Abstract
Legionnaires' disease is a potentially lethal pneumonia that is primarily due to infection by the species Legionella pneumophila, although more than 40 other species are known. Certain L. pneumophila subgroups, particularly serogroup 1, are associated with the majority of the epidemics. The genetic bases for these differences in virulence have not been determined. Three strains, AA100, JR32, and Lp01, have been used in many molecular pathogenesis studies of L. pneumophila. We found genetic differences between these strains by PCR and Southern analyses that may be related to their ability to cause disease. We also examined the distribution of these genetic loci in clinical and environmental isolates of Legionella and found a correlation between the presence of two of these loci, rtxA and lvh, and the ability to cause disease in humans. Examination of the interactions of these strains with host cells suggested that they differ in important phenotypic characteristics including adherence, entry, and intracellular replication. Furthermore, in the mouse model of infection they display differing levels of replication in lungs. These studies emphasize the importance of further investigation into the genetic makeup of these strains, which is likely to lead to the identification of additional factors involved in Legionella pathogenesis.
Legionella strains are ubiquitous inhabitants of biofilms in aquatic environments or moist soil, replicating as intracellular parasites of protozoa (48, 49). When aerosols containing Legionella pneumophila are inhaled, the bacteria enter alveolar macrophages by coiling or conventional phagocytosis (15, 33). The bacterial phagosome neither acidifies nor fuses with the lysosomes, yet interacts with smooth vesicles, mitochondria, and ribosomes in the host cell via unknown mechanisms (32). Although more than 40 species of Legionella are known (3), the majority of Legionnaires' disease cases have been attributed to L. pneumophila serogroup 1 (8, 37, 47). Membrane protein profiles and pulsed-field gel electrophoresis of epidemic-associated serogroup 1 isolates of L. pneumophila result in very distinct patterns (24). Certain subgroups of L. pneumophila serogroup 1 are associated with human disease (21, 34, 56), but the genetic bases for these differences in virulence have not been determined.
Recently, two sets of L. pneumophila genes, designated dot (defective in organelle trafficking) and icm (intracellular multiplication), have been identified that are required for intracellular growth and trafficking (4, 7, 54). Interestingly, some of the proteins encoded by these loci contain significant sequence similarity to IncI plasmid ColIb-P9 Tra/Trb proteins involved in DNA transfer and type IV secretion (10, 55). In line with this similarity, it has been shown that these loci play a role in conjugation (52, 61). A second type IV secretion system, named lvh for Legionella vir homologues (53), has been found that is similar to the Agrobacterium tumefaciens virB locus. This type IV secretion system is crucial for delivery of DNA and proteins to plant cells by Agrobacterium, leading to the neoplastic disease crown gall (28, 29, 60, 67), and has been adapted by Bordetella pertussis for secretion of proteins involved in pathogenesis (65). The L. pneumophila lvh genes can substitute for some components of the dot/icm system in plasmid conjugation but not for intracellular growth (53). However, it has not been determined whether the lvh genes play a role in the virulence of L. pneumophila.
We have identified another locus in L. pneumophila strain AA100 involved in type IV secretion, designated tra1. We found that this locus is not present in two other strains of L. pneumophila serogroup 1 commonly used for molecular pathogenesis studies, JR32 and Lp01. In addition, we examined the distribution of this locus, lvh, rtxA, and enhC, in L. pneumophila laboratory strains and clinical isolates. The lvh and rtxA regions are found more frequently in strains associated with human disease, while the tra1 locus is not. The presence of loci that correlate with disease also correlates with several traits associated with virulence including adherence, entry, trafficking, and growth in mice. These studies suggest that important differences exist between L. pneumophila serogroup 1 strains, which may affect their ability to survive in the environment and cause disease.
MATERIALS AND METHODS
Strains and growth conditions.
L. pneumophila AA100 serogroup 1 is a naturally arising streptomycin-resistant mutant (41) of clinical isolate 130b, from the Wadsworth Veterans Administration Hospital in Los Angeles, Calif. (23).
L. pneumophila strain JR32 was a gift from Howard Shuman (Columbia University College of Physicians and Surgeons, New York, N.Y.). JR32 is a homogeneous salt-sensitive Philadelphia 1 (64) variant isolated from the streptomycin-resistant and restriction-deficient strain AM511 (36). L. pneumophila Philadelphia 1 and its derivative Lp01 were kindly provided by Ralph Isberg (Tufts University School of Medicine, Boston, Mass.) and Michele Swanson (University of Michigan Medical School, Ann Arbor). L. pneumophila Lp01 is a streptomycin-resistant variant and a presumptive restriction-minus mutant constructed by conjugative introduction of plasmid pAM40 into this strain and its subsequent curing (4). Legionella strains were grown either on buffered charcoal yeast extract (BCYE) agar in the standard manner (22) at 37°C in 5% CO2 or with shaking in buffered yeast extract broth and passaged no more than twice in the laboratory before use as described previously (18).
Cell lines and culture conditions.
HEp-2 cells (ATCC CCL23), established from a human epidermoid carcinoma, were grown at 37°C with 5% CO2 in RPMI 1640 plus 5% heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine. U-937 cells (ATCC CRL1593.2), a human monocyte/macrophage-like cell line, were maintained in RPMI 1640 plus 10% fetal bovine serum at 37°C. These cells were differentiated with 100 nM phorbol 12-myristate 13-acetate (PMA; Sigma) 48 h prior to use. Acanthamoeba castellanii cells (ATCC 30234), an environmental host for Legionella, were grown at room temperature in M712 broth in 75-cm2 tissue culture flasks, seeded in 24-well tissue culture dishes (Nunclon) at 2 × 105 cells/well for 24 h, washed once with high salt-medium before use, and incubated in this medium for 1 h at 37°C before infection (16, 42).
Entry and adherence assays.
Entry assays were carried out essentially as described previously (15, 16). U-937 cells were seeded in 24-well tissue culture dishes (Nunclon) at a concentration of 106 cells per well with 100 nM PMA and incubated for 48 h at 37°C in 5% CO2. HEp-2 cells were seeded in the same dishes at a concentration of 1.5 × 106 cells/well and allowed to adhere overnight at 37°C. Cell culture medium was replaced with fresh medium just prior to infection. BCYE agar-grown bacteria were suspended and diluted in the same medium as the cells that were to be infected. Cultures of buffered yeast extract-grown bacteria with an optical density at 600 nm of 0.2 to 0.5 (exponential) or 2.3 to 2.5 (postexponential) were diluted in the same manner. The bacteria were added and incubated with the cells for 30 min (for U-937 cells) and 90 min (for HEp-2 cells) at 37°C, washed with warm phosphate-buffered saline (PBS), and incubated in the appropriate culture medium plus 100 μg of gentamicin/ml for 2 h. The cells were then washed twice with PBS and lysed in sterile water. The number of intracellular bacteria was determined by plating for CFU on BCYE. Adherence assays with HEp-2 cells were performed in a similar manner. After adding bacteria to the monolayer, the cells were washed five times and lysed with sterile water. Entry and adherence levels were calculated as % entry = 100 × (CFU of gentamicin-resistant bacteria/CFU of inoculum) and % adherence = 100 × (CFU of cell-associated bacteria/CFU of inoculum). To correct for variation between experiments, entry and adherence are reported relative to L. pneumophila strain AA100, i.e., relative entry = 100 × (% entry of test strain/% entry of AA100).
Intracellular growth assays.
The growth kinetics of L. pneumophila in U-937 cells and A. castellanii were determined as described previously (15, 16). Briefly, bacteria were added to a monolayer of 106 cells/well of U-937 cells or A. castellanii in 24-well tissue culture dishes at a multiplicity of infection of 0.1 or 1, respectively, and incubated for 1 h at 37°C. Extracellular bacteria were killed by gentamicin treatment for 2 h, and the cells were washed twice and incubated in fresh medium at 37°C for various times before lysis in water. Survival is expressed as the portion of CFU present at each time compared to that at time zero, T0 = 1 h of infection plus 2 h of gentamicin treatment, i.e., mean CFU Tx/T0.
Mouse infections.
The virulence of different L. pneumophila strains was examined in A/J and C57BL/6J mice as described previously (15, 17). C57BL/6J and A/J mice were infected by intratracheal or intranasal inoculation with 107 bacteria suspended in PBS. At various time points the lungs were harvested and bacteria in the lungs were quantitated as described previously (5, 15). Data presented are the means and standard deviations of bacterial counts (CFU per gram of lung) for 12 mice in each experimental group.
DNA manipulations.
Chromosomal DNA preparation, isolation of plasmid DNA, and cloning techniques were performed as previously described (17, 51). Escherichia coli XL1-Blue (Stratagene) and ψec47 (18) were used for propagation of plasmids. When necessary, kanamycin was added at 25 μg/ml to bacterial media.
PCR analysis.
The primers for PCR analysis were designed based on the DNA sequence from L. pneumophila strain AA100 and are summarized in Table 1. After denaturation of the bacterial chromosomal DNA template at 94°C for 3 min, 30 cycles of PCR amplification were performed. In each reaction 0.1 μM (each) primer, 5 mM deoxynucleotide triphosphate mix, 1× PCR buffer, 25 mM MgCl2, and 2.5 U of Taq DNA polymerase (Boehringer Mannheim-La Roche) were used.
TABLE 1.
Oligonucleotides
| Name/regiona | Sequence (5′→3′) |
|---|---|
| O1/traH upstream | CGAAGGGTATCAACCCCGGT |
| O2/traH upstream | TGCGGTCCCAGAAGTTAGCC |
| O3/traH upstream | CCATAGCTATTAGCCGCATC |
| O4/traH | ATGAGACTATTGGAGCAGCC |
| O4R/traH | GGCTGCTCCAATAGTCTCAT |
| O5/traI | CAACCAATCAGCCCAGGTTT |
| O5R/traI | AAACCTGGGCTGATTGGTTG |
| O6/traI | ACCATCCTACTCAGAGGGCG |
| O7/traJ upstream | CTTTACCCACTCGCTCCTGT |
| O8/traJ | AAGCTTAAAACACCCACCAG |
| O9/traK | GTAAAGCCCCGAATTTCGG |
| O10/traM | AATGCGGCATTAGCTAGTAG |
| O11/traM downstream | CCAGAGCTTAGTTGGACATT |
| lvh1/prpA | GTTTTAATCCCCCAGCAAGC |
| lvh2/prpA | AATATCCCTACTCATCCTCG |
| lvh3/lvhB3 | GGCTAGGAGGTTCTTGTG |
| lvh4/lvhB4 | ATTGGCCGAGATGTCCTT |
| lvh5/lvhB8 | CCTCTACGCATTACAACGCC |
| lvh6/lvhB9 | GTGGTGGTAAAGGGAATGCC |
| lvr1/lvrE | GGTCCAATGGGTCCAGCAGG |
| lvr2/lvrE | AGTGGCTGATTCTGGAGTGG |
| enh1/enhC | AATGCTTTGTATGCCCTCGG |
| enh2/enhC | CATATCAGCGCTTTGGCCATC |
| rtx1/rtxA | GATCCGCAAGTAGCGCTCAC |
| rtx2/rtxA | TGTAATGCTGGCATTAGGCG |
| rtx3/rtxA | CTGATGCTGCTACGGAACAC |
| rtx4/rtxA | CCGCAGTCATTACACCTGCG |
Each oligonucleotide was given a designation as shown in Fig. 1. Region corresponds to the gene that the oligonucleotide is located within, 5′ of (upstream) or 3′ of (downstream).
Southern analysis and colony hybridization.
Bacterial chromosomal DNA was cut with EcoRI or PstI and electrophoresed on a 0.8% agarose gel. After electrophoresis, the DNA was transferred to a nylon membrane. Probes were labeled by PCR with digoxigenin, and subsequent steps for both colony hybridization and Southern analysis were carried out by the manufacturer's methods described for the Genius System (Boehringer Mannheim-La Roche). Oligonucleotides used to make the probes are indicated in Table 1. Hybridizations were carried out at low or high stringency (51).
DNA sequence analysis.
DNA sequence analysis was carried out initially using a forward primer, lac1, GGCACGACAGGTTTCCCGAC, from the plasmid pWKS130 (62). The sequence was continued by primer walking directly on a cosmid that carries the region of interest. These cosmids were isolated by colony hybridization of an L. pneumophila AA100 total genomic DNA library described previously (18). All regions described were sequenced completely on both strands using Big Dye Terminator (Applied Biosystems) cycle sequencing and subsequent analysis on an ABI 310 automated sequencing apparatus (Applied Biosystems). Sequence analysis and assembly were carried out using Gene Construction Kit 2 (Textco) and comparison with known sequences using Blast (1). Sequence alignment was done by using Lasergene (DNASTAR II) software.
Phagosome-lysosome fusion.
For quantitation of the frequency of lysosome fusion, U-937 cells were seeded on coverslips as described previously (17), infected for 15 min at 37°C as described for entry assays, washed three times with PBS, and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After fixation the bacteria were detected with polyclonal anti-L. pneumophila rabbit antisera (16) and a secondary anti-rabbit Cy2-conjugated antibody (Amersham Pharmacia Biotech), and lysosome-associated membrane protein 1 (LAMP-1) was detected with anti-human LAMP-1 monoclonal antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City) and a secondary anti-mouse Cy5-conjugated antibody (Amersham Pharmacia Biotech). All antibody incubations and washes were carried out as described previously (16). Dual images of labeled coverslips were captured on a Bio-Rad MRC1024ES confocal microscope and analyzed using Adobe Photoshop.
Statistical analysis.
All in vitro experiments were carried out in triplicate and repeated at least three times. The in vivo experiments were carried out with 12 mice per experimental group. The significance of the results was determined by analysis of variance. P values of <0.05 were considered significant.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study have been deposited in GenBank with accession numbers AF410854 (lvh region) and AY053454 (tra1 region).
RESULTS
Identification of new genes involved in type IV secretion.
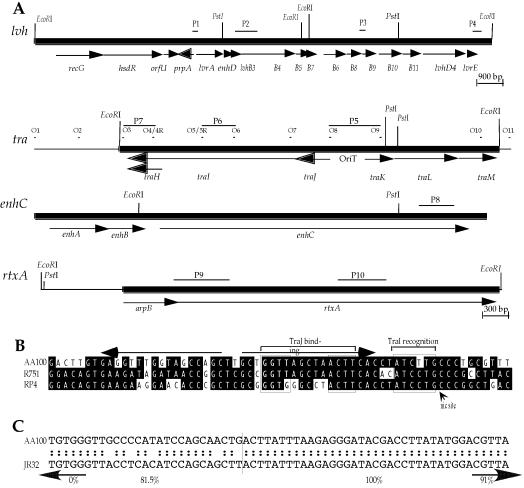
We have recently identified several loci that confer enhanced entry on wild-type L. pneumophila strain AA100 (18). Adjacent to one of these regions, a locus was identified that is predicted to encode proteins similar to components of a type IV secretion system and was designated tra1. Since the original construct is noncontiguous, we examined whether the tra1 and entry locus are contiguous on the L. pneumophila chromosome. PCR analyses in these regions demonstrated that tra1 and the entry locus are on different noncontiguous L. pneumophila chromosomal fragments, separated by a Sau3AI site (data not shown). A contiguous cosmid containing the L. pneumophila tra1 locus was isolated by colony hybridization of a total genomic DNA library from strain AA100 (18) and used for sequence analysis of the entire locus. We determined the nucleotide sequence of both strands of a 4,949-bp EcoRI fragment from this cosmid containing the tra1 region (Fig. 1). The guanine-plus-cytosine content of this fragment is only slightly lower (37.6%) than that of the L. pneumophila chromosome (39%). Six putative open reading frames (ORFs) were identified with similarity to TraHIJKLM proteins of R751 and RP4 plasmids of Enterobacter aerogenes and E. coli, respectively (Table 2). Both of these plasmids belong to the IncP incompatibility group (43, 59). These genes are organized as two putative operons that appear to be divergently transcribed from a 398-bp intergenic region where a potential origin of transfer (OriT) is located (Fig. 1B). A similar divergently transcribed organization is found in IncP plasmids (43, 59, 68). The nick region of the putative OriT is highly conserved and conforms to the consensus YAWCYTG, commonly found in IncP plasmids, some phage, and origins that replicate by a rolling-circle mechanism (43). We also identified an imperfect 19-bp inverted repeat sequence (Fig. 1B) that is thought to be recognized by TraJ, allowing initiation of the DNA transfer process during conjugation (43).
FIG. 1.
(A) Genetic organization of putative virulence loci in L. pneumophila strain AA100. Arrows indicate coding regions and the direction of transcription. Probes (P1 to P10) and oligonucleotides (O1 to O11) used in Southern and PCR analyses, respectively, are shown above their binding sites. The scale for the lvh locus is shown below its map, and the scale for the other regions is below rtxA. (B) Alignment of the L. pneumophila tra1 locus with the OriT regions from the R751 and RP4 plasmids. The conserved binding regions for TraJ and TraI are indicated by boxes, and the nick region is indicated with a labeled arrow. (C) Alignment of L. pneumophila strain AA100 and JR32 lvh regions at the DNA level beginning 42 bp downstream from the putative translational start of lvrA. The percent identities of the AA100 and JR32 sequences for the left and right (separated by the vertical line) regions are indicated below the sequence. Arrows indicate the percent identities of the AA100 and JR32 sequences for the upstream and downstream regions out to the flanking EcoRI sites of lvh shown in panel A.
TABLE 2.
Similarity between L. pneumophila Tra and Inc plasmids
| Protein | Plasmid | % Ident/Sima | No. of aa | Functionb | Accession no.c |
|---|---|---|---|---|---|
| TraH | R751d | 62/81 | 68 | Relaxosome stability | S22993 |
| RP4e | 58/78 | S23000 | |||
| TraI | R751 | 38/58 | 480 | DNA relaxase | S22994 |
| RP4 | 40/60 | S23001 | |||
| ColI-P9f | 25/42 | AB021078 | |||
| TraJ | R751 | 39/55 | 114 | OriT binding | S22996 |
| RP4 | 37/52 | A34172 | |||
| TraK | R751 | 33/51 | 104 | OriT binding | S22997 |
| RP4 | 32/50 | S23003 | |||
| TraL | R751 | 70/80 | 241 | ATP/GTP binding | T08536 |
| RP4 | 71/82 | S23003 | |||
| TraM | R751 | 36/68 | 141 | Regulation | T08537 |
| RP4 | 38/69 | S23005 |
% Ident/Sim is the percent identity/similarity between the L. pneumophila protein and its homologue.
Putative function as predicted from similarity.
Accession number for the protein used.
IncPβ plasmid.
IncPα plasmid.
IncI plasmid.
We recently identified another type IV secretion locus during the isolation of a gene, designated enhD, involved in entry by L. pneumophila strain AA100 (D. A. Ridenour et al., unpublished observations). This type IV secretion locus has also been identified by another laboratory through sequence analysis of the L. pneumophila strain JR32 genome and designated lvh (53). The L. pneumophila lvh locus contains 11 genes that encode homologues of the A. tumefaciens Vir virulence proteins, designated lvhB2-B11 and lvhD4 (53). Upstream of these genes are three genes, lvrA, lvrB, and lvrC, that do not encode proteins with similarity to the Vir proteins, and it has been proposed that they play a role in regulation of type IV secretion. Most of the lvh locus from L. pneumophila strain AA100 is nearly identical to that previously sequenced from strain JR32. However, no similarity exists at the nucleotide level between these two L. pneumophila strains in the upstream region, beginning within lvrA (Fig. 1C). The first putative ORF upstream of lvrA in strain JR32, which would be transcribed in the opposite direction, is similar to lambda repressor cII (53) and the putative cI repressor from the E. coli O157:H7 prophage CP-933V (27% identity and 40% similarity; accession no. AE005443) at the amino acid level over the entire protein. In the case of strain AA100, a similar ORF exists and is most similar to the JR32 ORF (35% identity and 56% similarity) at the amino acid level with no similarity at the nucleotide level. Interestingly, the AA100 ORF is more similar to the phage P22 repressor protein C2 (27% identity and 42% similarity; accession no. V01153) than it is to the cI repressor from the E. coli O157:H7 prophage CP-933V (24% identity and 40% similarity). Based on these similarities, we have designated this ORF prpA, for putative phage repressor. These observations suggest that the sequence divergence between strains AA100 and JR32 observed within and upstream of lvrA is due to a recombination event between two related phage. Three additional putative ORFs were identified near prpA that encode proteins with similarity to OrfU of Erwinia amylovora (44% identity and 71% similarity over 185 amino acids [aa]; accession no. AF083877_5), HsdR of Helicobacter pylori (29% identity and 47% similarity over 947 aa; accession no. AE001565), and RecG-related protein of Deinococcus radiodurans (27% identity and 39% similarity over 350 aa; accession no. AE002053_3).
Presence of putative virulence loci in different L. pneumophila strains.
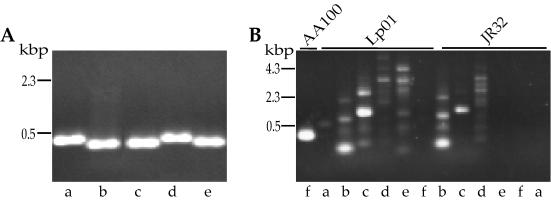
Since differences were observed in the lvh region of L. pneumophila strains AA100 and JR32, we investigated whether other genetic differences exist that may be related to pathogenesis. We first examined the presence of the tra1 locus in AA100, JR32, and Lp01 by PCR analyses using six different sets of oligonucleotides that produce a specific product in strain AA100 (Fig. 2). These oligonucleotides were tested under different annealing temperatures and template concentrations, but only those within traHI (O4R and O5) result in an apparently specific single band for JR32. The potentially specific band obtained with JR32 is 100 bp larger than that obtained with strain AA100, suggesting that it may be nonspecific. In order to test this possibility we sequenced the product and found that it contains at least two and possibly more sequences, resulting in multiple overlapping bands in sequencing gels (data not shown). Thus, none of the oligonucleotide pairs within the tra1 locus produce a specific product in JR32 or Lp01 by PCR. These data suggest that the entire tra1 locus is not present in either JR32 or Lp01.
FIG. 2.
A representative sample of the PCR analyses used to compare L. pneumophila strain AA100 (panel A for oligonucleotide pairs a to e and panel B for pair f) with JR32 and Lp01. Oligonucleotides were designed for PCR within the AA100 tra locus and used under the same conditions for all three strains. Oligonucleotide pairs and their locations are as follows: a, O1 and O2, downstream of traI; b, O3 and O4, carboxy terminus of traHI; c, O4R and O5, within traHI; d, O5R and O6, within traI; e, O7 and O8, spanning traIJ; and f, O10 and O11, spanning the carboxy terminus of traM.
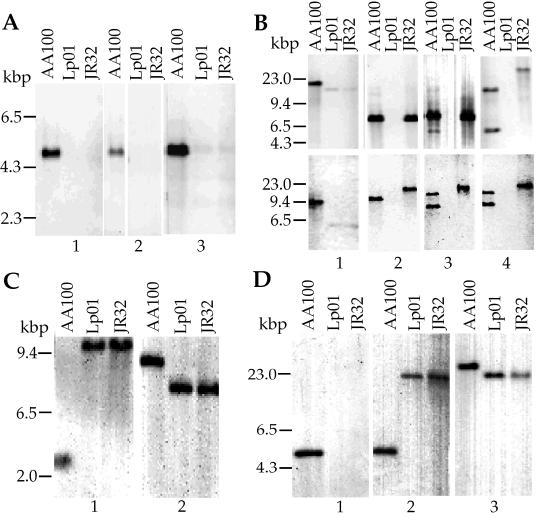
In order to confirm the absence of the tra1 locus in these strains, we carried out Southern analysis with probes specific for this region from strain AA100. In addition, we examined differences in the lvh, rtxA, and enhC loci in AA100, JR32, and Lp01 by Southern analysis (Fig. 3). Under low-stringency conditions, all tra1 probes hybridized to a single DNA fragment in AA100 but did not hybridize to DNA from JR32 or Lp01. Both Southern and PCR analyses indicate that the tra1 locus is absent from JR32 and Lp01. As expected from sequence analyses, JR32 has the lvh locus by Southern analyses. In addition, the probe just upstream of lvrA, P1, does not hybridize to JR32 even at low stringency, confirming the sequence divergence from AA100 in this region. Interestingly, DNA from strain Lp01 does not hybridize to any of the four probes from within the lvh locus of AA100, indicating that this locus is also absent. This is a particularly surprising observation since Lp01 and JR32 are thought to be derived from the same epidemic and thus would be expected to be very closely related strains. The passage histories of strains Lp01 and JR32 are different and may have resulted in the acquisition of some genetic differences. This is particularly important since both strains were separately selected for streptomycin resistance and restriction-minus phenotypes (4, 36). We examined the parental strain of Lp01 for the presence of the lvh and tra1 loci by PCR. Although the lvh locus was present in this strain, the tra1 locus was not (data not shown). Possibly, the lvh locus was lost from the Lp01 parental strain during the selection for a restriction-minus phenotype, since the upstream region appears to carry hsdR and potentially other components of a restriction system. Interestingly, AA100 displays two bands that hybridize to the probes P3 and P4 from the downstream genes, lvhB9 and lvrE of the lvh region. This observation suggests that there may be at least some components of another type IV secretion system in AA100. This second band is not observed in JR32 or Lp01. The enhC and rtxA loci that have been associated with entry and virulence of L. pneumophila (17, 18) are present in both JR32 and Lp01. However, the amino-terminal probe for rtxA does not hybridize to either JR32 or Lp01, suggesting that there is significant sequence divergence in the amino terminus of this gene, similar to that observed within lvrA of the lvh locus. Thus, the lvh, enhC, and rtxA loci are present in the three L. pneumophila strains examined, though the parental strain of Lp01 appears to have lost lvh during manipulation in the laboratory. In contrast, the tra1 locus is present only in strain AA100, suggesting that it is not required for pathogenesis.
FIG. 3.
Southern analysis of the L. pneumophila tra1 (A), lvh (B), enhC (C), and rtxA (D) loci in strains AA100, JR32, and Lp01. Total bacterial DNA was digested with either EcoRI (A; B, bottom; C, lane 2; and D, lanes 1 and 2) or PstI (B, top; C, lane 1; and D, lane 3) and hybridized with probes against tra1 (P5 [A1], P6 [A2], and P7 [A3]), lvh (P1 [B1], P2 [B2], P3 [B3], and P4 [B4]), enhC (P8 [C]), and rtxA (P9 [D1] and P10 [D2 and D3]). The positions of probes within each locus are shown in Fig. 1. In panel A2 irrelevant lanes were removed and the gap between strains AA100 and Lp01 was closed.
Correlation of putative virulence loci with disease.
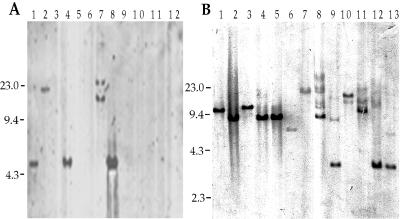
In order to further examine the importance of these loci in the production of disease by Legionella, we examined the presence of these loci in a number of clinical and environmental isolates. We carried out Southern analyses using the conserved regions of the tra1, lvh, rtxA, and enhC loci. Examples of some of the data obtained are shown in Fig. 4, and the complete data are summarized in Table 3. There was no correlation between the presence of the tra1 locus and the ability to cause disease, since 70% of the disease-associated isolates do not carry this locus. The enhC locus was present in all Legionella isolates, irrespective of their association with disease, suggesting that it is essential for the ability of the bacteria to survive and/or replicate in the environment. Since we have previously constructed an L. pneumophila strain with an in-frame deletion in this gene, it is not essential under laboratory growth conditions (18). Both the presence of the lvh locus and that of the rtxA locus correlate with the ability to cause disease in humans (P < 0.01). Since the genetic differences that we observed among AA100, JR32, and Lp01 may be related to their ability to cause disease, we examined these strains for differences in their interactions with host cells.
FIG. 4.
Southern analysis of the tra1 (A) and enhC (B) loci in environmental and clinical isolates of Legionella. Total bacterial DNA was digested with EcoRI and hybridized with the probes P5 against tra1 (A) and P8 against enhC (B). Minor bands in some lanes are thought to be due to partial digestion but could be due to sequences distantly related to the probe present in these strains. The strains shown are L. pneumophila AA100 (A, lane 1), L. pneumophila Allentown (A, lane 2, and B, lane 1), L. pneumophila Camperdown (A, lane 3, and B, lane 2), L. pneumophila Benidorm (A, lane 4, and B, lane 3), L. pneumophila Olda (A, lane 5, and B, lane 4), L. pneumophila Chicago 8 (A, lane 6, and B, lane 5), L. anisa (A, lane 7, and B, lane 6), L. dumoffii (A, lane 8, and B, lane 12), L. feeleii (A, lane 9, and B, lane 8), L. longbeachae (A, lane 10, and B, lane 9), L. gormanii (A, lane 11, and B, lane 10), L. pneumophila Philadelphia 1 Lp01 (A, lane 12), L. brunensi (B, lane 7), L. moravica (B, lane 11), and L. sainthelensi (B, lane 13). Numbers to the left of each panel are molecular sizes in kilobase pairs.
TABLE 3.
Presence of loci in different Legionella strains
| Strain | Diseasea | Locusb
|
Source | |||
|---|---|---|---|---|---|---|
| lvh | tra | rtxA | enhC | |||
| L. pneumophila 130b AA100 sg 1 | + | + | + | + | + | ATCC BAA-74 |
| L. pneumophila Philadelphia 1 JR32 sg 1 | + | + | − | + | + | H. Shumand |
| L. pneumophila Philadelphia 1 Lp01 sg 1 | + | −c | − | + | + | R. Isberge/M. Swansonf |
| L. pneumophila Allentown 1 sg 1 | + | + | + | + | + | ATCC 43106 |
| L. pneumophila Camperdown sg 1 | + | + | − | + | + | ATCC 43113 |
| L. pneumophila Benidorm 03E sg1 | + | + | + | + | + | ATCC 43108 |
| L. pneumophila Olda sg 1 | + | + | − | + | + | ATCC 43109 |
| L. pneumophila Chicago 8 sg 7 | + | + | − | + | + | ATCC 33823 |
| L. pneumophila Heysham 1 sg 1 | − | − | − | ND | + | ATCC 43107 |
| L. anisa | − | − | + | − | + | ATCC 35290 |
| L. dumoffii | − | − | + | − | + | ATCC 33279 |
| L. feelii | + | + | − | + | + | ATCC 35072 |
| L. longbeachae | + | + | − | − | + | ATCC 33484 |
| L. gormanii | − | + | − | − | + | ATCC 43769 |
| L. sainthelensi | − | − | ND | − | + | ATCC 35248 |
| L. moravica | − | ND | ND | − | + | ATCC 43877 |
| L. brunensis | − | − | ND | − | + | ATCC 43878 |
| Legionella genomospecies I | − | − | ND | ND | + | ATCC 51913 |
| L. waltersii | − | − | ND | ND | + | ATCC 51914 |
Associated (+) or not (−) with disease in humans.
+, presence of the locus; −, absence of the locus; ND, not determined.
In contrast to Lp01, the parental strain of Lp01 was positive for lvh and negative for tra loci by PCR.
Columbia University College of Physicians and Surgeons.
Tufts University School of Medicine.
University of Michigan Medical School.
Differences in adherence and entry.
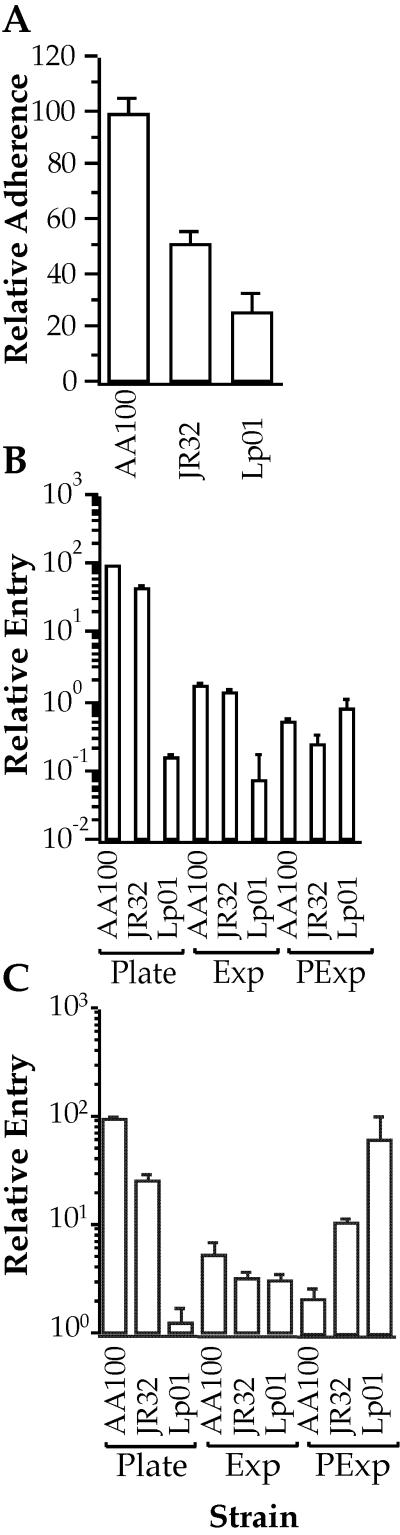
We have previously found that the mechanisms of L. pneumophila adherence and entry correlate with virulence in animal models (15, 17). In addition, studies indicate that the abilities of Lp01 to enter host cells and express other virulence-associated characteristics depend upon its stage of growth in liquid medium (11). Thus, we examined the adherence and entry of AA100, JR32, and Lp01 strains to monocytes and epithelial cells and the effects of the stage of growth in liquid medium on their entry phenotype (Fig. 5). Adherence to epithelial cells by plate-grown bacteria is lower with JR32 and Lp01 than with AA100 (P < 0.01), though the levels of adherence are lower for Lp01 than for JR32 (P < 0.01). Entry into both monocytes and epithelial cells is lower for JR32 and Lp01 than for AA100 when grown on solid media (P < 0.01). In addition, both JR32 and Lp01 enter monocytes and epithelial cells at either the same levels as or lower levels than that of AA100 when grown in liquid media, with the exception of post-exponential-phase bacteria into monocytes, where both strains enter at higher levels (P < 0.01) than that of AA100. However, even under this growth condition, where the highest levels of entry are obtained for Lp01, they are similar to those seen for plate-grown AA100. Interestingly, entry into epithelial cells and monocytes by AA100 is not enhanced but reduced (P < 0.03) by post-exponential growth in liquid media, and entry of JR32 is not significantly affected by growth phase in epithelial cells but is enhanced in monocytes. Although these observations confirm results from previous studies regarding the effects of growth phase on entry of strain Lp01 (11), they suggest that there are significant differences in the efficiency and potentially the mechanisms of adherence and entry by different L. pneumophila serogroup 1 strains, JR32 and AA100.
FIG. 5.
Adherence to epithelial cells (A) and entry into epithelial cells (B) and monocyte-derived macrophages (C) by L. pneumophila strains AA100, JR32, and Lp01 grown on BCYE agar plates (A to C) or in liquid media (B and C) to exponential (Exp) or post-exponential (PExp) phase. Adherence and entry by plate-grown strain AA100 were arbitrarily set to 100. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
Intracellular survival and replication.
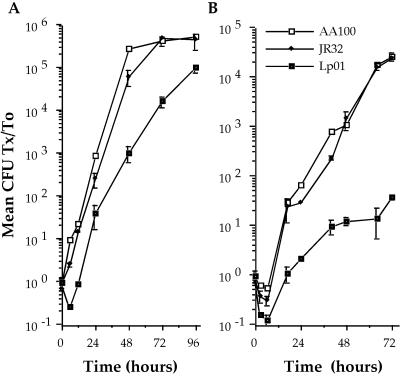
Since differences in the mechanisms of L. pneumophila adherence and entry correlate with the ability to survive and replicate in host cells (15, 17), we examined growth of AA100, JR32, and Lp01 in monocytes and amoebae. We found that AA100 and JR32 survive and replicate equally well in both monocytes and amoebae, whereas Lp01 is both killed more readily at early time points and replicates more slowly (P < 0.01) in both cell types (Fig. 6). All three strains are killed more efficiently by amoebae than by monocytes during early time points, suggesting that amoebae are more bactericidal for L. pneumophila than are human monocytes. Interestingly, Lp01 exhibits only a 12-fold increase in CFU over 72 h in amoebae compared with the typically greater-than-10,000-fold growth of AA100 and JR32. These data indicate that, while the genetic differences between AA100 and JR32 affect adherence and entry, they do not affect intracellular survival and replication in vitro, whereas these or potentially other genetic differences significantly impact the intracellular survival and replication of strain Lp01.
FIG. 6.
Intracellular survival and replication of L. pneumophila strains AA100, JR32, and Lp01 in human monocyte-derived macrophages (A) and the amoeba A. castellanii (B). Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate. Results shown are for a typical experiment.
Intracellular trafficking.
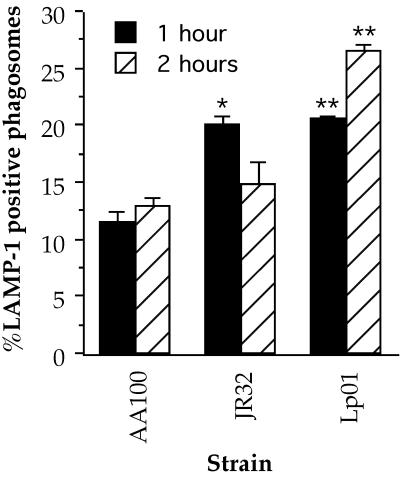
Since inhibition of lysosome fusion with the bacterial phagosome is thought to be important for survival of L. pneumophila (32), it is possible that the differences that we observed in intracellular survival are related to trafficking. The presence of LAMP-1, a membrane glycoprotein found in lysosomes and late endosomes, in phagosomes has been widely used to indicate fusion with lysosomes (12, 46, 50, 58). Thus, we compared the frequencies of colocalization of this lysosomal marker with the bacterial phagosome in cells infected with AA100, JR32, and Lp01 (Fig. 7). At the early time point, 1 h after addition of the bacteria, both JR32 and Lp01 display slightly higher frequencies of lysosomal fusion than that of strain AA100. Interestingly, whereas the frequency of lysosomal fusion for JR32 is similar to that of AA100 at 2 h, the frequency of fusion for Lp01 is even greater than that at 1 h and more than twofold that of AA100 at the same time point. These data indicate that the differences in intracellular survival observed for strain Lp01 may, at least in part, be related to differences in the ability to inhibit lysosomal fusion. The higher frequencies of fusion for strain JR32 only at early time points may be responsible for at least a portion of the differences measured for entry, since the bacteria must survive intracellularly in order to be quantitated with CFU-based assays. It would be interesting to determine whether these differences in trafficking are the result of the mechanism of entry used or a separate defect that affects events immediately after uptake.
FIG. 7.
Fusion of the L. pneumophila phagosome with lysosomes in human monocyte-derived macrophages as measured by colocalization of the lysosomal marker LAMP-1 with the bacterial vacuole. The single and double asterisks indicate data points that are significantly different (P < 0.02 and P < 0.01, respectively) from those for AA100 at the same time point. Data points and error bars represent the means and standard deviations, respectively, of three separate counts of 30 phagosomes. Results shown are for a typical experiment.
Differences in survival and replication in mice.
In order to test whether the genetic and phenotypic differences observed in vitro correlate with differences in the ability to survive and replicate in vivo, we examined the ability of AA100, JR32, and Lp01 to infect mice. We used A/J mice, which are thought to be susceptible to infection by L. pneumophila (9), and C57BL/6J mice, which are thought to be relatively resistant to infection with L. pneumophila (66), for these studies. Although the initial numbers of bacteria are similar for all three bacterial strains in both A/J and C57BL/6J, Lp01 does not replicate in mouse lungs as well as does AA100 or JR32 after either intranasal or intratracheal inoculation (Table 4). Interestingly, JR32 replicates in a manner similar to that of AA100 when inoculated intranasally but does not replicate as well after intratracheal inoculation. This difference may be at least partially due to differences in the bacterial load, since the numbers of initial CFU in the lung are 100-fold lower after intranasal inoculation than after intratracheal inoculation. Overall, these data indicate differences in the growth of these strains in mice that correlate well with the genetic and phenotypic variations observed.
TABLE 4.
Replication of different L. pneumophila strains in mouse lungs
| Route of inoculation,a mouse strain, and experimental groupc | No. of bacteria in lungs (CFU/g)b after:
|
||
|---|---|---|---|
| 4 h | 1 day | 2 days | |
| Intranasal | |||
| C57BL/6J | |||
| AA100 | (6.2 ± 0.4) × 103 | (8.2 ± 0.2) × 103 | (1.3 ± 0.2) × 104d |
| JR32 | (5.1 ± 0.4) × 103 | (7.1 ± 0.3) × 103 | (9.5 ± 0.4) × 103d |
| Lp01 | (4.8 ± 0.4) × 103 | (5.5 ± 0.3) × 103 e | (3.4 ± 0.2) × 103e |
| A/J | |||
| AA100 | (3.9 ± 0.3) × 103 | (5.3 ± 0.5) × 103 | (1.2 ± 0.2) × 104d |
| JR32 | (3.5 ± 0.4) × 103 | (7.1 ± 0.2) × 103 | (9.8 ± 0.3) × 103d |
| Lp01 | (1.1 ± 0.3) × 103 | (3.3 ± 0.4) × 103 e | (5.1 ± 0.3) × 103d,e |
| Intratracheal | |||
| C57BL/6J | |||
| AA100 | (5.6 ± 0.2) × 105 | (6.1 ± 0.4) × 106 d | (9.2 ± 0.2) × 106d |
| JR32 | (2.3 ± 0.3) × 105 | (3.1 ± 0.2) × 105 e | (6.5 ± 0.4) × 105d,e |
| Lp01 | (4.3 ± 0.3) × 105 | (6.7 ± 0.2) × 105 e | (3.3 ± 0.4) × 105e |
| A/J | |||
| AA100 | (3.5 ± 0.4) × 105 | (7.1 ± 0.2) × 105 d | (5.3 ± 0.3) × 106d |
| JR32 | (1.3 ± 0.3) × 105 | (6.3 ± 0.4) × 105 d | (2.4 ± 0.2) × 106d,e |
| Lp01 | (2.6 ± 0.2) × 105 | (4.7 ± 0.4) × 105 e | (7.2 ± 0.3) × 105d,e |
An inoculum of 107 bacteria was used for all experimental groups.
Data represent the means ± standard deviations of duplicate platings from all mice in each group.
There are 12 mice in each experimental group.
Significantly different (P < 0.05) from the number of bacteria at 4 h.
Significantly different (P < 0.05) from strain AA100.
DISCUSSION
Pathogenesis involves the ability to interact with host cells and tissues in a manner that allows avoidance of host defenses, survival, and replication (26, 27). Many of the bacterial factors involved in this process are not required and often not expressed in other growth environments including laboratory media (20, 39, 40). Since they are not required, mutations in these genes can accumulate or the genes may be lost upon passage in the laboratory. This phenomenon is particularly problematic in the case of L. pneumophila, where multiple passage on laboratory media results in the complete loss of virulence (25, 38). Thus, it is critical that the passage history of the L. pneumophila strains involved be well understood when analyzing data obtained from pathogenesis studies. In the present study, we examined genetic differences among three commonly studied L. pneumophila strains, AA100, JR32, and Lp01. In addition to differences at the genetic level, these strains also differ in a number of phenotypic characteristics that are likely to be important for their ability to cause disease. This is the first description of any differences among these important laboratory strains, which may have been considered nearly identical isolates of L. pneumophila serogroup 1. These data indicate that observations made with a particular strain of L. pneumophila may not be directly applicable to all other strains. Therefore, it is important to compare multiple strains of L. pneumophila prior to making concrete conclusions regarding the more subtle aspects of pathogenesis and gene regulation.
One of the loci present in L. pneumophila strain AA100 but absent from JR32 and Lp01 is the tra1 locus, which contains genes involved in type IV secretion. Type IV secretion systems are built from core components of conjugation machinery (14). The tra1 locus is a new example to add to the growing list of these components in L. pneumophila and other bacteria (13, 14, 19). Interestingly, we found that this locus is present in only approximately 30% of the Legionella strains examined, and there is no significant correlation with the ability to cause disease. These observations may indicate that the tra1 locus is not required for pathogenesis. However, an E. coli mutant with a mutation in traJ, an F+ plasmid regulatory gene, is less invasive and less able to cause meningitis in the neonatal rat than is the wild type (2). In addition, an L. pneumophila mutant with a mutation in traA, involved in DNA processing during conjugation, does not replicate in amoebae and macrophages (44). Since this and other loci involved in type IV secretion affect L. pneumophila virulence (45, 52, 61), examination of a larger number of Legionella strains for the presence of this locus as well as evaluation of the effects of a specific tra1 mutant are mandated to clarify its role, if any, in pathogenesis.
The L. pneumophila lvh locus is present primarily in strains associated with disease, which suggests a role in pathogenesis that could be related to the observed differences in levels of virulence. This locus is present in AA100 and JR32 but absent from Lp01; thus, the loss of this locus may account for at least a portion of the reduced virulence-associated phenotypes observed with Lp01. Our data suggest that the lvh locus may have been lost from this strain during selection for a restriction-minus phenotype. This hypothesis could be tested by transforming Lp01 with this locus and evaluating the resulting strain's restriction and virulence phenotypes. Since strain Lp01 inhibits lysosomal fusion even in the absence of the lvh locus (35, 50, 57), though to a somewhat lesser extent than does strain AA100, other bacterial factors are most likely involved in the prevention of lysosomal fusion. Since type IV secretion systems are thought to be involved in the proper trafficking of L. pneumophila (14, 35, 50), Lp01 offers the opportunity to examine the factors involved in the absence of background inhibition of phagolysosome fusion that might result from the presence of two type IV secretion systems. It is unclear exactly why the upstream region of the lvh locus is different in AA100 than in JR32 and whether this difference affects virulence. It appears that a recombination event within the lvrA genes of two related phage produced this variation, but the time frame for this event and its importance in virulence are unclear without the characterization of this region in a number of different strains. Insight into how this region is acquired by L. pneumophila may be gained by the identification of the other recombinational junction and the site(s) for integration of this putative prophage. In addition, analysis of mutations in the other genes carried by this putative phage may be worthwhile, since this region appears to correspond to at least a portion of a potentially transferable pathogenicity island (6, 30, 31), which often contains a number of important virulence determinants.
The rtxA and enhC loci are present in all three L. pneumophila strains examined, but only the rtxA locus correlates with the ability to cause disease in humans. These data fit well with our observations that this locus is important for entry into host cells and virulence in mice (17, 18). Variability in the amino-terminal portion of rtxA was observed, but the nine amino acid repeats associated with host cell binding and activity of RTX proteins (63) are present in the carboxy-terminal region and conserved in all three strains. Although the enhC gene has been shown elsewhere to play a role in entry into host cells (18), its exact function in uptake remains unclear. The presence of this gene in all Legionella strains, regardless of their association with human disease, suggests that this locus is important for the ability of these bacteria to survive and/or replicate in the environment. Possibly, this gene is required for proper entry into and survival within protozoa, which are thought to be the primary hosts for Legionella in the environment. This concept implies that loci that correlate with disease in humans may not be absolutely necessary for survival and replication in protozoa but increase disease symptoms in humans sufficiently to cause infections to present clinically, rather than allowing natural recovery in the absence of intervention.
In conclusion, these studies report genetic variability in three important L. pneumophila serogroup 1 strains, AA100, JR32, and Lp01, commonly used for genetic analysis of pathogenesis. In addition, we report significant differences in several virulence-associated phenotypes in these strains. Although all three strains have the ability to adhere to, enter into, inhibit lysosomal fusion in, and replicate within monocytes as well as replicate in both resistant and susceptible mouse strains, they vary in the degree of their ability to accomplish each step in the infection process. Furthermore, the regulation of entry into host cells under different growth conditions also differs between these strains. These differences should be taken into consideration when analyzing data generated from different laboratories and may have important implications in interpretation of the results obtained. Total genomic analysis of each of these strains should lead to identification of the molecular basis for these differences and is likely to greatly improve our understanding of Legionella pathogenesis.
Acknowledgments
We thank Ralph Isberg for his insightful suggestion that Lp01 may have lost the lvh locus during selection for a restriction-minus strain, resulting in the observation that the lvh locus is present in the Lp01 parental strain. We also thank Joseph Vogel for sharing his observation that tra1 is absent from strain Lp01 with us, resulting in the initiation of this study.
This work was supported by grant AI40165 from the National Institutes of Health.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J. L., C. A. Wass, S. J. Weissman, and K. S. Kim. 2000. Application of signature-tagged mutagenesis for identification of Escherichia coli K1 genes that contribute to invasion of human brain microvascular endothelial cells. Infect. Immun. 68:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 4.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 5.Bermudez, L. E., M. Petrofsky, P. Kolonoski, and L. S. Young. 1992. An animal model of Mycobacterium avium complex disseminated infection after colonization of the intestinal tract. J. Infect. Dis. 165:75-79. [DOI] [PubMed] [Google Scholar]
- 6.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 8.Breiman, R. F. 1993. Modes of transmission in epidemic and nonepidemic Legionella infection: directions for further study, p. 30-35. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 9.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and C. Engleberg. 1994. Replicative Legionella pneumophila lung infections in intratracheally inoculated A/J mice: a murine model of human Legionnaires' disease. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 10.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 11.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. W., W. Pan, M. P. D'Souza, and J. T. August. 1985. Lysosome-associated membrane proteins: characterization of LAMP-1 of macrophage P388 and mouse embryo 3T3 cultured cells. Arch. Biochem. Biophys. 239:574-586. [DOI] [PubMed] [Google Scholar]
- 13.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 19.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 20.DiRita, V. J., and J. J. Mekalanos. 1989. Genetic regulation of bacterial virulence. Annu. Rev. Genet. 23:455-482. [DOI] [PubMed] [Google Scholar]
- 21.Dournon, E., W. F. Bibb, P. Rajagopalan, N. Desplaces, and R. M. McKinney. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157:496-501. [DOI] [PubMed] [Google Scholar]
- 22.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein, P. H., C. Nakahama, J. O. Tobin, K. Calarco, K. B. Beer, J. R. Joly, and R. K. Selander. 1986. Paleoepidemiologic investigation of Legionnaires disease at Wadsworth Veterans Administration Hospital by using three typing methods for comparison of legionellae from clinical and environmental sources. J. Clin. Microbiol. 23:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehret, W., G. Anding, I. Tartakovskii, and G. Ruckdeschel. 1993. Molecular epidemiology of outbreak-associated serogroup 1 isolates of Legionella pneumophila, p. 30-35. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 25.Elliot, J. A., and W. Johnson. 1982. Virulence conversion of Legionella pneumophila serogroup 1 by passage in guinea pigs and embryonated eggs. Infect. Immun. 35:943-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finlay, B. B., and S. Falkow. 1989. Common themes in microbial pathogenicity. Microbiol. Rev. 53:210-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fullner, K. J. 1998. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J. Bacteriol. 180:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullner, K. J., J. C. Lara, and E. W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107-1109. [DOI] [PubMed] [Google Scholar]
- 30.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 31.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 32.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 34.Joly, J. R., R. M. McKinney, J. O. Tobin, W. F. Bibb, I. D. Watkins, and D. Ramsay. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23:768-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi, A. D., S. Sturgill-Koszycki, and M. S. Swanson. 2001. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell. Microbiol. 3:99-114. [DOI] [PubMed] [Google Scholar]
- 36.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 89:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marston, B. J., J. F. Plouffe, R. F. Breiman, T. M. File, Jr., R. F. Benson, M. Moyenudden, W. L. Thacker, K.-H. Wong, S. Skelton, B. Hackman, S. J. Salstrom, and J. M. Barbaree. 1993. Preliminary findings of a community-based pneumonia incidence study, p. 36-37. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 38.McDade, J. E., and C. C. Shepard. 1979. Virulent to avirulent conversion of Legionnaires' disease bacterium (Legionella pneumophila)--its effect on isolation techniques. J. Infect. Dis. 139:707-711. [DOI] [PubMed] [Google Scholar]
- 39.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. F., J. J. Mekalanos, and S. Falkow. 1989. Coordinate regulation and sensory transduction in the control of bacterial virulence. Science 243:916-922. [DOI] [PubMed] [Google Scholar]
- 41.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 42.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 44.Polesky, A. H., J. T. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinowitz, S., H. Horstmann, S. Gordon, and G. Griffiths. 1992. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J. Cell Biol. 116:95-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reingold, A. L., B. M. Thomason, B. J. Brake, L. Thacker, H. W. Wilkinson, and J. N. Kuritsky. 1984. Legionella pneumonia in the United States: the distribution of serogroups and species causing human illness. J. Infect. Dis. 149:819.. [DOI] [PubMed] [Google Scholar]
- 48.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoeba. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 52.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 54.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal, G., and H. A. Shuman. 1999. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol. Microbiol. 33:669-670. [DOI] [PubMed] [Google Scholar]
- 56.Stout, J. E., J. Joly, M. Para, J. Plouffe, C. Ciesielski, M. J. Blaser, and V. L. Yu. 1988. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila. J. Infect. Dis. 157:486-495. [DOI] [PubMed] [Google Scholar]
- 57.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson, M. S., and R. R. Isberg. 1996. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect. Immun. 64:2585-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 60.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 61.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 62.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 63.Welch, R. A. 1991. Pore-forming cytolysin of gram-negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 64.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 65.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida, S., and Y. Mizuguchi. 1986. Multiplication of Legionella pneumophila Philadelphia 1 in cultured peritoneal macrophages and its correlation to susceptibility of animals. Can. J. Microbiol. 32:438-442. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziegelin, G., W. Pansegrau, B. Strack, D. Balzer, M. Kroger, V. Kruft, and E. Lanka. 1991. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Sequence 1:303-327. [DOI] [PubMed] [Google Scholar]