lysed by phages of groups I and III (phages 29, 52, 80, 6, 84, 85, 52A+, and 79+ (Fig. 1).
The seven strains of the outbreak that occurred in H5 belong to phage type a.
The majority of strains (23, 92%) susceptible to group III phages were multiresistant and displayed a homogeneous methicillin resistance phenotype. All strains lysed by group I phages were only resistant to β-lactam antibiotics and gentamicin; they showed a heterogeneous methicillin resistance phenotype without associated multiresistance.
Genotypic characterization of the isolates and phenotypic analysis of the different pulsotypes.
PFGE analysis of the 53 MRSA isolates revealed six major pulsotypes, A to F, and 25 subtypes (Fig. 2). The main pulsotypes were A, B, and C; 20 (38%), 18 (34%), and 9 (17%) of the 53 isolates, respectively, belonged to these pulsotypes.
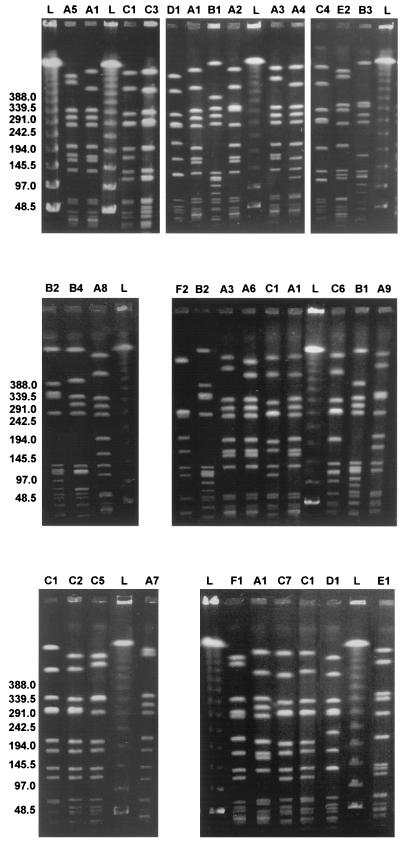
FIG. 2.
PFGE of the SmaI-digested genomic DNA of representative MRSA strains of each pulsotype and related subtypes. A, B, and C, DNA patterns of the Cordobes MRSA epidemic clone and the known South American and pediatric clones, respectively (numbers indicate subtypes); L, DNA molecular size markers (lambda DNA ladder; Promega).
The distribution of the two major DNA pulsotypes and the related subtypes of the MRSA strains among the six hospitals is listed in Table 2.
TABLE 2.
Distributions of the two most frequent pulsotypes and related subtypes of MRSA strains in six hospitals of Córdobaa
| PFGE major pulsotype (n, %)a | PFGE subtype | No.c (%) of isolates from hospitald:
|
||||||
|---|---|---|---|---|---|---|---|---|
| HT (53) | H1 (18) | H2 (8) | H3 (12) | H4 (8) | H5 (4, 7) | H6 (3) | ||
| A (20, 38) | A1 | 10 (19) | 2 (11) | 1* (12.5) | 4 (33) | 2 (25) | 1 (25, 100) | |
| A2 | 1 (2) | 1 (6) | ||||||
| A3 | 2 (4) | 2 (11) | ||||||
| A4 | 1 (2) | 1 (6) | ||||||
| A5 | 1 (2) | 1 (6) | ||||||
| A6 | 1 (2) | 1 (6) | ||||||
| A7 | 2 (4) | 2 (25) | ||||||
| A8 | 1 (2) | 1 (25) | ||||||
| A9 | 1 (2) | 1 (12.5) | ||||||
| B (18, 34) | B1 | 11 (21) | 3 (17) | 1* (25) | 1 (8) | 2 (50) | 3 (100) | |
| B2 | 5 (9) | 4 (22) | 1 (2.5) | |||||
| B3 | 1 (2) | 1 (8) | ||||||
| B4 | 1 (2) | 1 (6) | ||||||
Data in boldface are for MRSA isolates involved in an outbreak that occurred August to September 1999.
n, number of isolates obtained from May to July 1999.
Asterisks indicate community-acquired infections.
HT, all hospitals. Total numbers of isolates from hospitals are in parentheses.
The A major DNA pulsotype was detected in five of the six hospitals, where it was the predominant clonal family. This major clonal type is composed of nine subtypes (A1 to A9). The A1 subtype was identified in 10 (19%) of the 53 isolates from five hospitals and was one of the principal clones in three (H1, H3, and H4) of them (Table 2). Moreover, the A1 subtype was associated with micro-outbreaks observed in these hospitals at the ICU (H3, H4) and in an SS (H1).
Only one strain showing the A1 pulsotype was isolated in hospital H5 from May to July 1999. However seven strains sharing this subtype were identified at the ICU and in a surgical ward during the next 2 months, indicating the clonal spreading of the A1 subtype in these areas of the hospital (Table 2 and Fig. 3B).
The A3 and A7 subtypes also produced micro-outbreaks in the ICUs of hospitals H1 and H4, respectively.
The PFGE-DNA restriction pattern of strains with the A pulsotype differ from those of strains with the B and C pulsotypes by more than six bands, demonstrating that these strains are genetically unrelated (Fig. 2). Thus, the Dice coefficient of similarity among major patterns had a average value of 69% (Fig. 1).
Among the 20 MRSA isolates that have the A pulsotype, 14 (70%) were classified as phage type a, whereas 6 (30%) were nontypeable with the Basic International Set of Typing Phages for S. aureus.
As shown in the Fig. 1, 17 of 20 isolates belonging to the A pulsotype have a multiresistant phenotype. Moreover, all members of this most frequent pulsotype were susceptible to trimethoprim-sulfamethoxazole and minocycline (Fig. 4).
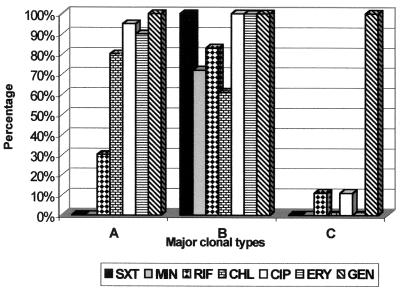
FIG. 4.
Resistance patterns of the most frequent major clonal types, A, B, and C. The resistance to the indicated antibiotics was determined for all isolates, and the resistance profiles for pulsotypes A, B, and C are shown. The results are percentages of the total number of strains analyzed. SXT, trimethoprim-sulfamethoxazole; MIN, minocycline; RIF, rifampin; CHL, chloramphenicol; CIP, ciprofloxacin; ERY, erythromycin; GEN, gentamicin.
Isolates with the A3 subtype (n = 2) were resistant only to ciprofloxacin and chloramphenicol, whereas that with the A9 subtype (n = 1) was resistant only to clindamycin and erythromycin in addition to β-lactam antibiotics. All isolates that have the A pulsotype showing >1% of cells with high-level methicillin resistance in the EOP assay were considered homoresistant.
The B pulsotype comprised four PFGE DNA subtypes, which were recovered from 18 patients from all hospitals, accounting for 34% of the total isolates. The B1 subtype was identified for 11 isolates distributed in the six hospitals and was associated with micro-outbreaks in the medical service in two hospitals (H1 and H6).
The B2 subtype was recovered from five patients, of which four were inpatients at the SS of H1, suggesting the clonal spreading of the B2 subtype in this area of the hospital (Table 2).
Isolates with the B pulsotype had a PFGE-DNA pattern highly similar to that of MRSA isolates representative of the South American clone already detected in other cities of Argentina such as Buenos Aires and Tucuman. Additionally, isolates with the B1 subtype had a PFGE-DNA pattern indistinguishable from that of isolates with the more prevalent subtype identified previously in hospitals from these cities, designated the B1 pulsotype in that work (9). This isolate was included for comparative analysis in the present study (Fig. 3A).
More than half (n = 10) of the 18 MRSA isolates having the B pulsotype were identified as phage type a, whereas 8 (44%) were nontypeable with the Basic International Set of Typing Phages.
All isolates with the B pulsotype had a multiresistant phenotype and were also resistant to trimethoprim-sulfamethoxazole (Fig. 1). For all isolates >1% of cells had high-level methicillin resistance on the EOP essay; thus they were considered homoresistant.
In addition, a similar investigation of infections due to MRSA strains carried out recently (April to June 2001) in the same hospitals revealed prevalences of 45 (n = 30) and 21% (n = 14) for isolates belonging to major clones A and B, respectively. Moreover, 93% of these isolates were obtained from nosocomial infections (data not shown).
The third-most-frequent clonal family, the C pulsotype, represented 17% (nine) of the isolates. They were recovered from seven patients with nosocomial infections (five patients were hospitalized in private hospitals) and two patients with community-acquired infections. This pulsotype comprises seven subtypes (C1 to C7). The C1 subtype was observed for three isolates (33%) recovered from three different hospitals.
The C pulsotype showed a high degree of genetic similarity to a MRSA isolate that is representative of the so-called pediatric clone already detected in two Argentinean cities, Buenos Aires and Tucuman (9). Thus, the C1 subtype has a PFGE pattern 96% similar to that of a representative strain of the pediatric clone, used as a control (Fig. 3A, compare lanes C1 and P).
Four isolates (44%) of the C pulsotype were identified as phage type c, whereas two isolates were nontypeable with the Basic International Set of Typing Phages. The three remaining isolates show phage type b (Fig. 1).
Six isolates of the C clonal family were resistant only to β-lactam antibiotics and gentamicin (MR−), two strains were also resistant to rifampin (MR−), and one was to resistant to clindamycin and erythromycin (MR*). All isolates were considered heteroresistant because yielded cells displayed high-level methicillin resistance in the EOP assay at a frequency ranging from 10−5 to 10−7 (Fig. 1).
Dendrogram of PFGE.
The genetic relationship of the isolates exhibiting PFGE type A with the representative isolates of the South American clone (B), as well as with isolates exhibiting PFGE type C and pediatric clone isolates is shown as a dendrogram (Fig. 1).
As indicated, the Dice coefficient of similarity among the major patterns had an average valve of 69%, and the subtype variants showed similarity values higher than 73%.
DISCUSSION
The emergence of resistant pathogens, particularly gram-positive pathogens, is an important factor in the morbidity and mortality of hospitalized patients. In the face of this growing resistance among these organisms, the selection of the correct antimicrobial and nonpharmacological interventions, based on correct identification and susceptibility test data, has become increasingly challenging. MRSA and, more recently, glycopeptide-resistant enterococci and staphylococci represent a significant danger for the patient. As a consequence, earlier and more precise identification of the pathogens most frequently associated with nosocomial infection is essential (28).
Molecular typing, complemented by conventional methods, provides a sensitive and specific approach for outbreak tracking, and its usefulness in nosocomial epidemiology is very well documented (3, 6, 22, 23, 24, 25, 30, 40, 44, 50, 53).
We analyzed the prevalence of MRSA and the phenotypic and genotypic characteristics of MRSA isolates from patients assisted in six hospitals of Córdoba, Argentina, from May to July 1999. The mean percentage of staphylococcal isolates that were MRSA was 39%, which is in agreement with previous national reports (4, 56).
The most frequent MRSA-induced infections were vascular or septic (25%), infections after surgical treatment (19%), and respiratory infections (15%). The occurrence of surgical wound infections and catheter-associated bacteremia has important infection control implications, as were described in several analyses (28, 31, 32, 58).
The results obtained in this work demonstrate that two genetically distant major clones of MRSA, designated A (38%) and B (34%) and showing epidemic characteristics, coexist in the environment of hospitals in Córdoba, the second-most-populous city of Argentina. The observed distribution is substantially different from that previously reported in other Argentinean cities such as Buenos Aires (9, 10), Tucuman (9), and Posadas (10). In all them the South American clone is the single most frequent clone identified so far. In addition, this clone was also found in other countries of South America and Europe (1, 9, 13, 38, 46, 47).
The A pulsotype was isolated from patients hospitalized in five of the six hospitals, where it was the predominant major clonal family. The A1, A3, and A7 subtypes were associated with micro-outbreaks in the ICU and/or SS in three hospitals (H1, H3, and H4). The A1 subtype, which accounted for 50% of the A clone isolates, was detected in five hospitals. Moreover, the same subtype triggered an outbreak that occurred in the H5 hospital and that affected the inpatients of the surgical ward and of the ICU during the 2 months following the period covered by this study. Thus, the outbreak started, in July, from an index case of MRSA infection detected in a surgical ward and rapidly spread to the ICU. Few medical wards were affected, and currently the outbreak has been partially controlled due to the reinforcement of the appropriate infection control measures. During these months, no significant increase in the incidence of MRSA infections in the other hospitals was observed.
The ability to spread observed for the overall A clone (intra- and interhospital), its easy transmission, and its categorization as clinically or epidemiologically significant were in complete agreement with the guidelines used to design strains as epidemic (42).
The second major clone in terms of frequency (34%), the B pulsotype, which showed a PFGE pattern of DNA similar to that for the South American clone, was detected in all hospitals. The epidemic characteristics of this clone were described in various studies (1, 9, 10, 30, 38, 46, 47).
A recent investigation carried out in the same hospitals from April to June 2001 further suggests that clone A has a greater potential to cause epidemics than the South American clone, as indicated by a twofold-higher prevalence (45 versus 21%). These results strongly suggest that clone A is replacing clone B in the hospital environment.
These epidemic strains had homogeneous methicillin resistance and were typically multiresistant. The majority of the strains were typeable with the Basic International Set of Typing Phages for S. aureus and were included in group III. This phage susceptibility pattern, together with a homogeneous methicillin resistance phenotype, has been described for many epidemic strains (42, 52).
The A clone, characterized by greater susceptibility to a number of antibiotics than the South American clone, is considered clinically and epidemiologically important, allowing antibiotic therapy not involving vancomycin. Avoiding the overuse of glycopeptide antibiotics for treatment of infections produced by the A clone would contribute to controlling the emergence of strains with decreased susceptibility to glycopeptides. The emergence of vancomycin resistance of both sporadic and epidemic MRSA strains in Japan, the United States, and Europe has already been described (7, 18, 20, 21, 37, 59).
The majority of the patients affected by MRSA nosocomial infections during this study were hospitalized mainly at the ICU and SS. Several studies have confirmed the central role of these wards as a source of intra- and interhospital spread of epidemic MRSA. Many elements are thought to contribute to the high occurrence of MRSA infections in patients confined to these special care units, such as the relatively high antibiotic pressure, movement of personnel and/or patients between wards of one hospital or among different hospitals, and weak immune system defense (25, 31, 34, 58). However, the observation that the sporadic MRSA strains neither spread between patients nor became established in the hospitals strengthens the hypothesis that epidemic MRSA strains have intrinsic features that allow them to disseminate in the hospital environment (15, 22, 35, 42). Nevertheless, the specific phenotypic and genetic factors contributing to the epidemic behavior of this bacterium have not been identified so far. A recent study concluded that it appears unlikely that a collection of different epidemic strains will collectively exhibit significant differences in specific phenotypic traits relative to sporadic MRSA. Rather, different combinations of phenotypic and genotypic traits could contribute to the epidemicity of individual strains, favoring the colonization phase of infection as opposed to enhanced expression of toxins and tissue-damaging enzymes (30).
The third clonal family in terms of frequency (17%) described here, the C pulsotype or pediatric clone, was not associated with a suspected outbreak. In contrast to the epidemic clones, this pulsotype showed heterogeneous methicillin resistance as already described. All of the isolates were included in group I or mixed groups I and III, a feature not previously described for this clone. The existence of homogeneously and heterogeneously resistant MRSA strains that belong to different lytic phage groups as well as to different genetically defined clusters has been previously reported (1, 45, 52). Seventy-one percent of nosocomial infections produced by clone C were isolated from inpatients in private hospitals, where the use of antibiotics is perhaps more controlled than in the public hospitals and thus a low antimicrobial pressure is established. The majority of the patients were hospitalized in general wards or medical services where there is a low patient turnover.
These characteristic are in agreement with the theory proposed in a recent study that the continued high prevalence of the pediatric clone is seen in epidemiological settings characterized by low antimicrobial pressure and patient turnover (39).
In conclusion, these results demonstrate that two main epidemic MRSA clones coexist in the hospital environment of Córdoba, Argentina, the South American clone and a clone not previously described in Argentina, which we named the Cordobes clone. This clone was not detected in other cities of Argentine or in Brazil, at least until 1998 (2, 9, 10, 13). It was not identified in a previous study carried out during the first trimester of 1998 in the two hospitals that have the highest number of beds, H1 and H2, suggesting that it emerged recently, at least in our country, and shows a relatively high dissemination rate. After this work was concluded, a new clone was described in Chile (2). The PFGE-DNA pattern of the so-called Chilean clone is similar that of clone A described here, suggesting that they are closely related at the genetic level.
Finally, it will be epidemiologically relevant to analyze the evolution of this clone during the following years in order to better understand the coexistence and/or the eventual replacement of the South American clone by the Cordobes/Chilean clone. A detailed knowledge of its epidemic behavior should contribute to successful strategies to control the dissemination of MRSA infections.
Acknowledgments
This work was supported by the National Council for Scientific Research and Technology of Argentina (CONICET), the Secretaría de Ciencia y Técnica-Universidad Nacional de Córdoba (SECyT-UNC), and Agencia Córdoba Ciencia. C.S. is a fellow recipient of the SECyT-UNC. J.L.B. is a career investigator member of CONICET.
The Argentinean MRSA isolates representative of two major international clonal families were kindly provided by Alejandra Corso and coworkers, Instituto Nacional de Enfermedades Infecciosas, ANLIS, and C. Malbrán, Buenos Aires, Argentina.
REFERENCES
- 1.Aires de Sousa, M., I. Santos-Sanchez, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. De Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., M. Miragaia, I. Santos Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., I. S. Sanchez, A. van Belkum, W. van Leeuwen, H. Verbrugh, and H. De Lencastre. 1996. Characterization of methicillin-resistant Staphylococcus aureus from Portuguese hospitals by multiple genotyping methods. Microb. Drug Resist. 2:331-341. [DOI] [PubMed] [Google Scholar]
- 4.Bantar, C., A. Famiglietti, M. Golberg, M. Radice, and the Subcomisión de Antimicrobianos SADEBAC, Asociación Argentina de Microbiología. 2000. SIR: Sistema Informático de Resistencia. Comparative analysis during two prevalence periods during 1999. Bulletin 144. Asociacíon Argentina de Microbiología, Buenos Aires, Argentina.
- 5.Blair, J. E., and R. E. O. Williams. 1961. Phage typing of staphylococci. Bull. W. H. O. 24:771-784. [PMC free article] [PubMed] [Google Scholar]
- 6.Brun-Buisson, C. 1998. Methicillin-resistant Staphylococcus aureus. Trends, epidemiology, clinical impact and prevention. Pathol. Biol. 46:227-234. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1997. Staphylococcus aureus with reduced susceptibility to vancomycin. Morbid. Mortal. Wkly. Rep. 46:765-766. [PubMed] [Google Scholar]
- 8.Cookson. B. 1995. Aspects of the epidemiology of MRSA in Europe. J. Chemother. 7(Suppl. 3):93-98. [PubMed] [Google Scholar]
- 9.Corso, A., I. Santos Sanches, M. Aires de Sousa, A. Rossi, and H. De Lencastre. 1998. Spread of a methicillin-resistant and multiresistant epidemic clone of Staphylococcus aureus in Argentina. Microb. Drug Resist. 4:277-288. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva Coimbra, M. V., A. Teixeira, R. L. B. Ramos, S. C. Predari, L. Castello, A. Famiglietti, C. Vay, L. Klan, and S. Figueiredo. 2000. Spread of the Brazilian epidemic clone of a multiresistant MRSA in two cities in Argentina. J. Med. Microbiol. 49:187-192. [DOI] [PubMed] [Google Scholar]
- 11.De Lencastre, H., I. Couto, I. Santos, A. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 13:64-73. [DOI] [PubMed] [Google Scholar]
- 12.Del Valle, O., P. Trincado, M. T. Martin, E. Gomez, A. Cano, and A. Vindel. 1999. The prevalence of methicillin-resistant Staphylococcus aureus phagotype 95 in the Hospitales Vall d'Hebron of Barcelona. Enferm. Infecc. Microbiol. Clin. 7:498-505. [PubMed] [Google Scholar]
- 13.Diekema, D. J., M. A. Pfaller, J. Turnidge, J. Verhoef, J. Bell, A. D. C. Fluit, G. V. Doern, R. N. Jones, and the SENTRY Participants Group. 2000. Genetic relatedness of multidrug-resistant, methicillin (oxacillin)-resistant Staphylococcus aureus bloodstream isolates from SENTRY antimicrobial resistance surveillance centers worldwide, 1998. Microb. Drug Resist. 6:213-221. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez, M. A., H. De Lencastre, J. Linares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frenay, H. M. E., J. P. G. Theelen, L. M. Schouls, C. M. J. E. Vandenbroucke-Grauls, J. Verhoef, W. J. van Leeuwen, and F. R. Mooi. 1994. Discrimination of epidemic and nonepidemic methicillin-resistant. Staphylococcus aureus strains on the basis of protein A gene polymorphism. J. Clin. Microbiol. 32:846-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner, J. S., W. R. Jarvis, T. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 17.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerin, F., A. Buu-Hoi, J. L. Mainardi, G. Kac, N. Colardelle, S. Vaupre, L. Gutmann, and I. Podglajen. 2000. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in Parisian hospital. J. Clin. Microbiol. 38:2985-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman, B. J., and A. Tomasz. 1986. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 29:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, K. Yabuta, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 22.Hoefnages-Schuermans, A., A. Borremans, W. Peetermans, S. Van Lierde, G. Reybrouck, and J. Van Eldere. 1997. Origin and transmission of methicillin-resistant Staphylococcus aureus in an endemic situation: differences between geriatric and intensive-care patients. J. Hosp. Infect. 36:209-222. [DOI] [PubMed] [Google Scholar]
- 23.Hookey, J., J. Richardson, and B. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemaitre, N., W. Sougakoff, A. Masmoudi, M. H. Fievet, R. Bismuth, and V. Jarlier. 1998. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J. Clin. Microbiol. 36:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mato, R., I. Santos Sanchez, M. Venditti, D. J. Platt, A. Brown, M. Chung, and H. De Lencastre. 1998. Spread of multiresistant Iberian clone of methicillin-resistant Staphylococcus aureus to Italy and Scotland. Microb. Drug Resist. 4:107-112. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk susceptibility test. Approved standard M2-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Nichols, R. L., and I. I. Raad. 1999. Management of bacterial complications in critically ill patients: surgical wound and catheter-related infections. Diagn. Microbiol. Infect. Dis. 33:121-130. [DOI] [PubMed] [Google Scholar]
- 29.Nicola, F., C. Bantar, L. F. Canigia, S. Relloso, H. Bianchini, and J. Smayesvsky. 2000. Comparison of several methods to determine methicillin-resistance in Staphylococcus aureus with focus on borderline strains. Diagn. Microbiol. Infect. Dis. 36:91-93. [DOI] [PubMed] [Google Scholar]
- 30.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 31.Peltroche-Lllacsahuanga, H., G. Haase, and R. Lutticken. 1998. Methicillin-resistant Staphylococcus aureus (MRSA), clinical implications. Chirurg 69:801-805. [DOI] [PubMed] [Google Scholar]
- 32.Pujol, M., C. Pena, R. Pallares, J. Ayats, and J. Ariza, and F. Gudiol. 1994. Risk factors for nosocomial bacteremia due to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 13:96-102. [DOI] [PubMed] [Google Scholar]
- 33.Ramos, R. L., L. A. Teixeira, L. R. Ormonde, P. L. Siqueira, M. S. Santos, D. Marangoni, and A. M. Figueiredo. 1999. Emergence of mupirocin resistance in multiresistant Staphylococcus aureus clinical isolates belonging to Brazilian epidemic clone III::B:A. J. Med. Microbiol. 48:303-307. [DOI] [PubMed] [Google Scholar]
- 34.Richard, B. R., A. De Lencastre, W. Eisner, E. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 35.Roberts, J. I., and M. A. Gaston. 1987. Protein A and coagulase expression in epidemic and nonepidemic Staphylococcus aureus. J. Clin. Pathol. 40:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, R. B., A. M. Tennenberg, W. Eisner, E. Severina, B. Shopsin, and B. N. Kreiswirth. 1998. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb. Drug Resist. 4:175-183. [DOI] [PubMed] [Google Scholar]
- 37.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sader, H., A. Pignatari, R. Hollis, and R. Jones. 1994. Evaluation of interhospital spread of methicillin-resistant Staphylococcus aureus in Sao Paulo, Brazil, using pulsed-field gel electrophoresis of chromosomal DNA. Infect. Control Hosp. Epidemiol. 15:320-323. [DOI] [PubMed] [Google Scholar]
- 39.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. De Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmenlinna, S., O. Lyytikäinen, P. Kotilainnen, R. Scotford, E. Siren, and J. Vuopio-Varkila. 2000. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Finland. Eur. J. Clin. Microbiol. Infect. Dis. 19:101-107. [DOI] [PubMed] [Google Scholar]
- 41.SENTRY Antimicrobial Program Participants Group. 2001. Global aspects of antimicrobial resistance among key bacterial pathogens. Results from the 1997-1999 SENTRY antimicrobial program. Clin. Infect. Dis. 32(Suppl. 2.):81-155. [Google Scholar]
- 42.Simor, A., D. Boyd, L. Louie, A. McGeer, M. Mulvey, and B. Willey. 1999. Characterization and proposed nomenclature of epidemic strains of methicillin-resistant Staphylococcus aureus in Canada. Can. Commun. Dis. Rep. 25:105-108. [PubMed] [Google Scholar]
- 43.Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys. 1992. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang, Y.-W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Konher, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiological data for molecular typing of methicillin-resistant Staphylococcus aureus J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tassios, P. T., A. C. Vatopoulos, A. Xanthaki, E. Mainas, R. V. Goering, and N. J. Legakis. 1997. Distinct genotypic clusters of heterogeneously methicillin resistant Staphylococcus aureus from a Greek hospital. Eur. J. Clin. Microbiol. Infect. Dis. 16:170-173. [DOI] [PubMed] [Google Scholar]
- 46.Teixeira, L. A., M. C. Lourenco, and A. Figueiredo. 1996. Emergence of a methicillin-resistant Staphylococcus aureus clone related to the Brazilian epidemic clone III::B:A causing invasive disease among AIDS patients in a Brazilian hospital. Microb. Drug Resist. 2:393-399. [DOI] [PubMed] [Google Scholar]
- 47.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. S. Figueiredo, H. De Lencastre, and A. S. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenover, F., R. Arbeit, P. Goering, and the Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for the healthcare epidemiologist. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 49.Tenover, F., R. Arbeit, P. Goering, P. Michelsen, D. Murray, D. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hébert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Mills, M. Mulligan, and M. A. Pfaller. 1994. Comparison of traditional and molecular method of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trzcinski, K., W. Van Leeuwen, A. Van Belkum, P. Grzesiowski, J. Kluytmans, M. Sijmons, H. Verbrugh, W. Witte, and W. Hryniewicz. 1996. Two clones of methicillin-resistant Staphylococcus aureus in Poland. Clin. Microbiol. Infect. 3:198-207. [DOI] [PubMed] [Google Scholar]
- 52.Trzcinski, K., W. Hryniewicz, H. Claus, and W. Witte. 1994. Characterization of two different clusters of clonally related methicillin-resistant Staphylococcus aureus strains by conventional and molecular typing. J. Hosp. Infect. 28:113-126. [DOI] [PubMed] [Google Scholar]
- 53.Van Belkum, A., R. Bax, P. Peerbooms, W. H. F. Goessens, N. van Leeuwen, and W. G. V. Quint. 1993. Comparison of phage typing and fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 31:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Belkum, A. 2000. Molecular epidemiology of methicillin resistant Staphylococcus aureus strains: state of affairs and tomorrow's possibilities. Microb. Drug Resist. 6:173-187. [DOI] [PubMed] [Google Scholar]
- 55.Vindel, A., C. Martin-Bourgon, and J. A. Saez Nieto. 1987. Characterization of non-typeable strains of Staphylococcus aureus from cases of hospital infection. Epidemiol. Infect. 99:91-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whonet International Programme. 1999. Survey of the levels of antimicrobial resistance in Argentine. Whonet Program 1998. Medicina 59:8-16.10436549 [Google Scholar]
- 57.Williams, R. E. O., and J. E. Rippon. 1952. Bacteriophage typing of Staphylococcus aureus. J. Hyg. (Cambridge) 50:320-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, A. P. 1999. Emerging antimicrobial resistance in the surgical compromised host. J. Chemother. 11:518-523. [DOI] [PubMed] [Google Scholar]
- 59.Woodford, N., M. Warner, and H. M. Aucken. 2000. Vancomycin resistance among epidemic strains of methicillin-resistant Staphylococcus aureus in England and Wales. J. Antimicrob. Chemother. 45:258-259. [DOI] [PubMed] [Google Scholar]