Abstract
Nasopharyngeal carcinoma (NPC) is a highly prevalent disease in Southeast Asia and its prevalence is clearly affected by genetic background. Various theories have been suggested for its high incidence in this geographical region but to these days no conclusive explanation has been identified. Chromosomal imbalances identifiable through comparative genomic hybridization may shed some light on common genetic alterations that may be of relevance to the onset and progression of NPC. Review of the literature, however, reveals contradictory results among reported findings possibly related to factors associated with patient selection, stage of disease, differences in methodological details etc. To increase the power of the analysis and attempt to identify commonalities among the reported findings, we performed a meta-analysis of results described in NPC tissues based on chromosomal comparative genomic hybridization (CGH). This meta-analysis revealed consistent patters in chromosomal abnormalities that appeared to cluster in specific "hot spots" along the genome following a stage-dependent progression.
Background
Nasopharyngeal carcinoma (NPC) is a Epstein-Barr virus-associated cancer [1] highly prevalent in Southeast Asia and especially southern China where it occurs at a prevalence about a 100-fold higher compared with other populations not at risk [2,3]. Although environmental factors may contribute to this geographical association [4], the high prevalence of NPC in U.S. immigrants suggests a genetic influence [5]. Indeed, genetic markers associated with the disease have been proposed including an enhanced or reduced prevalence of some HLA haplotypes [6] or, more recently, a genomic region within chromosome 4 identified through genome-wide scanning of case control studies of familiar NPC from the Guangdong region [7].
Independently of the genetic root at the basis of its geographical prevalence, NPC is progressively characterized by widespread genomic imbalances possibly occurring before and during carcinogenesis [8]. Different strategies of comparative genomic hybridization (CGH) have been utilized to screen NPC for evidence of consistent chromosomal gains and losses along the progression of the disease [9-14]. However, most studies included a relatively small number of cases applied to different experimental models including the analysis of cell lines, xenografts or neoplastic tissues impairing the ability to interpreter the significance and evaluate the true frequency of chromosome imbalances in NPC. In particular, stratification of cases according to staging could not be performed at a power that could yield statistically meaningful information due to the limited sample population included in individual studies. Therefore, we entertained a meta-analysis of available information based on CGH of NPC samples derived directly from neoplastic material and excluding information derived from cell lines which might skew the true in vivo prevalence although others have shown a reasonable correlation between genetic alterations observed in vivo and those identifiable through the study of cell lines [15,16].
Six studies were identified that utilized directly tissue samples and contained sufficient information to allow a direct cross comparison of the results among them [8-12,17]. In this fashion, 188 NPC cases from Southern Asia were evaluated for which the CGH-derived information could be related to clinical staging.
Results
Chromosomal imbalances in patients with NPC
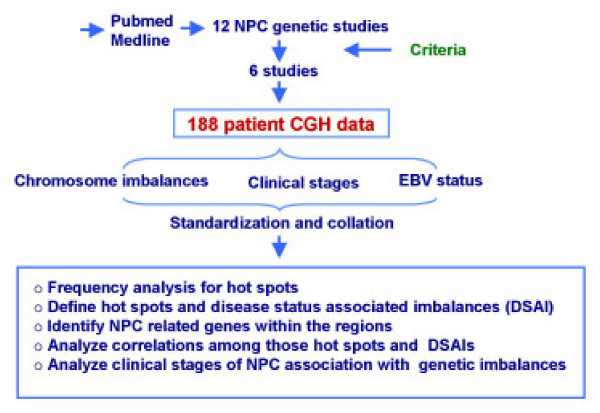
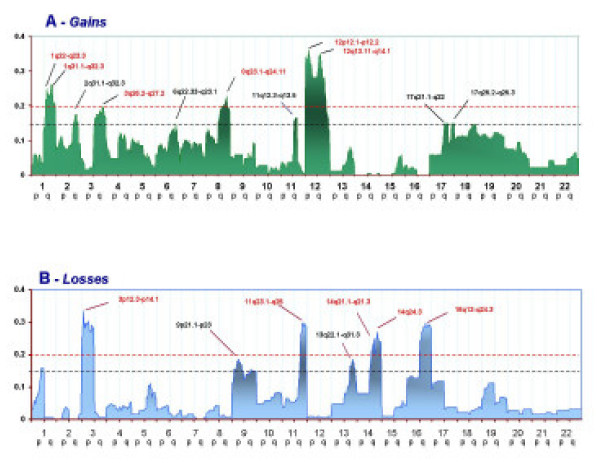
The flow chart of the strategy applied for the meta-analysis is summarized in Figure 1 (see also materials and methods). The complete meta-analysis of all available CGH data revealed that chromosome gains were prevalent in 1q, 2q, 3q, 6q, 8q, 11q, 12p, 12q, 17p, and 17q (Figure 2). Hot spots could be identified in several regions: the ones with highest prevalence were in 1q, 3q, 8q, 12p, and 12q displaying more than 20% frequency. Hot spots with slightly lower prevalence (15 to 20%) could be identified in, 2q, 6q, 11q, 17p, and 17q. Chromosome losses occurred mainly on 3p, 8p, 11q, 13q, 14q, and16q. Among them, losses in 3p, 11q, 14q, and 16q occurred with more than 20% frequency while in 8p and 13q occurred with 15%-20% frequency. In total 12 hot spots were identified by this meta-analysis. Known oncogenes, tumor suppressor genes or other NPC related genes located in these hot-spots are presented in Table 1.
Figure 1.
Flow chart summarizing the strategy adopted for the meta-analysis of chromosomal imbalances associated with NPC. The analysis was based on 6 studies whose information was comparable and reported on tissue samples eliminating studies reporting on cell lines or xenograft information [9-12, 17, 51]. See Materials and Methods for details.
Figure 2.
Frequency of chromosomal gains (panel A) or losses (panel B) in 188 samples derived from patients with NPC. The Y axis reports the frequency of chromosomal imbalances for individuals chromosome regions clustered into sections as described in the Materials and Methods. The X axis represents and ordered distribution of the chromosome analyzed. A cut-off of 0.15 corresponds to a frequency of imbalances higher than 15% arbitrarily selected to define "hot spots". A cut-off of .20 defines the predominant hot-spots used for clustering analysis.
Table 1.
Genes located in "hot spots" identified by the present meta-analysis
| Ch | Arm | Host spot, DSAI | Freq (%) | Candidate NPC-related Genes | references |
| 1 | +1q | + 1q22-q23.3 | 25.00 | LAMC2 | [13] |
| + 1q31.1-q32.3 | 26.06 | ||||
| 2 | +2q | + 2q31.1-q32.3 | 17.55 | ||
| 3 | -3p | - 3p12.3-p14.1 | 33.51 | FHIT | [31] |
| - 3p21.2-p21.33 | 30.32 | RASSF1A,, Blu | [32,33] | ||
| - 3p24.2-p26.3 | 29.26 | RARβ2, RAF1 | [13,34] | ||
| +3q | + 3q26.2-q27.2 | 19.68 | deltaN-p63 | [35] | |
| 6 | +6p | +6p21.2-p23 | 23.33 E | HSP70-2 | [36] |
| 8 | +8q | + 8q23.1-q24.11 | 22.87 | MYC | [37] |
| 9 | -9P | - 9p21.1-p23 | 18.61 | P14, p16, UBAPI, NGX6 | [38–41] |
| 11 | +11q | + 11q12.3-q13.5 | 16.49 | Cyclin D1 | [14] |
| -11q | - 11q23.1-q25 | 29.79 | TSLC1/IGSF4, | [42] | |
| 12 | +12p | + 12p12.1-p12.2 | 36.17 | ||
| +12p11.21-p11.22 | 34.04 | ||||
| +12q | + 12q13.11-q14.1 | 35.11 | MDM2, STAT2 | [43,44] | |
| 13 | -13q | - 13q22.1-q31.3 | 18.61 | EDNRB, LIG4 | [45,46] |
| 14 | -14q | - 14q21.1-q21.3 | 25.00 | ||
| - 14q24.3 | 27.13 | AKTI/PKB, SIVA | [43] | ||
| 15 | +15q | +15q21.3-q25.3 | 26.67 E | DAPK2 (?) | [47] |
| 16 | -16q | - 16q13-q24.3 | 29.79 | E-cadherin, BRD7 | [48,49] |
| 18 | +18p | +18p | 20.00 S | ||
| +18q | +18q21.1-q21.33 | 21.25 S | |||
| 20 | +20q | +20q11.21-q13.13 | 30.00 E | PLUNC (?) | [50] |
Two studies reported the EBV status of a tumor for a total of 30 cases [10,17] In all cases, the tumors displayed evidence of EBV presence. Because this information supports with higher confidence the categorization of NPC to the WHO EBV-related tumors, we performed a separate meta-analysis only on these samples. The results were largely similar to those observed in the whole population analyzed (Figure 3). However, additional regions were identified with high frequency of imbalances that could be best explained by a higher sensitivity of the methodologies applied in these particular studies. Alternatively, it is possible that the other studies included cases of tumors not harboring EBV and, therefore, not canonically EBV-related. In this particular though unlikely case, these 4 hot spots could be considered DSA specifically linked to the EBV etiopathogenesis.
Figure 3.
Frequency of chromosomal in 30 samples derived from patients with NPC known to contain EBV virus. The Y axis reports the frequency of chromosomal imbalances for individuals chromosome regions clustered into sections as described in the Materials and Methods. The X axis represents and ordered distribution of the chromosome analyzed.
Chromosomal imbalances according to WHO staging
When the information was available, cases of NPC were grouped according to WHO staging [18]. One hundred-eight cases could be collected of which 28 were grouped as stage I-II and 80 as stage III-IV. Comparison of frequencies in chromosomal imbalances was carried between the two categories and their significance was examined based on a Fisher's exact test. In general, the same patterns of chromosomal gains or losses were seen in both categories with a trend to higher frequencies in advanced stages (Figure 4). Statistical analysis identified specific areas whose frequency of imbalances was significantly related to stage (Table 2). Of interest were also areas of gain such as 3p, 8q, 12p, 12q or loss such as 14p and 14q where a similar frequency was noted independently of stage suggesting that, possibly, this represent earlier and more fundamental chromosomal aberrations that occur early during NPC oncogenesis.
Figure 4.
Frequency of chromosomal gains (panel A) or losses (panel B) in 108 samples derived from patients with NPC of which 28 were grouped as WHO stage I-II (red line) and 80 as WHO stage III-IV (green line). The Y axis reports the frequency of chromosomal imbalances for individuals chromosome regions clustered into sections as described in the Materials and Methods. The X axis represents and ordered distribution of the chromosome analyzed.
Table 2.
Significantly different frequencies of chromosomal imbalances in early compared to advanced NPC.
| Selected Region | Stage (%) | Fisher's exact | ||
| I-II (n = 28) | III-IV (n = 80) | |||
| Gain | +1q22-q23.3 | 10.71 | 36.25 | 0.015 |
| +1q31.1-q32.3 | 10.71 | 32.50 | 0.015 | |
| +18p | 00.00 | 21.25 | 0.005 | |
| +18q21.1-q21.33 | 00.00 | 20.00 | 0.010 | |
| Loss | -3p12.3-p14.1 | 14.29 | 42.50 | 0.010 |
| -3p25.1-p26.3 | 10.71 | 37.50 | 0.008 | |
| -11q23.1-q25 | 10.71 | 33.75 | 0.026 | |
| -16q13-q24.3 | 14.29 | 37.50 | 0.032 | |
Putative sub-grouping of NPC cases according to relatedness of chromosomal imbalances
Clustering analysis was performed by combining gain and loss data according to a binomial mathematical measure in which lack of imbalances was assigned a value of 0 and gain or losses were indiscriminately assigned a value of one across the data set. Hot spots containing imbalances occurring more that 20% of the times in the population tested were then evaluated by selecting the single representative region within each hot spot portraying the highest frequency value. This data set was, then, combined and cluster analysis was performed (Figure 5). This explorative test identified two sub groups of NPC predominantly characterized by gains and the other by losses. In each category two subgroups could be observed one in which imbalances occurring in any stage of NPC were clustered together (green boxes) and those predominantly occurring in advanced stage NPC (blue boxes). Interestingly, a sub group of NPC was identified that included chromosomal imbalanced detected only in NPC cases for which there knowledge about EBV presence was suggesting that either these studies were characterized by a different level of resolution in identifying chromosomal imbalances or that the NPC cases included in these studies represented a biologically distinct subtype of cancer tightly linked to a EBV-related etiology while the cases included in the other studies represented a more heterogeneous population.
Figure 5.
Correlation analysis based on clustering programs comparing the relatedness of the most frequently chromosomal imbalances identified in this study (frequency at least > 20%). Chromosomal regions representative of individual hot sports were selected as described in the material and methods. Distances are presented according to the φ 4 point correlation coefficient. Green boxes underline clusters including chromosomal aberrations found in early (Stage I-II) and late (Stage III-IV) NPC; blue boxes define clusters enriched with aberrations specific for late stage NPC and the red box defines chromosomal imbalances identified only by the studies in which the EBV status of the tumor was reported.
Discussion
Meta-analyses are a powerful tool that allows the identification of patterns consistent in independent studies that, therefore, permits segregating true biological entities from artifactual findings related to biases associated with individual studies. The present meta-analysis of NPC cases explored the frequencies reported of chromosomal aberrations associated with NPC that could be detected with a low resolution assay such as CGH. Consistently present gains and losses could be identified which were defined as "hot spots". Although some of them appeared to occur with some reasonable frequency, none of them appeared to be particularly predominant with the most frequent chromosomal imbalances occurring at most in 30% of the cases evaluated. This phenomenon implies that NPC develops through the accumulation of multiple low-frequency genetic events. The presence of several genes known to be related to the etiopathogenesis of NPC in the hot spots identified further supports the conclusion that alterations in these genomic regions may play a prominent role in the oncogenesis. In addition, the identification of hot spots in which not know NPC-specific biomarker as been reported (Table 1) proposes other genomic regions of possible interest to be investigated for the identification of unknown cancer inducing factors.
Stage specific and incremental imbalances were observed with no hot spot identified in early cases (Stage I-II) that could not be seen in later cases (Stage III-IV) but several examples of incremental imbalances associated with advanced NPC. Surprisingly, unique imbalances were identified in cases in which the EBV status of the tumor was reported. Although it is conceivable that those cases in which EBV status was not reported may have represent disease taxonomy un-related to the EBV etiopathogenesis this possibility seems quite unlikely. First the cases analyzed were from an area of the World where EBV-associated NPC is endemic. Second, all cases in which the EBV status was evaluated, its presence turned out to be evident suggesting that also in the other cases the same results could be expected. It is, therefore, possible, that the two studies presenting this information [10,17]. were carried out adopting methodologies characterized by different level of sensitivity in detecting chromosomal imbalances. This question will need to be further addressed in future studies.
Interestingly, NPC appeared to segregate, based on cluster algorithms of relatedness, according to a general tendency toward chromosomal loss or gain. It is only, within category of loss or gain that imbalances present in early and late stage NPC segregated from those occurring at a later phase. This is reminiscent of the "tree model" of NPC pathogenesis that we previously reported [4]. Whether, this finding has a biological foundation or is related to a different sensitivity of CGH in reporting gain or losses will need also to be further explored.
Recently, Yan W et al [19] reported a novel study addressing chromosomal abnormalities associated with cervical nodal metastases in NPC. Since this information was not available at the time of our studies, we utilized this study to validate the prediction power of our results on an independent data set. In Yan's study, 23 samples of primary NPC were compared with 9 samples derived from lymph node metastases. A similar pattern of genomic imbalances was seen in primary and metastatic tumors reflecting their common clonal origin. Primary tumors were characterized by frequent gains of 5p, 12p, 12q and 18p and losses of 1p, 3p, 9q that identified hot-spots (frequency = 15%) as gains 12 12p and 12q and as losses in 1p, 3p, 9q, 14q and 16 q (Figure 2). In addition, the areas of frequent aberrations that did not appear as hot spots in the meta-analysis (< 15% frequency) were still characterized by higher frequency than in the direction (gain or loss) indicated by Yan's study. Thus, it appears that the results compiled by the meta-analysis could predict with high accuracy the findings observed in an independent set of individuals.
Chromosomal aberrations observed specifically in lymph nodal metastases in Yan's study included also losses in 9p, 16p, 17q, 20q, 21p, 21q and 22q and gains in 8q and 8p [19]. These findings diverged from the results of the meta-analysis based on primary NPC samples with only loss of 9p and gain of 8q that were represented by hot spots. In addition, loss of 16p was found in 100% on lymph nodal metastases, the meta-analysis results demonstrated that loss of 16p is more common in advanced stage primary NPC with a frequency below 15% in early stages and above 20% in stage III-IV. Thus, this region, may be a potential area of interest when looking at genes that might be associated with the metastatic potential of NPC and other cancers [20-24]. However, also in this case, when the results did not collimate the trends noted were in the same direction. Overall, this meta-analysis aimed at unifying other's results about chromosomal imbalances associated with NPC development and progression, identified consistent aberrations that could be identified in an independent study. This suggests that these chromosomal imbalances are strongly specific for the disease.
Materials and methods
Literature search
Searches were performed on PubMed and Medline using the following combination of search terms: "NPC", "Nasopharyngeal carcinoma", "and "CGH" and "comparative genome hybridization". In total, 12 studies were identified [8-13,17,25-29]., however, only 6 [8-12,17] were eligible for further analysis based on the following inclusion criteria: only NPC tumor biopsies were evaluated (studies using NPC cell lines and xenograft cell lines were excluded), the studies involved exclusively Southern Asian NPC patients, applied comparable Chromosome CGH analysis platforms, reported detailed CGH values using similar thresholds for definition of chromosomal loss or gain (if the normal DNA vs tumor DNA ratio was < 0.75 the region was considered as a chromosomal gain; if >1.20 it was considered a loss). A total of 188 NPC cases were identified from the 6 studies including 3 studies from Hong Kong, 2 from China, and 1 from Taiwan. Significant chromosomal imbalances were detected in 155 patients. World Health Organization (WHO) staging [18] was available in 108 cases; 28 belonged to early stage (stage I and II), 80 belonged to advanced stage (stage III and IV). EBV positive status was reported only in 30 patients from two studies.
Data integration
CGH data derived from the 6 studies was standardized by dividing each chromosome arm into 20 sub-regions from the centromere toward each telomere. Chromosome gains and losses reported by the 6 studies were assigned to the relevant chromosomal region and gains or losses were recorded into two separate excel files reducing the information into a binary model (1 = chromosome imbalance, 0 = no imbalance). Chromosomal loss or gain were defined as previously stated according to a threshold signal intensity ratio between normal DNA over tumor DNA < 0.75 (gain) or > 1.20 (loss). WHO staging of NPC and EBV status were also entered in the data file.
Definition of hot spot and Disease Status-Associated Imbalance (DSAI)
Regions with high frequency of chromosomal imbalances were considered 'hot spots". These were defined as chromosome regions with frequency imbalance observed in at least 15% of cases while the adjacent regions (boundaries of the hot spot) displayed a drop if frequency of at least 2%. The 15% value was arbitrarily selected based on the observation that the median frequency of chromosomal gains was 5.8% (1st quartile = 2.7 and 3rd quartile = 11.2) with an average of 7.7 ± 7.0 (Standard deviation, SD) and of losses was 3.7% (1st quartile = 1.6 and 3rd quartile = 8) with an average of 6.4 ± 7.4 (SD). A value of 15% included, therefore, chromosomal regions with a frequency of imbalances at least more than one SD above the mean and above at least the 3rd quartile across the all data set.
Disease status associated imbalance (DSAI) corresponded to a chromosome region with a significantly high frequency of imbalances in clinically relevant subgroups of cases such as advanced versus early stage patients. The 1p, 16p, 19p, 19q, 22q regions and the whole Y chromosome were not analyzed because they have been described to yield false positives results in CGH analysis [8,11]. Finally, the X chromosome was also not analyzed due to a rare prevalence of events associated with it and lack of information about the patient's gender.
Statistical analysis
Chromosomal gains and losses were separately profiled for all chromosomal regions previously defined arms by assigning a binary value of 1 or 0 according to presence or absence of imbalance for that region in each sample studied. Frequency of imbalances for each chromosomal region analyzed was then calculated and graphically visualized. Hot spots were selected for further analysis. Additionally, the frequency of chromosomal imbalances observed in studies reporting the EBV-status of tumors was also separately analyzed because of the higher confidence that these tumors represented classical EBV-induced NPC. A two-tailed Fisher's exact test was applied to explore the strength of relationships between given chromosomal imbalances and stages of NPC in order to identify DSAI. Based on hot spots and DSAI, a hierarchical clustering analysis [30] was performed to explore possible correlations among distinct chromosomal regions where imbalances were frequent. A flow chart representing the procedure of the meta-analysis is shown in Figure 1.
Clustering algorithms
Hot spots with a frequency above 205 were selected for clustering analysis to identify potential subgroups of NPC with a defined pattern of genetic imbalances. Within in each spot a representative region was selected by choosing the region associated with the highest frequency on imbalances. Data from each of the selected region was gathered in a separate data base assigning a value of 0 to individual cases where no genetic imbalances were observed and of 1 to either losses or gains. In this fashion chromosomal imbalances could be clustered independently of their genetic tendency toward loss or gain. Hierarchical clustering was then carried on SPSS 10.0 based on hot-spot ranking values giving an approximation of relationship among various imbalances measured according to the Φ 4 point correlation coefficient.
This article was partly supported by a grant (30371535/C030310) from National Natural Science Foundation of China.
Contributor Information
Xin Li, Email: lixin2@cc.nih.gov.
Ena Wang, Email: Ewang@cc.nih.gov.
Ying-dong Zhao, Email: Zhaoy@mail.nih.gov.
Jia-Qiang Ren, Email: renj@cc.nih.gov.
Ping Jin, Email: pingj@cc.nih.gov.
Kai-Tai Yao, Email: ktyao@fimmu.com.
Francesco M Marincola, Email: FMarincola@cc.nih.gov.
References
- Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10:803–821. doi: 10.1158/1078-0432.CCR-0670-3. [DOI] [PubMed] [Google Scholar]
- Chan ATC, Teo PML, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol. 2002;13:1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:421–429. doi: 10.1016/S1044579X02000858. [DOI] [PubMed] [Google Scholar]
- Huang Z, Desper R, Schaffer AA, Yin Z, Li X, Yao K. Construction of tree models for pathogenesis of nasopharyngeal carcinoma. Genes Chromosomes Cancer. 2004;40:307–315. doi: 10.1002/gcc.20036. [DOI] [PubMed] [Google Scholar]
- McDermott AL, Dutt SN, Watkinson JC. The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol. 2001;26:82–92. doi: 10.1046/j.1365-2273.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- Goldsmith DB, West TM, Morton R. HLA associations with nasopharyngeal carconoma in Southern Chinese: a meta-analysis. Clin Otolaryngol. 2002;27:61–67. doi: 10.1046/j.0307-7772.2001.00529.x. [DOI] [PubMed] [Google Scholar]
- Feng BJ, Huang W, Shugart YY, Lee MK, Zhang F, Xia JC, et al. Genome-wide scan for familial nasopharyngeal carcinoma reveals evidence of linkage to chromosome 4. Nat Genet. 2002;31:395–399. doi: 10.1038/ng932. [DOI] [PubMed] [Google Scholar]
- Fang Y, Guan X, Guo Y, Sham J, Deng M, Liang Q, et al. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30:254–260. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1086>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Ko JY, Chen PJ, Shu CH, Hsu MT, Tsai SF, et al. Chromosomal aberrations in nasopharyngeal carcinoma analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;25:169–175. doi: 10.1002/(SICI)1098-2264(199906)25:2<169::AID-GCC13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Hui AB, Lo KW, Leung SF, Teo P, Fung MK, To KF, et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int J Cancer. 1999;82:498–503. doi: 10.1002/(SICI)1097-0215(19990812)82:4<498::AID-IJC5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Chien G, Yuen PW, Kwong D, Kwong YL. Comparative genomic hybridization analysis of nasopharygeal carcinoma: consistent patterns of genetic aberrations and clinicopathological correlations. Cancer Genet Cytogenet. 2001;126:63–67. doi: 10.1016/S0165-4608(00)00392-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang L, Zhang X, Zhang L, Zhang B, Yu Y, et al. [Chromosomal aberration analyzed by comparative genomic hybridization in nasopharyngeal carcinoma] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2001;18:338–342. [PubMed] [Google Scholar]
- Hui AB, Lo KW, Teo PM, To KF, Huang DP. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol. 2002;20:467–473. [PubMed] [Google Scholar]
- Hui AB, Or YY, Takano H, Tsang RK, To KF, Guan XY, et al. Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res. 2005;65:8125–8133. doi: 10.1158/0008-5472.CAN-05-0648. [DOI] [PubMed] [Google Scholar]
- Roschke AV, Stover K, Tonon G, Schaffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- Fan CS, Wong N, Leung SF, To KF, Lo KW, Lee SW, et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma. Hum Pathol. 2000;31:169–178. doi: 10.1016/S0046-8177(00)80216-6. [DOI] [PubMed] [Google Scholar]
- Shanmugaratnam K, Sobin LH. World Health Organization International Histological Classification: Histological typing of tumors of the upper respiratory tract and ear. 2. Berlin: Springer-Verlag; 1991. [Google Scholar]
- Yan W, Song L, Wei W, Li A, Liu J, Fang Y. Chromosomal Abnormalities Associated with Neck Nodal Metastasis in Nasopharyngeal Carcinoma. Tumour Biol. 2005;26:306–312. doi: 10.1159/000089289. [DOI] [PubMed] [Google Scholar]
- Chujo M, Noguchi T, Miura T, Arinaga M, Uchida Y, Tagawa Y. Comparative genomic hybridization analysis detected frequent overrepresentation of chromosome 3q in squamous cell carcinoma of the lung. Lung Cancer. 2002;38:23–29. doi: 10.1016/S0169-5002(02)00151-4. [DOI] [PubMed] [Google Scholar]
- Alcock HE, Stephenson TJ, Royds JA, Hammond DW. Analysis of colorectal tumor progression by microdissection and comparative genomic hybridization. Genes Chromosomes Cancer. 2003;37:369–380. doi: 10.1002/gcc.10201. [DOI] [PubMed] [Google Scholar]
- Chu LW, Troncoso P, Johnston DA, Liang JC. Genetic markers useful for distinguishing between organ-confined and locally advanced prostate cancer. Genes Chromosomes Cancer. 2003;36:303–312. doi: 10.1002/gcc.10171. [DOI] [PubMed] [Google Scholar]
- Kadota M, Tamaki Y, Sekimoto M, Fujiwara Y, Aritake N, Hasegawa S, et al. Loss of heterozygosity on chromosome 16p and 18q in anaplastic thyroid carcinoma. Oncol Rep. 2003;10:35–38. [PubMed] [Google Scholar]
- Noguchi T, Kimura Y, Takeno S, Chujo M, Uchida Y, Mueller W, et al. Chromosomal imbalance in esophageal squamous cell carcinoma: 3q gain correlates with tumor progression but not prognostic significance. Oncol Rep. 2003;10:1393–1400. [PubMed] [Google Scholar]
- Yan J, Fang Y, Liang Q, Huang Y, Zeng Y. Novel chromosomal alterations detected in primary nasopharyngeal carcinoma by comparative genomic hybridization. Chin Med J (Engl) 2001;114:418–421. [PubMed] [Google Scholar]
- Song LB, Yan J, Jian SW, Zhang L, Li MZ, Li D, et al. [Molecular mechanisms of tumorgenesis and metastasis in nasopharyngeal carcinoma cell sublines] Ai Zheng. 2002;21:158–162. [PubMed] [Google Scholar]
- Wong N, Hui AB, Fan B, Lo KW, Pang E, Leung SF, et al. Molecular cytogenetic characterization of nasopharyngeal carcinoma cell lines and xenografts by comparative genomic hybridization and spectral karyotyping. Cancer Genet Cytogenet. 2003;140:124–132. doi: 10.1016/S0165-4608(02)00657-X. [DOI] [PubMed] [Google Scholar]
- Or YY, Hui AB, Tam KY, Huang DP, Lo KW. Characterization of chromosome 3q and 12q amplicons in nasopharyngeal carcinoma cell lines. Int J Oncol. 2005;26:49–56. [PubMed] [Google Scholar]
- Rodriguez S, Khabir A, Keryer C, Perrot C, Drira M, Ghorbel A, et al. Conventional and array-based comparative genomic hybridization analysis of nasopharyngeal carcinomas from the Mediterranean area. Cancer Genet Cytogenet. 2005;157:140–147. doi: 10.1016/j.cancergencyto.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JY, Lee TC, Hsiao CF, Lin GL, Yen SH, Chen KY, et al. Definition of three minimal deleted regions by comprehensive allelotyping and mutational screening of FHIT, p16(INK4A), and p19(ARF) genes in nasopharyngeal carcinoma. Cancer. 2002;94:1987–1996. doi: 10.1002/cncr.10406. [DOI] [PubMed] [Google Scholar]
- Chow LS, Lo KW, Kwong J, To KF, Tsang KS, Lam CW, et al. RASSF1A is a target tumor suppressor from 3p21.3 in nasopharyngeal carcinoma. Int J Cancer. 2004;109:839–847. doi: 10.1002/ijc.20079. [DOI] [PubMed] [Google Scholar]
- Qiu GH, Tan LK, Loh KS, Lim CY, Srivastava G, Tsai ST, et al. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene. 2004;23:4793–4806. doi: 10.1038/sj.onc.1207632. [DOI] [PubMed] [Google Scholar]
- Kwong J, Lo KW, Chow LS, To KF, Choy KW, Chan FL, et al. Epigenetic silencing of cellular retinol-binding proteins in nasopharyngeal carcinoma. Neoplasia. 2005;7:67–74. doi: 10.1593/neo.04370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook T, Nicholls JM, Brooks L, O'Nions J, Allday MJ. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene. 2000;19:3439–3444. doi: 10.1038/sj.onc.1203656. [DOI] [PubMed] [Google Scholar]
- Jalbout M, Bouaouina N, Gargouri J, Corbex M, Ben Ahmed S, Chouchane L. Polymorphism of the stress protein HSP70-2 gene is associated with the susceptibility to the nasopharyngeal carcinoma. Cancer Lett. 2003;193:75–81. doi: 10.1016/S0304-3835(02)00697-3. [DOI] [PubMed] [Google Scholar]
- Yu Y, Dong W, Li X, Yu E, Zhou X, Li S. Significance of c-Myc and Bcl-2 protein expression in nasopharyngeal carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129:1322–1326. doi: 10.1001/archotol.129.12.1322. [DOI] [PubMed] [Google Scholar]
- Kwong J, Lo KW, To KF, Teo PM, Johnson PJ, Huang DP. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res. 2002;8:131–137. [PubMed] [Google Scholar]
- Zhang L, Fang Y, Huang B, Hou J, Zhao M, Li H, et al. Rapid tissue microarray assay of p16 protein expression for different stage nasopharyngeal carcinoma. Zhonghua Bing Li Xue Za Zhi. 2002;31:132–134. [PubMed] [Google Scholar]
- Wang L, Ma J, Li J, Li X, Zhang Q, Peng S, et al. NGX6 gene inhibits cell proliferation and plays a negative role in EGFR pathway in nasopharyngeal carcinoma cells. J Cell Biochem. 2005;95:64–73. doi: 10.1002/jcb.20393. [DOI] [PubMed] [Google Scholar]
- Xiao B, Fan S, Zeng Z, Xiong W, Cao L, Yang Y, et al. Purification of novel UBAP1 protein and its decreased expression on nasopharyngeal carcinoma tissue microarray. Protein Expr Purif. 2005 doi: 10.1016/j.pep.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Hui AB, Lo KW, Kwong J, Lam EC, Chan SY, Chow LS, et al. Epigenetic inactivation of TSLC1 gene in nasopharyngeal carcinoma. Mol Carcinog. 2003;38:170–178. doi: 10.1002/mc.10156. [DOI] [PubMed] [Google Scholar]
- Xie L, Xu L, He Z, Zhou W, Wang L, Zhang L, et al. Identification of differentially expressed genes in nasopharyngeal carcinoma by means of the Atlas human cancer cDNA expression array. J Cancer Res Clin Oncol. 2000;126:400–406. doi: 10.1007/PL00008488. [DOI] [PubMed] [Google Scholar]
- Wu HC, Lu TY, Lee JJ, Hwang JK, Lin YJ, Wang CK, et al. MDM2 expression in EBV-infected nasopharyngeal carcinoma cells. Lab Invest. 2004;84:1547–1556. doi: 10.1038/labinvest.3700183. [DOI] [PubMed] [Google Scholar]
- Han W, Li H, Xie L, Xu L, Zhang L, Yao K. [Plerosis of cDNA array of normal human nasopharyngeal tissue and nasopharyngeal carcinoma] Zhonghua Zhong Liu Za Zhi. 2002;24:114–117. [PubMed] [Google Scholar]
- Lo KW, Tsang YS, Kwong J, To KF, Teo PM, Huang DP. Promoter hypermethylation of the EDNRB gene in nasopharyngeal carcinoma. Int J Cancer. 2002;98:651–655. doi: 10.1002/ijc.10271. [DOI] [PubMed] [Google Scholar]
- Wong TS, Chang HW, Tang KC, Wei WI, Kwong DL, Sham JS, et al. High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients. Clin Cancer Res. 2002;8:433–437. [PubMed] [Google Scholar]
- Wang S, Jiang Y, Huang H, Yang Q. [Expression and correlation with metastasis of E-cadherin, CD44H, matrix metalloproteinase-3, nm23H1 and VEGF in nasopharyngeal carcinoma] Lin Chuang Er Bi Yan Hou Ke Za Zhi. 2004;18:470–472. [PubMed] [Google Scholar]
- Zhou J, Ma J, Zhang BC, Li XL, Shen SR, Zhu SG, et al. BRD7, a novel bromodomain gene, inhibits G1-S progression by transcriptionally regulating some important molecules involved in ras/MEK/ERK and Rb/E2F pathways. J Cell Physiol. 2004;200:89–98. doi: 10.1002/jcp.20013. [DOI] [PubMed] [Google Scholar]
- Zhang B, Nie X, Xiao B, Xiang J, Shen S, Gong J, et al. Identification of tissue-specific genes in nasopharyngeal epithelial tissue and differentially expressed genes in nasopharyngeal carcinoma by suppression subtractive hybridization and cDNA microarray. Genes Chromosomes Cancer. 2003;38:80–90. doi: 10.1002/gcc.10247. [DOI] [PubMed] [Google Scholar]
- Faint JM, Annels NE, Curnow SJ, Shields P, Pilling D, Hislop AD, et al. Memory T cells constitute a subset of the human CD8+CD45RA+ pool with distinct phenotypic and migratory characteristics. J Immunol. 2001;167:212–220. doi: 10.4049/jimmunol.167.1.212. [DOI] [PubMed] [Google Scholar]