Abstract
Invasive infections caused by non-type b encapsulated Haemophilus influenzae have increased in frequency in the last decade. This change prompted us to characterize the genetic relationships of 48 recently isolated invasive H. influenzae type a (Hia), e (Hie), and f (Hif) strains by comparison of restriction digest patterns (RDPs). Recent Hia isolates exhibited moderate genetic diversity, with the majority segregating into two major clonotypes. Recent Hie and, especially, Hif strains displayed considerably restricted genetic diversity. In particular, all but one Hif strain segregated into a single clonotype, and half of these isolates had identical RDPs. These results are consistent with the hypothesis that the increased incidence of disease due to non-type b encapsulated H. influenzae reflects the emergence of hypervirulent clones, especially in the case of Hif. Alternatively, it is possible that non-type b encapsulated H. influenzae strains have limited overall genetic diversity.
Encapsulated Haemophilus influenzae strains elaborate one of six structurally and antigenically distinct capsular polysaccharides, designated a to f (21). Prior to the introduction of highly immunogenic polysaccharide-protein conjugate vaccines in the last decade, H. influenzae type b (Hib) was the most common cause of serious bacterial infections in infants in the United States. As a consequence, considerable attention has focused on the epidemiology and molecular pathogenesis of Hib disease.
In contrast to the situation with Hib, invasive infections caused by non-type b encapsulated H. influenzae strains have been reported uncommonly in the past and usually in the elderly or in immunocompromised persons (19). However, we recently described a cluster of invasive infections due to Hia in previously healthy young children (1). In addition, active surveillance studies suggest that invasive non-type b encapsulated H. influenzae infections are increasingly common. Perdue and coworkers reported that the incidence of invasive non-type b encapsulated H. influenzae infections in Alaskan residents increased by 120% between 1980 and 1990 and between 1991 and 1996 (20). The number of invasive Hif infections reported in the United States and non-type b infections reported in Britain has increased over a similar time period (10, 25). The small but progressive increase in serious disease due to non-type b encapsulated H. influenzae prompted us to examine the molecular epidemiology of these bacteria.
Understanding the population structure of pathogenic bacteria may provide insights into the pathogenesis of human infections (24). Studies in the 1980s correlating biochemical typing, outer membrane protein expression, and multilocus enzyme electrophoresis demonstrated nonrandom associations between these characteristics, suggesting that the population structure of encapsulated H. influenzae was clonal (17). Furthermore, this work showed that most invasive Hib disease worldwide was caused by a limited number of bacterial clonotypes. In the United States, the majority of infections were caused by strains of only three major outer membrane protein or multilocus enzyme electrophoresis electrophoretic types (17).
No recent studies have examined the genetic variation of non-type b encapsulated H. influenzae. Here, we characterize the genetic relationship of a collection of recently isolated invasive non-type b encapsulated H. influenzae strains.
(Presented at the Pediatric Academic Societies Annual Meeting, 29 April 2001, Baltimore, Md., abstract 1386.)
MATERIALS AND METHODS
Bacterial strains.
Invasive isolates of encapsulated H. influenzae were obtained from the collections of A. Smith, University of Missouri, Columbia; J. Kellner, Alberta Children's Hospital; E. Shapiro, Yale University School of Medicine; and the State Public Health Laboratories of Alabama, California, Colorado, Idaho, Iowa, Minnesota, Missouri, Oklahoma, North Carolina, Tennessee, Utah, and Wisconsin. Aside from the site and date of isolation or date of referral to the public health laboratory, for most isolates little clinical information was available in order to protect patient confidentiality. In most instances, isolates were obtained at distinct time points, and referring laboratories believed that these isolates were unrelated.
Five bacterial strains were identified over an 8-month period in an outbreak of invasive Hia disease in Utah, but these bacteria were isolated from children with no obvious epidemiological connection to one another (1). More than one isolate per subject (such as from separate body sites of the same patient) were not referred. Bacteria were grown on chocolate II agar (Edge Biological, Memphis, Tenn.) or in brain heart infusion broth supplemented with NAD (2 μg/ml) and hemin (10 μg/ml) (BHIs).
PFGE.
Chromosomal DNA was prepared for pulsed-field gel electrophoresis (PFGE) by purification in agarose plugs. Bacteria were grown in BHIs to an optical density at 600 nm (OD600) of 0.300. A 1.5-ml aliquot of the suspension was centrifuged briefly to pellet cells. Bacteria were washed twice in 1 ml of 10 mM Tris (pH 8.0)-1 mM EDTA (pH 8.0)-100 mM NaCl (SE) and then resuspended in SE to an OD600 of 1.200. An equal volume of bacterial suspension and 1.6% low-melting-point SeaPlaque agarose (BioWhittaker Molecular Applications, Rockland, Maine) were combined and added to plug molds. Plugs were incubated with proteinase K (10 μg/ml) in 50 mM Tris (pH 7.8)-50 mM EDTA (pH 8.0)-1% Sarkosine at 55°C overnight, rinsed with sterile water, and then incubated for 30 min at 25°C with 0.05 mg of phenylmethylsulfonyl fluoride per ml in 10 mM Tris-HCl (pH 7.4)-1 mM EDTA. Plugs were rinsed with sterile water, washed three times with 10 mM Tris-HCl (pH 7.4)-1 mM EDTA, and stored at 4°C.
Plugs were equilibrated in 250 μl of 1× restriction buffer at 25°C for 30 min. The buffer was then replaced with 250 μl of fresh buffer, and 15 U of SmaI or ApaI (Life Technologies, Rockville, Md.) was added. Plugs were digested at 30°C overnight and then equilibrated by incubating for 10 min in 0.045 M Tris-borate-0.001 M EDTA (0.5× TE) buffer.
Restriction fragments were separated with a clamped homogeneous electric field device using a modification of a protocol developed by A. Smith, University of Missouri, Columbia. Gels were run in a CHEF DRII apparatus (Bio-Rad Laboratories, Hercules, Calif.) for 28 h at 180 V in a 1% Seakem agarose (BioWhittaker Molecular Applications) gel in 0.5× TE buffer at 12°C. Pulse parameters included a ramp time of 1 to 26 s. Gels were stained for 50 min with 0.5 μg of ethidium bromide per ml in 0.5× TE, and images were captured with a still video camera (Eagle Eye II; Stratagene, La Jolla, Calif.) using a UV transilluminator without destaining.
Analysis of genetic relatedness.
Restriction digest patterns (RDPs) of H. influenzae isolates were determined by quantitation of the distance of migration of restriction fragments from the gel origin for each strain. Similarity coefficients between all possible pairs of RDPs were calculated as follows: similarity = (number of shared bands × 2)/total number of bands. Restriction fragments of <40 kb were excluded from analysis because of the increased difficulty of visualizing these smaller fragments on stained agarose gels. Cluster analysis of the RDPs was then performed by the unweighted pair group method with average linkage using locally developed software as previously described (24).
RESULTS
Bacterial strains.
Forty-eight invasive non-type b H. influenzae isolates were obtained from 12 states and included 18 Hia, 10 Hie, and 20 Hif strains. Isolates were recovered between 1998 and 2000, mostly as part of active surveillance studies by public health departments in the 12 states.
PFGE typing.
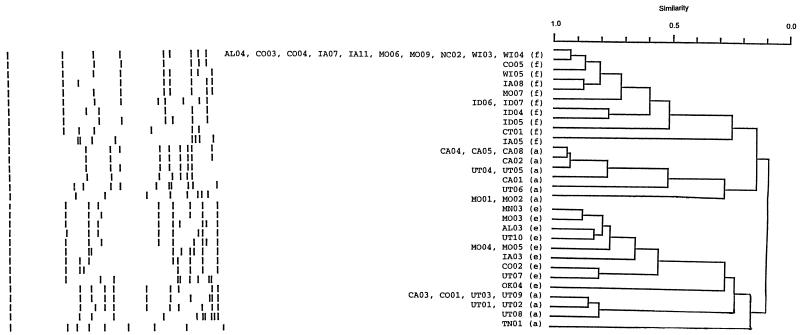
Strains expressing the same serotypes resembled each other with a similarity of ≥0.22 for ApaI and ≥0.36 for SmaI. Hia had the greatest overall genetic diversity of the serotypes studied. Nevertheless, based on ApaI digestion patterns, 15 of 24 Hia isolates segregated into two major lineages at a similarity level of 0.77 (Fig. 1). One lineage included strains with four distinct RDPs, represented by one to two strains each. The second lineage contained strains with three unique RDPs that were shared by one to four strains.
FIG. 1.
Dendrogram of encapsulated H. influenzae isolates based on ApaI restriction digest patterns. Shown on the right are serotypes (in parentheses) and strain designations. Strains expressing the same serotypes resemble each other with a similarity of >0.22. To the left are schematic representations of restriction digest patterns.
SmaI digestion also divided invasive Hia strains into two major lineages, in this case at a genetic similarity of 0.5 (Fig. 2 and Fig. 3). SmaI divisions were generally comparable to those determined by ApaI digestion. As in the ApaI classification, with SmaI digestion strains CA03, CO01, UT03, and UT09 had identical RDPs and clustered to strains UT01, UT02, and UT08 at a similarity of ≥0.73. In the second major lineage, strains CA01, CA05, and CA08 had identical RDPs, which clustered with the RDPs of CA02, CA04, UT04, and UT05 at a similarity of ≥0.84.
FIG. 2.
Dendrogram of encapsulated H. influenzae isolates based on SmaI restriction digest patterns. Shown on the right are serotypes (in parentheses) and strain designations. Strains expressing the same serotypes resemble each other with a similarity of >0.36. To the left are schematic representations of restriction digest patterns.
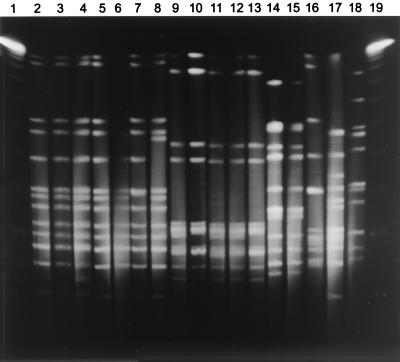
FIG. 3.
SmaI RDPs of serotype a strains. Shown is genomic DNA from 17 Hia isolates after digestion with SmaI and separation of restriction fragments by PFGE. Lanes 1 and 19, molecular size standards. Lanes 2 to 8, major lineage strains. Lanes 9 to 13, second major lineage strains. Lanes 14 to 18, isolates of minor lineages.
Most RDPs from Hie belonged to a single genetic lineage, with 9 of 10 strains clustering at a similarity level of 0.55 in the ApaI classification (Fig. 1) and at a similarity of 0.68 in the SmaI classification (Fig. 2). Whereas only strains MO04 and MO05 had the same RDP in the ApaI classification, in the SmaI classification only one of these nine strains, AL03, had a unique RDP. In both classifications, strain OK04 was less related to other Hie strains than these were to one another.
Ten of 20 Hif strains from very diverse geographic locations had an identical RDP following ApaI digestion, and nine additional strains were closely related to this group, at a similarity level of 0.51 (Fig. 1). Together these 19 strains constituted a single major lineage. SmaI digests of invasive Hif strains confirmed the considerable restriction in genetic diversity of these isolates, with 11 strains having an identical RDP and an additional eight strains clustering to them at a similarity of 0.55 (Fig. 2 and Fig. 4). Strain IA05, in both classifications, was less related to other Hif strains than these were to one another. In the SmaI classification, in fact, this strain appeared to be as closely related to some Hia strains as to other Hif strains.
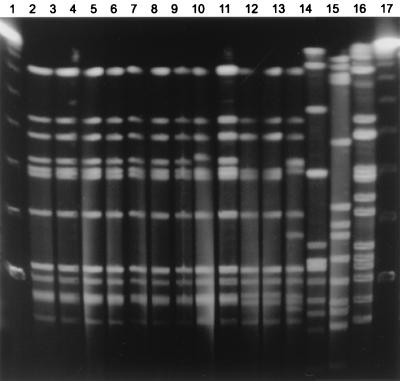
FIG. 4.
SmaI RDP of serotype f strains. Shown is genomic DNA from 14 Hif and one nontypeable H. influenzae isolate after digestion with SmaI and separation of restriction fragments by PFGE. Lanes 1 and 17, molecular size standards. Lanes 2 to 13 and 16, major lineage strains. Lane 14, IA05. Lane 15, a nontypeable H. influenzae isolate.
The mean total molecular size of ApaI restriction fragments, excluding fragments of ≤40 kb, varied slightly between different serotypes (Hia, 1,401 ± 201 kb; Hie, 1,443 ± 51 kb; and Hif, 1,348 ± 170 kb). The mean total molecular size of Hif SmaI restriction fragments was similar to that of ApaI fragments (1,487 ± 175 kb and 1,178 ± 327 kb, respectively). However, the sizes of Hia and Hie strains were considerably smaller (802 ± 92 kb and 878 ± 92 kb, respectively), suggesting that large comigrating restriction fragments are present in these strains. The size of the genome of the acapsular type d strain Rd has been reported to be 1,830 kb, and the size of a representative type b strain is estimated to be 1,950 kb (3, 7).
DISCUSSION
Encapsulated H. influenzae strains have been subclassified by serotyping, biochemical testing, outer membrane protein typing, lipopolysaccharide analysis, multilocus enzyme electrophoresis, and multilocus sequence typing (2, 6, 11, 18, 21). Both serotyping and biotyping have limited discriminatory power, but other subtyping systems permit greater resolution of phenotypic relationships. This work has demonstrated that type b H. influenzae strains have limited phenotypic diversity and a clonal population structure, whereas nontypeable H. influenzae strains are much more heterogeneous and appear to lack clonality (9, 16, 18).
More recently, molecular genetic techniques such as 16S ribotyping, random amplified polymorphic DNA analysis, multilocus sequence typing (MLST), and RDP analysis by PFGE have been used to study the epidemiology of Hib and nontypeable H. influenzae (5, 6, 22, 23). Direct comparison of individual molecular techniques has demonstrated excellent concordance between these methods, with RDP typing and MLST providing the greatest discriminatory power (5, 6, 8, 23). These genotypic techniques have confirmed phenotypic studies with respect to the degree of diversity of Hib and nonencapsulated strains. For example, in a 1999 study of Hib isolates, Exekiel Moor and coworkers found that 187 Australian strains had 67 distinct RDPs that could be clustered into seven groups at a genetic distance of 0.5 (5). Two RDP types predominated, however, and accounted for 20 and 22% of strains. Other investigators noted that the number of unique RDP genotypes found among nontypeable Italian and Japanese strains closely approximated the total number of isolates, whereas shared RDPs were common among Hib strains (4, 23).
Bacterial population structures have provided insights into factors contributing to the pathogenicity of other bacterial species (24). As a consequence, in this study we sought to identify similar relationships in non-type b encapsulated H. influenzae. Our data suggest that recent invasive isolates of Hia in the United States belong to two major clonotypes and have a degree of genetic diversity similar to that of Hib. In contrast, most recent invasive Hie and Hif infections are caused by a more restricted group of bacteria.
Musser and coworkers reported the only other comprehensive analysis of the population structure of non-type b encapsulated H. influenzae (18). In Musser's analysis, 52 Hia isolates had 20 electrophoretic types, clustering into two major groups at a genetic distance of ≤0.65 (18). In the present study, we identified 14 to 15 Hia RDPs falling into two major lineages. Although our methods are not directly comparable, since the earlier study used multilocus enzyme electrophoresis rather than RDP genotyping and different collections of strains were studied, these two estimates of overall diversity and clonality are remarkably consistent.
The recent invasive Hif strains in this study appear much more genetically restricted than Hia strains. Despite diverse geographic origins, over half of these strains had identical RDPs, and the majority had RDPs that differed only by one to two bands. Urwin and coworkers also found that 86% of 43 invasive Hif isolates had a predominant ribotype, and Cerquetti and colleagues noted that three of four invasive Italian Hif isolates had very similar RDPs (4, 25). Collectively, these data suggest that the genetic restriction that we have observed in invasive Hif strains is authentic.
In Musser's study, 50 Hif strains had 28 different electrophoretic types clustering into six groups at a genetic distance of ≤0.50. Two clusters were common; cluster K1c, which contained 15 isolates with 13 similar electrophoretic types, and cluster K2, which contained 30 isolates with 11 distinct electrophoretic types. Overall, these data suggest a more limited genetic diversity in our Hif isolates than previously observed.
Several hypotheses may explain this observation. It is possible that the isolates described in this study are intrinsically more pathogenic than unrelated bacteria and therefore are overrepresented in our collection of invasive strains. In contrast, the Hif isolates examined by Musser et al. were mostly recovered from persons with asymptomatic or minor and superficial infections. Further studies of colonizing and invasive isolates will be required to determine if the overall diversity of Hif strains is greater than we have observed in invasive strains. Second, the bacteria studied by Musser and coworkers included isolates obtained from 30 countries over five decades and therefore may reflect the collective diversity of an evolving population.
Although some Hib electrophoretic types have been shown to have a global distribution, in general two to three electrophoretic types predominate in an individual geographic region (18). It is possible that similar geographic restrictions exist in the distribution of some non-type b H. influenzae clonotypes. This would be somewhat surprising, since it might be expected that the increased frequency of intercontinental travel and human migration in recent decades would efficiently introduce new bacterial strains to a locale. Rather than the greater genetic restriction that we observed in this study, we would predict that recent isolates would exhibit greater genetic diversity.
The increase in invasive non-type b H. influenzae infections in the past decade is unexplained. It is possible that dramatic changes in the epidemiology of Hib in the past quarter century may also have altered the ecology of non-type b strains. Most notably, the widespread implementation of the Hib vaccine in the United States has reduced both the incidence of invasive Hib disease and rates of nasopharyngeal colonization among children (14, 15). As previously suggested, these changes may open an ecological niche that may be filled by other serotypes if the prevalence of colonization is related to competition by Hib or if it is affected by the expression of bacteriocins active only against non-type b serotypes (12, 13).
Invasive disease due to H. influenzae is likely to result from the interaction of host and bacterial factors. Although limited, our data are consistent with the possibility that genetic mechanisms may contribute to the virulence of non-type b H. influenzae strains. It has been hypothesized that some invasive non-type b encapsulated H. influenzae strains might originate from Hib strains by capsular switching, thus acquiring virulence factors normally present in virulent Hib strains. This phenomenon might occur through homologous recombination facilitated by the common organization of the H. influenzae encapsulation locus (16, 22). Although in vitro experiments suggest that exchange of encapsulation loci is possible, the clonal population structure of these bacteria argues that this event is uncommon. Additionally, we found that hmcA, the structural gene encoding the type b-specific bacteriocin hemocin, was absent in all non-type b encapsulated isolates, suggesting that none of these strains was related to Hib (data not shown).
It seems more likely, given these findings, that invasive non-type b strains arose from the evolutionary divergence of ancestral strains, through the selection of specific virulent phenotypes. Recently recovered members of one of the two major Hia lineages have RDPs identical or highly similar to those of an invasive strain isolated in the 1970s, suggesting that this virulent clone of bacteria has persisted and remained genetically stable since the early 1980s (data not shown). An increase in virulence could also have resulted from the progressive selection of fitness mutations over the past two decades or more abruptly by the acquisition of novel virulence genes by horizontal exchange between more virulent H. influenzae strains or other respiratory bacteria. Intragenic exchange of non-encapsulation locus genes may occur between Haemophilus or other bacterial strains that share an ecological niche, such as Neisseria meningitidis, but the frequency of such events in nature is currently unclear (5).
This study provides a genetic framework for studies of the pathogenesis of invasive non-type b H. influenzae infections by identifying the associations between strains of bacteria expressing the same capsular types. Comparison of phenotypic and genetic features of these lineages to one another and to noninvasive isolates may identify common factors contributing to human disease.
Acknowledgments
This work was supported by Cancer Center Support Core Grant CA-21765, CA-23944 (E.E.A.), AI-44167 (J.W.S.), and DC-02873 (J.W.S.), the American Heart Association (J.W.S.), the March of Dimes (J.W.S.), the American Lebanese-Syrian Associated Charities (ALSAC) (E.E.A.), and a summer research fellowship from the American Pediatric Society (C.L.O.).
We thank A. Smith, University of Missouri-Columbia, for helpful discussion. We are most grateful to the following individuals and agencies who contributed bacterial isolates to this study: E. Shapiro, Yale University School of Medicine; J. Kelner, Alberta Children's Hospital, Canada; J. Wong, Microbial Diseases Laboratory, State of California Department of Health Services; S. Mottice, Utah State Public Health Laboratory; K. Penterman, Minnesota Department of Health; M. Lytle, Oklahoma State Department of Health, Public Health Laboratory; N. Bradley, State Laboratory of Public Health, North Carolina; J. Beebe, Laboratory and Radiological Services, Colorado Department of Public Health and Environment; L. Cranidiotis, State of Alabama Department of Public Health; M. DeMartino, Hygienic Laboratory, University of Iowa; H. Hardin, Tennessee Department of Health Laboratory Services; M. Polya, State Laboratory of Hygiene, Wisconsin; and D. Byrd, Missouri State Health Department.
REFERENCES
- 1.Adderson, E. E., C. L. Byington, L. Spencer, A. Kimball, M. Hindiyeh, K. Carroll, S. Mottice, E. K. Korgenski, J. C. Christenson, and A. T. Pavia. 2001. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling H. influenzae type b: emerging pathogen in the vaccine era? Pediatrics 180:18.. [DOI] [PubMed] [Google Scholar]
- 2.Barenkamp, S. J., R. S. Munson, Jr., and D. M. Granoff. 1981. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J. Infect. Dis. 143:668-676. [DOI] [PubMed] [Google Scholar]
- 3.Butler, P. D., and E. R. Moxon. 1990. A physical map of the genome of Haemophilus influenzae type b. J. Gen. Microbiol. 136:2333-2342. [DOI] [PubMed] [Google Scholar]
- 4.Cerquetti, M., M. L. Ciofi degli Atti, G. Renna, A. E. Tozzi, M. L. Garlaschi, and P. Mastrantonio. 2000. Characterization of non-type b Haemophilus influenzae strains isolated from patients with invasive disease. J. Clin. Microbiol. 38:4649-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezekiel Moor, P., P. C. Collignon, and G. L. Gilbert. 1999. Pulsed-field gel electrophoresis used to investigate genetic diversity of Haemophilus influenzae type b isolates in Australia shows differences between Aboriginal and non-Aboriginal isolates. J. Clin. Microbiol. 37:1524-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Coton, T. R. Uttervack, M. C. Hanna, D. T. Nyugen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 8.Gazagne, L., C. Delmas, E. Bingen, and H. Dabernat. 1998. Molecular epidemiology of ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae. J. Clin. Microbiol. 36:3629-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granoff, D. M., S. J. Barenkamp, and R. S. Munson, Jr. 1982. Outer membrane protein subtypes for epidemiologic investigation of Haemophilus influenzae type b disease, p. 43-55. In S. H. Sell (ed.), Haemophilus influenzae: epidemiology, immunology and prevention of disease. Elsevier/North-Holland, New York, N.Y.
- 10.Hargreaves, R. M., M. P. Slack, A. J. Howard, E. Anderson, and M. E. Ramsay. 1996. Changing patterns of invasive Haemophilus influenzae disease in England and Wales after introduction of the Hib vaccination programme. Br. Med. J. 312:160-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilian, M., I. Sorenson, and W. Frederiksen. 1979. Biochemical characteristics of 130 recent isolates from Haemophilus influenzae meningitis. J. Clin. Microbiol. 9:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipitch, M. 1997. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl. Acad. Sci. USA 94:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LiPuma, J. J., H. Richman, and T. L. Stull. 1990. Haemocin, the bacteriocin produced by Haemophilus influenzae: species distribution and role in colonization. Infect. Immun. 6:1600-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, T. V., P. Pastor, F. Medley, M. T. Osterholm, and D. M. Granoff. 1993. Decreased Haemophilus colonization in children vaccinated with Haemophilus influenzae type b conjugate vaccine. J. Pediatr. 122:517-523. [DOI] [PubMed] [Google Scholar]
- 15.Murphy, T. V., K. E. White, P. Pastor, L. Gabriel, F. Medley, D. M. Granoff, and M. T. Osterholm. 1993. Declining incidence of Haemophilus influenzae type b disease since introduction of vaccination. JAMA 13:246-248. [PubMed] [Google Scholar]
- 16.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypeable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser, J. M., D. M. Granoff, P. E. Pattison, and R. K. Selander. 1985. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 82:5078-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings, W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 19.Nitta, D. M., M. A. Jackson, V. F. Burry, and L. C. Olson. 1995. Invasive Haemophilus influenzae type f disease. Pediatr. Infect. Dis. 14:157-160. [PubMed] [Google Scholar]
- 20.Perdue, D. G., L. R. Bulkow, B. G. Gellin, M. Davidson, K. M. Petersen, R. J. Singleton, and A. J. Parkinson. 2000. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination programs. JAMA 283:3089-3094. [DOI] [PubMed] [Google Scholar]
- 21.Pittman, M. 1931. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 53:471-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quentin, R., R. Ruimy, A. Rosenau, J. M. Musser, and R. Christen. 1996. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J. Clin. Microbiol. 34:1380-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito, M., A. Umeda, and S.-I. Yoshida. 1999. Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 37:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi, S., E. E. Adderson, Y. Nagano, N. Nagano, M. Briesacher, and J. F. Bohnsack. 1998. Identification of a highly encapsulated, genetically related group of invasive type III GBS. J. Infect. Dis. 177:1116-1119. [DOI] [PubMed] [Google Scholar]
- 25.Urwin, G., J. A. Krohn, K. Deaver-Robinson, J. D. Wenger, and M. M. Farley. 1996. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiologic characteristics in the H. influenzae serotype b vaccine era. Clin. Infect. Dis. 22:1069-1076. [DOI] [PubMed] [Google Scholar]