Abstract
BACKGROUND
One of the first proteins synthesized by a conceptus, is human chorionic gonadotropin (hCG). The receptor-binding β-subunit of hCG (hCGβ) is encoded by highly homologous CGB, CGB5, CGB7 and CGB8 genes. The function of two additional gene copies, CGB1 and CGB2, is still unknown.
AIM
We aimed to compare the expression of individual CGB genes during the normal pregnancy and in cases of recurrent miscarriages (RM) and ectopic pregnancy (EP)
METHODS
A semi-quantitative RT-PCR with fluorescent-labeled primers coupled with the gene-specific restriction and quantification was used.
RESULTS
The summarized expression of hCGβ genes was high throughout the pregnancy, and moderately correlated with hCG in serum. In cases of RM, reduced hormone values were consistent with low mRNA levels, whereas for EP no reduction in transcriptional activity was detected. CGB1 and CGB2 showed a considerable expressional peak during the first trimester, both in normal and ectopic pregnancy, but not for RM.
CONCLUSIONS
In case of RM, low hCG could result from expressional failure of hCGβ genes, whereas in EP the problems other than the transcriptional failure contribute to reduced hormone levels. The expression patterns of CGB1 and CGB2 suggest their putative role in the implantation stage.
Keywords: CGB gene expression, ectopic pregnancy, human chorionic gonadotropin, implantation, recurrent miscarriage
Introduction
Human chorionic gonadotropin (hCG) or „the hormone of pregnancy” is secreted mainly by the syncytiocytotrophoblasts of the placenta. HCG is indispensable for successful progression of pregnancy. Its classical function is to maintain the production of steroid hormones and other growth factors in the corpus luteum. In addition, hCG has been reported to modulate the blastocyst implantation (Srisuparp et al., 2001), uterine vascularization and angiogenesis (Zygmunt et al., 2002; Toth et al., 2001) uterine quiescence and immunological adaptation during pregnancy (Rao, 2001). The synthesis of hCG begins shortly after fertilization, the β-subunit of the hormone has been detected in the two-cell stage embryo (Jurisicova et al., 1999). After implantation, hCG is transported into maternal bloodstream where the maximum level is reached at 9–10 weeks of pregnancy. Concentration decreases from the 10th to the 16th week of gestation, falling to 10% of peak first trimester value. During the third trimester hCG level rises in a gradual, yet significant, manner from 22 weeks until term (Hay, 1988). Low levels of hCG during the first trimester of pregnancy is related to miscarriage, ectopic pregnancy and failure of vitro fertilization procedure (Buyalos et al., 1992; Gerhard and Runnebaum, 1984; Letterie and Hibbert, 2000; Poikkeus et al., 2002). Conversely, high concentration of hCG, especially β-subunit and their metabolites refers to gestational trophoblastic disease. hCG expression in non-pregnant state is a sensitive and specific marker of trophoblastic tumors and many non-trophoblastic malignancies (Dawood et al. 1977; Gaspard et al., 1980; Stenman et al.,2004).
HCG, like other gonadotropic glycoproteins, is composed of two subunits: a common α - and a hormone-specific β-subunit. A single gene located on chromosome 6q12-q21 encodes the α-subunit; a cluster of genes localized on chromosome 19q13.3 determines the β-subunit of hCG. The whole cluster consists of seven homologous genes: one luteinizing hormone beta (LHB) gene and six chorionic gonadotropin beta (CGB) genes that all originate from LHB gene as a result of duplication during primate evolution (Talmadge et al., 1984). The β-subunit of hCG (163 aa protein) coding CGB, CGB5, CGB7 and CGB8 share 97–99% DNA sequence identity; and to the functionally distinct LHB gene 92–93%.
Although CGB1 and CGB2 genes are similar to the other genes in the cluster (85% identity), they encode a novel hypothetical protein (132 aa in length) that does not have any homology to the functional β-subunit; neither corresponds to any known protein GenBank database. This change has been caused by an inserted DNA fragment within the 5′untranslated region (UTR) of CGB1 and CGB2 providing a novel exon 1 (Fig.1), and one basepair (bp) shifted open reading frame for exons 2 and 3. Earlier studies have detected mRNA of CGB1 and CGB2 in the placenta as well in the pituarity, predicting the functionality of these genes (Bo and Boime, 1992; Dirnhofer et al., 1996). Previously, two reports (Bo and Boime, 1992; Miller-Lindholm et al., 1997) have studied the expressional pattern of individual hCG β-subunit determining genes (CGB, CGB5, CGB7 and CGB8) in the first trimester placentas, single cases of complicated pregnancies (spontaneous abortion, blighted ovum, hydatidiform mole), and cultured JAR choriocarcinoma cells.
Figure 1.
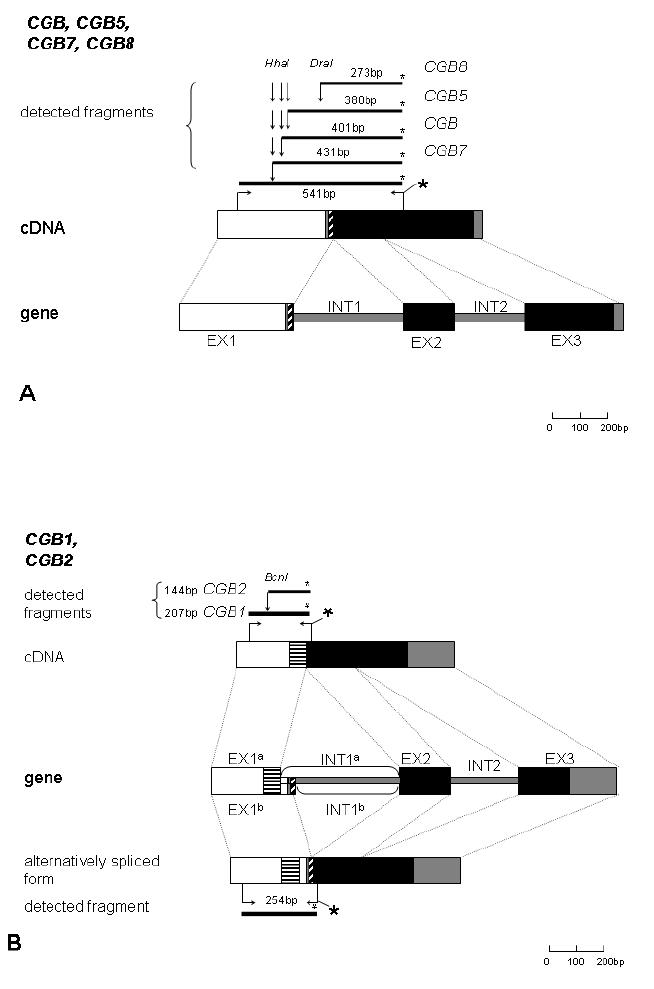
Schematic representation of the general organization of CGB genes, cDNA, amplification and the restriction producing the gene-specific fluorescent-labeled fragments: A, CGB, CGB5, CGB7, CGB8 and B, CGB1, CGB2.
The dark grey boxes represent non-coding segments with homologous DNA sequence, and the white boxes regions differing for the two sets of genes: CGB, CGB5, CGB7, CGB8 and CGB1, CGB2. Diagonally shaded boxes mark the coding region of the first exon that is included to mRNA of CGB, CGB5, CGB7, CGB8, and alternative spliced form of CGB1. Horisontally shaded boxes represent the coding region of the first exon specific to CGB1 and CGB2. Black boxes demonstrate the homologous coding area of exon 2 and 3 for all CGB genes. EX1a and INT1a mark the first exon and intron of CGB1 and CGB2, and EX1b and INT1b the same of alternatively spliced form of the CGB1. The horizontal arrows indicate the binding sites for the forward and the fluorescent-labeled (*) reverse primers. The bold lines represent the appropriate PCR product of length in basepairs. The vertical arrows indicate the restriction enzyme sites for HhaI, DraI and BsnI.
In the study reported here, we aimed to examine mRNA expression of all the six CGB genes, and the single LHB in parallel with hCG concentration during the course of normal pregnancy. As the changes in mRNA transcription of CGB genes may play a role in implantation failure, we also investigated the tissue samples from patients with recurrent (≥3) spontaneous or missed abortion, and patients with ectopic pregnancy.
Materials and Methods
Experimental Subjects
The study was approved by The Ethics Committee of the University of Tartu, Estonia (protocol no. 117/9, 16.06.03), and an informed consent was obtained from every participant.
Serum and chorionic villi/placental samples were obtained from females who underwent
elective therapeutic abortion during I trimester of pregnancy (4–12 weeks of gestational age, n=11);
therapeutic abortion during II trimester due to medical risks of pregnancy, no fetal anomalies were detected (17–21 weeks of gestational age, n=8);
normal delivery at term resulting from uncomplicated pregnancy (38–42 weeks of gestational age, n=12);
surgical treatment of ectopic pregnancy (6–14 weeks of gestational age, n=6);
cervical dilatation and uterine curettage because of recurrent (≥ third case in a row) incomplete or missed abortion (6–9 weeks of gestational age, n=7)
As chromosomal aberrations of an incidental origin contribute to first trimester miscarriage in nearly 70% of cases (Fritz et al., 2001), we included only the cases of recurrent miscarriages (having ≥2 spontaneous abortions preceeding current event) when the alternative mechanisms leading to pregnancy loss are more relevant. All couples had a normal karyotype and as a result of clinical evaluation (anatomical, immunological, hormonal, blood coagulation assessment) showed the unexplained cause for miscarriage.
The blood samples were taken on the day of abortion, delivery or surgery and frozen at −20°C. A total hCG in the serum was measured by chemiluminescent enzyme immunometric assay IMMULITE at United Laboratories, Tartu University Clinics. Chorionic villi/placental samples were snap-frozen in liquid nitrogen and stored at −80 °C until RNA isolation.
RNA extraction and cDNA synthesis
Total RNA from 90–1000 mg of tissue was extracted using TRIzol® reagent (Invitrogen, Carlsbad, CA) For purification of RNA, NucleoSpin® II (Macherey-Nagel) Isolation Kit was used. According to the protocol, RNAse-free DNAse I was used to remove contamination with genomic DNA. The products were quantified by measuring absorption ratios at 260/280 nm (Heλios Alpha&Beta Spectrophotometer, Thermo Spectronic).
RNA was reverse transcribed in a final volume of 20 μl containing total 1 μg of RNA, reaction buffer (50 mM Tris-HCl [pH 8.3 at 25 °C], 50 mM KCl, 4mM MgCl2, 10 mM DTT), 20U of RiboLock™ Ribonuclease Inhibitor, 0.5 mM of each dNTP, 0.5 μg of oligo(dT)18 primer, 40 U M-MuLV Reverse Transcriptase (First Strand cDNA Synthesis Kit, Fermentas Life Sciences). The samples were initially incubated at 70 °C for 5 min, the reverse transcription reaction was performed at 37 °C for 60 min, and stopped by heating at 70 °C for 10 min to inactivate the reverse transcriptase.
Oligonucleotide primer design
RT-PCR primers (Table I) were designed taking into account polymorphic positions in CGB genes defined by recent resequencing study (95 worldwide individuals) in our laboratory (Hallast, Nagirnaja, Margus and Laan, unpublished). The primers for co-amplification of the hCG β-subunit determining genes CGB, CGB5, CGB7 and CGB8 were positioned to the different exons with the criteria to include the restriction sites specific to each gene (Lazar et al., 1995; Miller-Lindholm et al., 1997) (Fig. 1). For co-amplification of CGB1 and CGB2 we used the mixture of two reverse primers differing in 1 bp (Giovangrandi et al., 2001), and designed a forward primer to obtain the PCR product covering the CGB2-specific BcnI restriction site. For normalization we amplified a widely used reference gene of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), with previously reported primers (Miller-Lindholm et al., 1997). An alternative reference gene, largest subunit of RNA polymerase II (RPII) was selected based on a recent methodological report comparing reference genes for expression studies (Radonic et al., 2004). For RPII amplification, we used the reported reverse primer (Radonic et al., 2004) coupled with a novel forward primer, in order to obtain a PCR product sized to be distinguishable during the GeneScan analysis. The RT-PCR of LHB was attempted with three primer pairs, including one reported (Giovangrandi et al., 2001) and two designed in this study to obtain a larger PCR product that could be distinguishable better during the GeneScan analysis.
Table 1.
The nucleotide sequences and fluorescent labels of the primers for RT-PCR of LHB/CGB and reference genes.
| Gene | Primer | ||||
|---|---|---|---|---|---|
| F/R | Sequence with Fluorescent Dye* | Tm (ºC) | Product size | ||
| CGB | } | F | 5′-GACCCCACCATAGGCAGAG-3′ | 61 | 541bp |
| CGB5 | |||||
| CGB7 | R | 5′-GGTAGTTGCACACCACCTGA-3′ FAM | 59.4 | ||
| CGB8 | |||||
| CGB1 | } | F | 5′-GGAGGGAGGAAGGGGAACT-3′ | 61 | 207bp |
| CGB2 | |||||
| CGB1 | RG | 5′-GCAACAGCAGCAGCCTCTTT-3′ FAM | 59.4 | ||
| CGB2 | RG | 5′-CAACAGCAGCAGCCCCTTT-3′ FAM | 58.8 | 206bp | |
| GAPDH | FML | 5′-CCATGGAGAAGGCTGGGG-3′ | 60.5 | 196bp | |
| RML | 5′-CCAAGTTGTCATGGATGACC-3′ HEX | 57.3 | |||
| RPII | F | 5′-CTTCACGGTGCTGGGCATT-3′ | 58.8 | 239bp | |
| RR | 5′-GTGCGGCTGCTTCCATAA-3′ TET | 56 | |||
| LHB** | FG | 5′-GCTACTGCCCCACCATGATG-3′ | 61.4 | 94bp | |
| R1G | 5′-ATGGACTCGAAGCGCACATC-3′ FAM | 59.4 | |||
| R2 | 5′-AGAGCCACAGGGAAGGAGAC-3′ FAM | 61.3 | 151bp | ||
| R3 | 5′-AGCTGAGAGCCACAGGGAAG-3′ FAM | 61.4 | 156bp | ||
Tm, melting temperature; F, forward primer; R, reverse primer; a primer originally published by
Radonic et al.
6-FAM, TET and HEX emit fluorescent signals at 532nm, 543nm, 557nm of wavelengths, respectively.
For LHB, one forward primer was combined with different reverse primers.
The 5′-end of reverse primers were labeled with an ABI compatible fluorescent dye (6-FAM, TET, HEX (Metabion International AG, Deutschland). All primer sequences are indicated in Table I.
RT-PCR amplification
The RT-PCR mixture in total volume of 25 μl consisted of 1 μl first-strand reaction product, 1 U Smart-Taq Hot DNA Polymerase (AppliChem GmbH), 2.5 mM MgCl2, 0.2 mM of each dNTP (Fermentas Life Sciences), 2.5 μl 10x PCR reaction buffer containing (NH4)2 SO4 and 400 nmol of forward and reverse primers. Each cDNA sample was also the amplification target for the two reference genes GAPDH and RPII under the same conditions. Amplification was attained by GeneAmp PCR System 2700 (Applied Biosystems) thermal cycling for 1 cycle of 95 °C for 15 min to denature, then 10 cycles of 95 °C for 20 sec to denature, and from 61 °C to 52 °C for 30 sec („touch-down”) to anneal, and 72 °C for 2 min to extend, followed by 20 cycles (GAPDH and CGB, CGB5, 7,8) or 25 cycles (RPII) or 30 cycles (CGB1/2 and LHB) of 95 °C for 20 sec, 51 °C for 30 sec and 72 °C for 2 min. The reaction was ended by final incubation of 72 °C for 10 min. The number of cycles needed for amplification for each assay was optimized to ensure that PCR remained in the logarithmic phase of amplification (data not shown).
Restriction and GeneScan analysis
The experimental scheme for discrimination of the RT-PCR products of CGB, CGB5, CGB7 and CGB8 was originally described by Lazar et al 1995. 5 μl of RT-PCR product containing a mixture of CGB, CGB5, CGB7 and CGB8 was digested with 1 U HhaI (Fermentas Life Sciences) and 1 U DraI (Fermentas Life Sciences), which discriminate divergence in the 5′UTR resulting in the gene-specific fluorescent-labeled products of 273bp for CGB8, 380bp for CGB5, 401bp for CGB and 431bp for CGB7. Discrimination of CGB1 and CGB2 was solved by a similar approach. The 5 μl of the combined product of CGB1/2 was treated with 1U BcnI (Fermentas Life Sciences) resulting in fluorescent fragments of 207 bp for CGB1, and restricted 144 bp for CGB2. The restriction analysis was repeated twice in independent reactions to confirm the full digestion.
The mixture of digested CGB genes and reference gene products (GAPDH and RPII) in equal volumes was combined with formamide containing fluorescently labeled GeneScan-500 TAMRA internal size standard (Applied Biosystems-Perkin Elmer), and electrophoresed through a denaturing 6% polyacrylamide gel in 0.5x TBE buffer in an Applied Biosystems™ 373 DNA Sequencer.
The gel was analyzed by GeneScan 2.1 software that allows determining the length of the fragment in the each lane. The intensity of the fluorescent dye derived from labeled primers and expressed the absorbance intensity in relative fluorescence units. The results on an electrophoretogram show both, height and area of the peaks corresponding to the gene-specific PCR product. Examples of different types of gene expression patterns are demonstrated in GeneScan electrophoretogram in Fig 2.
Figure 2.
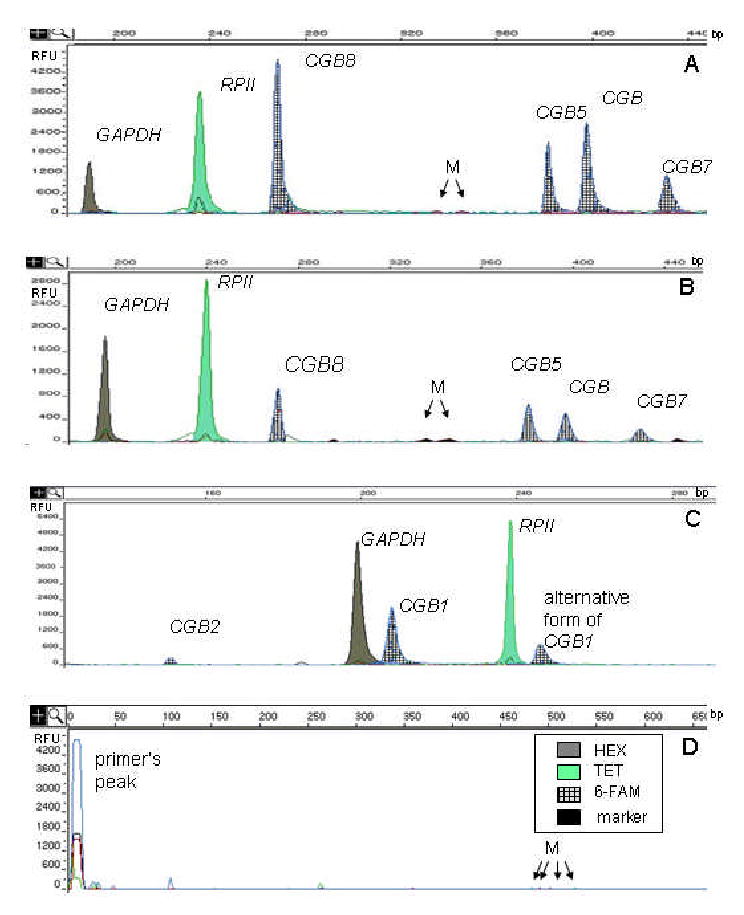
GeneScan electrophoretograms showing the amplified fluorescent-labeled (6-FAM, HEX or TET) products of reference genes and CGB genes after treatment by restriction enzymes. The x-axis shows the size of the detected fragments in base pairs (bp), and the y-axis represents the relative intensity of fluorescence (RFU). M, marks the peaks of the GeneScan-500 TAMRA internal size standard. The large peaks indicate the DNA fragments: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) at 196bp, RNA polymerase II largest subunit (RP II) at 239bp, CGB8 at 273bp, CGB5 at 380bp, CGB at 401bp, CGB 7 at 433bp, CGB1 at 207bp, CGB2 at 144bp, and alternatively spliced CGB1 at 254bp.
A, The expression pattern of hCG beta subunit determining genes in placenta from III trimester (41weeks) of normal pregnancy: CGB8>>CGB>CGB5>CGB7. B, The trophoblastic tissue from ectopic pregnancy (6weeks) expresses the genes: CGB8>CGB5>CGB>CGB7. C, The trophoblastic tissue from ectopic pregnancy (7 weeks) expresses CGB1, CGB2, and alternatively spliced CGB1. D, Negative control.
Sample target evaluation and statistical analysis
All samples were amplified in triplicate, and each PCR product combination (reference and target genes, internal markers) was electrophoresed at least twice in order to minimize the effect of researcher’s errors in manual loading of the sample, gel preparation or gel electrophoresis. As the correlation between values of the peak height and area (both in relative fluorescent units) from all experiments was high (r=0.82, p<0.0001), only values of peak height were used in analyses. The relative expressional level of each gene was calculated as followed: the value of the peak height (in relative fluorescent units) of each target gene was divided with the value of peak height of the reference gene (GAPDH or RPII) from the same lane of the gel.
For statistical analysis a Pearson correlation test and a random effects linear regression model, allowing a patient-level random intercept were used. All statistics were performed using R 2.0.1, a free software environment for statistical computing and graphics (http://www.r-project.org/).
Results
The expression of hCG beta-subunit determining genes
The summarized expression patterns of CGB, CGB5, CGB7, and CGB8 during normal pregnancy and two patient groups (ectopic pregnancy, EP; recurrent miscarriages, RM) are shown in Fig.3A,B and by each individual gene in Table II. mRNA of CGB, CGB5 and CGB8 were present in placental material of all studied individuals, whereas CGB7 expression was below detection limit in 28 cases of the total 210 of observations. None of the studied groups stood out dominantly in lacking CGB7 expression, the contribution of CGB7 to the hCG β-subunit determining transcription product varied 4.7–8.1% across study groups (Table II). In contrast to an extreme peak of hCG hormone production during the I trimester (Fig. 3C), the β-subunit coding genes are expressed continuously at the same level during the whole pregnancy, with only a small decrease in the II trimester. Thus, the expression level of β-subunit determining genes in placenta showed a moderate but significant correlation to hCG concentration in the serum (r=0.44, p<0.0001). Consistent with a previous report (Miller-Lindholm et al., 1997), the variation among individuals within the same study group was large for mRNA levels of all hCG β-subunit coding genes (Fig. 3A,B). In cases of RM, the expression levels of CGB, CGB5, CGB7 and CGB8 were significantly lower than expected for the normal first trimester placenta, being adjusted to gestational age and hCG concentration (p=0.017) (Fig. 3A,B). Notwithstanding the low values of hCG in the serum (Fig. 3C), the summarized level of mRNA of hCG β-subunit coding genes in cases of EP was comparable with the expression during normal implantation to uterus. The most prevalent contribution pattern of individual genes to hCG β-subunit production for both normal and complicated cases, was CGB8 > CGB5 ≈ CGB >> CGB7 (Table II). For the III trimester placentas, the contribution pattern of individual genes were slightly altered, CGB8≈CGB5>CGB>>CGB7 (Table II). Compared to the I trimester, the expression level of CGB5 was increased (p<0.05) and of CGB decreased (p<0.005).
Table 2.
Individual contribution (percentage and standard deviation) of CGB, CGB5, CGB7 and CGB8 into summarized chorionic gonadotropin beta subunit determining placental mRNA during the normal pregnancy, in cases of ectopic pregnancy (EP) and recurrent miscarriage (RM).
| Contribution (%) of an individual gene expression in total hCG beta-subunit mRNA
|
||||||||
|---|---|---|---|---|---|---|---|---|
| CGB8 | CGB5 | CGB | CGB7 | |||||
| mean ± SD | range | mean ± SD | range | mean ± SD | range | mean ± SD | range | |
| I trimester | 39.3±1.8 | 29–63 | 25.5±2.7 | 6–55 | 27.1±1.5 | 11–39 | 8.1±1.2 | 0–21 |
| II trimester | 48.1±2.8*** | 28–86 | 25.7±4.2 | 9–45 | 20.0±2.3*** | 1–29 | 6.2±1.8 | 0–16 |
| III trimester | 39.2±2.5 | 24–71 | 36.0±3.8** | 7–62 | 20.1±2.1*** | 2–32 | 4.7±1.6** | 0–19 |
| ectopic pregnancy | 48.0±3.2** | 31–62 | 18.7±4.7 | 7–28 | 25.3±2.6 | 14–54 | 8.0±2.1 | 0–17 |
| recurrent miscarriage | 47.1±3.7** | 28–56 | 23.3±4.9 | 13–34 | 22.0±2.8* | 17–27 | 7.6±2.2 | 0–13 |
p<0.005
p<0.05
p<0.08: Significant difference of the expression level of each gene during II and III trimester placentas as well as in cases of EP and RM compared to I trimester of normal pregnancy as a reference level.
Figure 3.
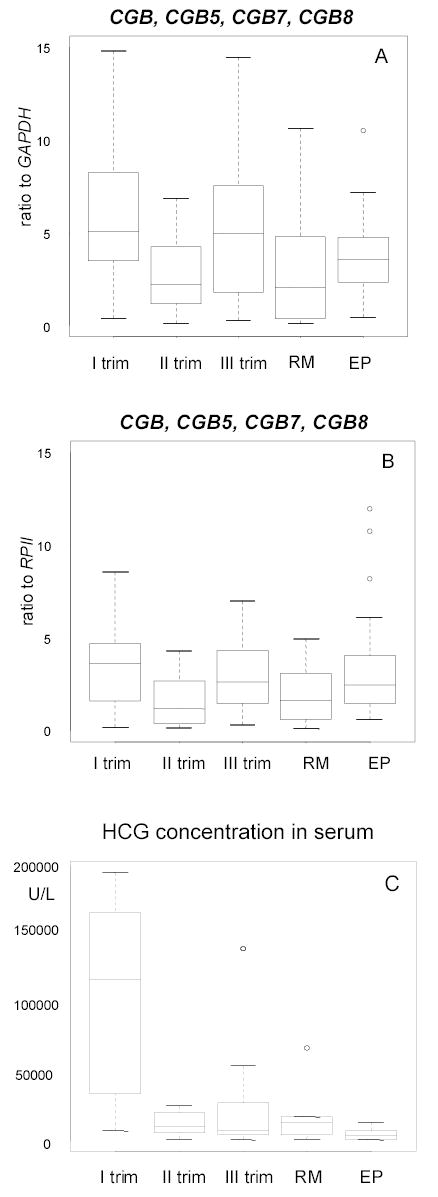
Notched boxplot for the distribution of the summarized expression level of hCG beta subunit mRNAs from CGB, CGB5, CGB7 and CGB8 in placenta during the first, second and trimester of normal pregnancy, in case of recurrent miscarriage (RM) and ectopic pregnancy (EP). The boxes represent the 25th and 75th percentiles. The median is denoted as the line that bisects the boxes. The whiskers are lines extending from each end of the box covering the extent of the data on 1.5 X interquartile range. Circles represent the outlier values.
A, Results in relative to reference gene GAPDH.
B, Results in relative to reference gene RPII. A strong correlation (r=0.6, p<0.001) was found between A and B.
C, hCG concentration in the serum (U/L) measured on the day of tissue sampling in same groups is shown. The level of hCG during the first trimester of normal pregnancy is significally higher compared to recurrent miscarriage and ectopic pregnancy (p<0.005).
The expression of CGB1 and CGB2 genes
The mRNA expression of CGB1 and CGB2 was low (Fig. 4A–D). A reliable peak above non-specific background for CGB1 was detectable in 80% of specimen, and for only in 50% of observations for CGB2. Although we did not succeed to demonstrate the statistically significant differences between the study groups, the results differed from the expression pattern of CGB, CGB5, CGB7 and CGB8. A clear expression peak was visible during the first trimester of normal pregnancy as well as for EP (Fig. 4A–D). For RM the levels of CGB1 and 2 were at the borderline of detection. In addition, for all study groups we detected an extra peak at 254bp that corresponds to an alternatively spliced product of CGB1, and similarly for RM it was close to the detection limit (Fig.1, 2, 4E,F). The expression of this alternative product correlated moderately with the major CGB1 product (r=0.44, p<0.0001), and in most cases the transcription level of the alternative product exceeded the level of ”original” CGB1. We could not detect an alternative splicing of CGB2 (expected fragment size 191 bp in length on GeneScan analysis).
Figure 4.
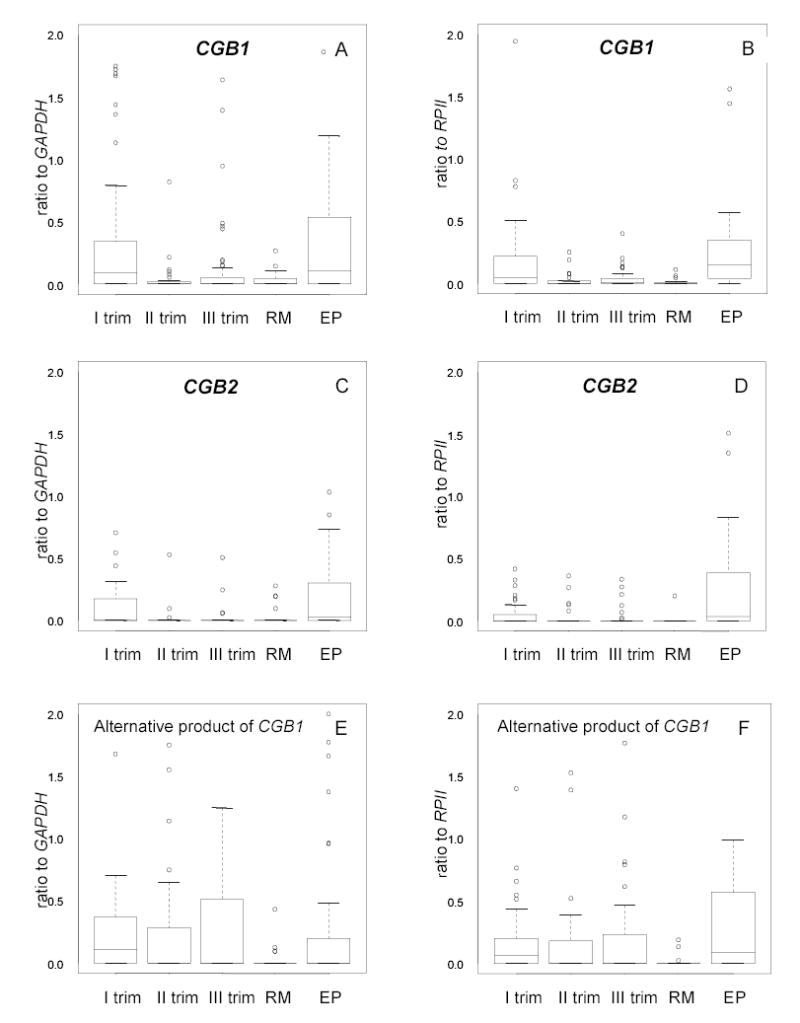
Notched boxplot for the distribution of expressional level of CGB1, CGB2 and alternatively spliced product of CGB1 in the placenta during the first, second and trimester of normal pregnancy, in case of recurrent miscarriage (RM) and ectopic pregnancy (EP).
The boxes represent the 25th and 75th percentiles. The median is denoted as the line that bisects the boxes. The whiskers are lines extending from each end of the box covering the extent of the data on 1.5 X interquartile range. Circles represent the outlier values.
A, C, E, Results in relative to reference gene GAPDH. B, D, F, Results relative to reference gene RPII. A strong correlation existed between the results obtained in experiments using GAPDH or RPII (r=0.71, p<0.0001).
The expression of LHB gene
We failed to demonstrate the presence of LHB during normal pregnancy as well as in complicated cases (EP, RM) using three alternative primer pairs. Therefore we conclude that the expression of LHB gene is fully down regulated during pregnancy, or remained undetectable by the semi-quantitative approach used in this study.
Discussion
The purpose of the study was to investigate the expressional profile of fertility and implantation related genes of LHB/CGB gene cluster in placenta during normal pregnancy, and in cases of ectopic pregnancy and recurrent miscarriage. We used and modified a semi-quantitative method (Miller-Lindholm et al., 1997) that employed RT-PCR followed by restriction enzyme digestion and GeneScan analysis of fluorescent-labeled products of CGB genes. The co-amplification of the cDNAs for a set of genes (here CGB, CGB5, CGB7, CGB8; and CGB1, CGB2) in one RT-PCR reaction guarantees equal amplification conditions and allows reliable comparison of expression levels of the target genes in a sample. Inclusion of two housekeeping reference genes (GAPDH and RPII) into the analysis enabled to make comparisons among the individuals and study groups. In order to overcome the variation in tissue sample quality, mRNA preparation and cDNA synthesis, the reference genes were amplified for each individual sample from the same cDNA preparation as the target genes, and co-electrophoresed during GeneScan analysis on the same lane with LHB/CGB genes.
Despite high DNA sequence similarity between individual hCG β-subunit coding genes, our study demonstrated high variation in expression levels, both among the genes themselves as well as among the individuals. The expressional variation between CGB, CGB5, CGB7 and CGB8 could result from a few divergent nucleotides located in the 5′-UTR or upstream region of the genes that serve as regulatory variants influencing the gene expression. The most divergent from the other genes, and also with the lowest expression level, is CGB7, with approximately 10 gene-discriminating nucleotides 330 bp upstream of exon 1 as well as 3 amino acid changes. The contribution of individual genes to hCG β-subunit production differed to some extent from previous studies (Bo and Boime, 1992; Miller-Lindholm et al., 1997), which reported CGB5 as the most actively transcribed gene in contrast to CGB8 in our study. The among-individual variation could be caused by either polymorphic variation of the studied genes and/or an individual’s differences in the process of trophoblast differentiation. A resequencing study of LHB/CGB genes has identified extremely high diversity level of these genes, most probably favored by the spread of polymorphisms by gene conversion between highly similar genes (Hallast, Nagirnaja, Margus and Laan, unpublished). Thus, there is an abundance of CGB, CGB5, CGB7 and CGB8 gene variants and their combinations carried by individuals in the population. On the other hand, the expression of hCG hormone β-subunit is associated with the fusion of cytotrophoblasts into a multinuclear syncytium (Maruo et al., 1992). During syncytialization selective promoter factors (Sp) and activating protein 2 (AP2) located in the proximal promoter region interact in increasing amounts with transcription factors Sp1, Sp3 and AP-2α, which enhance transcription of hCG β-subunit genes in differentiating term trophoblasts (Knofler et al., 2004; Maruo et al., 1992). In early placentas a different combination of factors may control β-subunit expression.
We detected an expression peak of both CGB1 and CGB2 during I trimester of normal pregnancy and EP, but not in placentas of the II, III trimester and RM. Thus, the pattern differed from that we found in expression of hCG β-subunit coding genes CGB, CGB5, CGB7 and CGB8. At the beginning of implantation and placentation, the invasion of the placental bed occurs in the tube in the same manner as it does in the uterus, suggesting that the ability to implant and form the normal placenta is determined by trophoblastic tissue of fetal origin rather than by maternal tissues (Randall et al., 1987). In contrast, miscarriaged pregnancies are related to poor trophoblast invasion (Lyall, 2002; Meegdes et al., 1988). By the second and third trimester the process of implantation has been replaced by the growth and maturation of the placenta. Our data on expression patterns of CGB1 and CGB2 suggest that these genes might have a role in implantation and placentation. As the protein product of CGB1 and CGB2 is still not characterized, further studies are needed to understand the function of these genes.
For CGB1 we detected an alternative spliced product, which arises when the splicing occurs at the consensus splice donor site of LHB, CGB and CGB5, CGB7 and CGB8 genes (Fig.1). The extra 47bp long sequence from the first intron is added into the transcript. In previous studies, the presence of alternatively spliced transcript of either CGB1 or CGB2 has been demonstrated in placenta (Bo and Boime, 1992) but not in pituitaries (Dirnhofer et al., 1996).
Is the expressional profile of LHB/CGB genes prognostic to pregnancy outcome? Indeed, recurrent miscarriages were characterized by a low level of mRNA of hCG β-subunit coding genes, and almost lacking CGB1 and CGB2 expression, supporting the hypothesis that miscarriages could be contributed by the expressional failure of CGB genes. Multiple scenarios for the expressional failure have been proposed such as polymorphisms affecting hormone glycosylation, as well as interaction with transcription factors, and genomic rearrangements of the LHB/CGB cluster due to highly homologous segments.hed data). For ectopic pregnancies, expression of CGB genes comparable with normal first trimester pregnancy contrasts with the low concentration of hCG in the serum, indicating the problems with stable hormone assembly and transport rather than pathological CGB variants and low transcriptional activity.
During implantation, trophoblasts exhibit the same behavior as tumor cells, such as tissue invasiveness, metastasis, loss of contact inhibition, escape from immune surveillance, and massive proliferative ability (Willey, 1999). The breast, ovarium, bladder, lung, renal and some other malignancies have been reported to express the hCG β-subunit coding genes CGB, CGB5 and CGB8 in contrast to poorly expressed CGB7 (Bellet et al., 1997; Giovangrandi et al., 2001; Lazar et al., 1995; Span et al., 2003; Span et al., 2002). As the prognostic value of CGB gene expression in predicting tumor progression is still controversial (Bieche et al., 1998; Hotakainen et al., 2002), detailed studies on the expression patterns of total mRNA levels of CGB genes as well as on the contribution of each individual gene, are needed.
In conclusion, as the main results of the study we would like to highlight the detection of high expression of CGB, CGB5 and CGB8 in the placenta throughout pregnancy with a minor decrease during the II trimester; significant reduction of CGB genes expression in case of recurrent miscarriage; low placental expressional activity of CGB7, possibly diverging from its current function; a putative role of CGB1 and CGB2 in implantation and placentation; and the presence of an alternatively spliced product of CGB1.
Acknowledgments
We thank all the patients, who participated in the study; dr. Helle Karro for providing facilities for patient material collection at the Tartu University Clinics, Women’s Clinic Estonia; Viljo Soo for his skillful technical assistance with running the GeneScan gels; drs. Tarmo Annilo, Gunnar Tasa, Elin Lõhmussaar and Laura Sedman for commenting the manuscript. We are also grateful to dr. Krista Fischer for helping with statistical analysis and graphics.
Footnotes
The study has been supported by Estonian Science Foundation (grant no. 5796) and Estonian Ministry of Education and Science (Core grant no. 0182641s04). K.R is a recipient of a stipend from the World Federation of Scientists’.M.L. is a recipient of Wellcome Trust International Senior Research Fellowship (grant no. 070191/Z/03/Z) in Biomedical Science in Central Europe.
References
- Bellet D, Lazar V, Bieche I, Paradis V, Giovangrandi Y, Paterlini P, Lidereau R, Bedossa P, Bidart JM, Vidaud M. Malignant transformation of nontrophoblastic cells is associated with the expression of chorionic gonadotropin beta genes normally transcribed in trophoblastic cells. Cancer Res. 1997;57:516–523. [PubMed] [Google Scholar]
- Bieche I, Lazar V, Nogues C, Poynard T, Giovangrandi Y, Bellet D, Lidereau R, Vidaud M. Prognostic value of chorionic gonadotropin beta gene transcripts in human breast carcinoma. Clin Cancer Res. 1998;4:671–676. [PubMed] [Google Scholar]
- Bo M, Boime I. Identification of the transcriptionally active genes of the chorionic gonadotropin beta gene cluster in vivo. J Biol Chem. 1992;267:3179–3184. [PubMed] [Google Scholar]
- Buyalos RP, Glassman LM, Rifka SM, Falk RJ, Macarthy PO, Tyson VJ, DiMattina M. Serum beta-human chorionic gonadotropin, estradiol and progesterone as early predictors of pathologic pregnancy. J Reprod Med. 1992;37:261–266. [PubMed] [Google Scholar]
- Dawood MY, Saxena BB, Landesman R. Human chorionic gonadotropin and its subunits in hydatidiform mole and choriocarcinoma. Obstet Gynecol. 1977;50:172–181. [PubMed] [Google Scholar]
- Dirnhofer S, Hermann M, Hittmair A, Hoermann R, Kapelari K, Berger P. Expression of the human chorionic gonadotropin-beta gene cluster in human pituitaries and alternate use of exon 1. J Clin Endocrinol Metab. 1996;81:4212–4217. doi: 10.1210/jcem.81.12.8954017. [DOI] [PubMed] [Google Scholar]
- Fritz B, Hallermann C, Olert J, Fuchs B, Bruns M, Aslan M, Schmidt S, Coerdt W, Muntefering H, Rehder H. Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)-Re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur J Hum Genet. 2001;9:539–547. doi: 10.1038/sj.ejhg.5200669. [DOI] [PubMed] [Google Scholar]
- Gaspard UJ, Reuter AM, Deville JL, Vrindts-Gevaert Y, Bagshawe KD, Franchimont P. Serum concentration of human chorionic gonadotropin and its alpha and beta subunits. 2. Trophoblastic tumours. Clin Endocrinol (Oxf) 1980;13:319–329. doi: 10.1111/j.1365-2265.1980.tb03391.x. [DOI] [PubMed] [Google Scholar]
- Gerhard I, Runnebaum B. Predictive value of hormone determinations in the first half of pregnancy. Eur J Obstet Gynecol Reprod Bio. 1984;17:1–17. doi: 10.1016/0028-2243(84)90075-3. [DOI] [PubMed] [Google Scholar]
- Giovangrandi Y, Parfait B, Asheuer M, Olivi M, Lidereau R, Vidaud M, Bieche I. Analysis of the human CGB/LHB gene cluster in breast tumors by real-time quantitative RT-PCR assays. Cancer Lett. 2001;168:93–100. doi: 10.1016/s0304-3835(01)00496-7. [DOI] [PubMed] [Google Scholar]
- Hay DL. Placental histology and the production of human choriogonadotrophin and its subunits in pregnancy. Br J Obstet Gynaecol. 1988;95:1268–1275. doi: 10.1111/j.1471-0528.1988.tb06817.x. [DOI] [PubMed] [Google Scholar]
- Hotakainen K, Ljungberg B, Paju A, Rasmuson T, Alfthan H, Stenman UH. The free beta-subunit of human chorionic gonadotropin as a prognostic factor in renal cell carcinoma. Br J Cancer. 2002;86:185–189. doi: 10.1038/sj.bjc.6600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurisicova A, Antenos M, Kapasi K, Meriano J, Casper RF. Variability in the expression of trophectodermal markers beta-human chorionic gonadotrophin, human leukocyte antigen-G and pregnancy specific beta-1 glycoprotein by the human blastocyst. Hum Reprod. 1999;14:1852–1858. doi: 10.1093/humrep/14.7.1852. [DOI] [PubMed] [Google Scholar]
- Knofler M, Saleh L, Bauer S, Galos B, Rotheneder H, Husslein P, Helmer H. Transcriptional regulation of the human chorionic gonadotropin beta gene during villous trophoblast differentiation. Endocrinology. 2004;145:1685–1694. doi: 10.1210/en.2003-0954. [DOI] [PubMed] [Google Scholar]
- Lazar V, Diez SG, Laurent A, Giovangrandi Y, Radvanyi F, Chopin D, Bidart JM, Bellet D, Vidaud M. Expression of human chorionic gonadotropin beta subunit genes in superficial and invasive bladder carcinomas. Cancer Res. 1995;55:3735–3738. [PubMed] [Google Scholar]
- Letterie GS, Hibbert M. Serial serum human chorionic gonadotropin (hCG) levels in ectopic pregnancy and first trimester miscarriage. Arch Gynecol Obstet. 2000;263:168–169. doi: 10.1007/s004040050275. [DOI] [PubMed] [Google Scholar]
- Lyall F. The human placental bed revisited. Placenta. 2002;23:555–562. doi: 10.1053/plac.2002.0850. [DOI] [PubMed] [Google Scholar]
- Maruo T, Ladines-Llave CA, Matsuo H, Manalo AS, Mochizuki M. A novel change in cytologic localization of human chorionic gonadotropin and human placental lactogen in first-trimester placenta in the course of gestation. Am J Obstet Gynecol. 1992;167:217–222. doi: 10.1016/s0002-9378(11)91661-5. [DOI] [PubMed] [Google Scholar]
- Meegdes BH, Ingenhoes R, Peeters LL, Exalto N. Early pregnancy wastage: relationship between chorionic vascularization and embryonic development. Fertil Steril. 1988;49:216–220. doi: 10.1016/s0015-0282(16)59704-0. [DOI] [PubMed] [Google Scholar]
- Miller-Lindholm AK, LaBenz CJ, Ramey J, Bedows E, Ruddon RW. Human chorionic gonadotropin-beta gene expression in first trimester placenta. Endocrinology. 1997;138:5459–5465. doi: 10.1210/endo.138.12.5618. [DOI] [PubMed] [Google Scholar]
- Poikkeus P, Hiilesmaa V, Tiitinen A. Serum HCG 12 days after embryo transfer in predicting pregnancy outcome. Hum Reprod. 2002;17:1901–1905. doi: 10.1093/humrep/17.7.1901. [DOI] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Randall S, Buckley CH, Fox H. Placentation in the fallopian tube. Int J Gynecol Pathol. 1987;6:132–139. doi: 10.1097/00004347-198706000-00005. [DOI] [PubMed] [Google Scholar]
- Rao CV. Tropic effects of LH and hCG on early pregnancy events in women’s reproductive tract. Early Pregnancy. 2001;5:18–19. [PubMed] [Google Scholar]
- Span PN, Manders P, Heuvel JJ, Thomas CM, Bosch RR, Beex LV, Sweep CG. Molecular beacon reverse transcription-PCR of human chorionic gonadotropin-beta-3, -5, and -8 mRNAs has prognostic value in breast cancer. Clin Chem. 2003;49:1074–1080. doi: 10.1373/49.7.1074. [DOI] [PubMed] [Google Scholar]
- Span PN, Thomas CM, Heuvel JJ, Bosch RR, Schalken JA, vd Locht L, Mensink EJ, Sweep CG. Analysis of expression of chorionic gonadotrophin transcripts in prostate cancer by quantitative Taqman and a modified molecular beacon RT-PCR. J Endocrinol. 2002;172:489–495. doi: 10.1677/joe.0.1720489. [DOI] [PubMed] [Google Scholar]
- Srisuparp S, Strakova Z, Fazleabas AT. The role of chorionic gonadotropin (CG) in blastocyst implantation. Arch Med Res. 2001;32:627–634. doi: 10.1016/s0188-4409(01)00330-7. [DOI] [PubMed] [Google Scholar]
- Stenman UH, Alfthan H, Hotakainen K. Human chorionic gonadotropin in cancer. Clin Biochem. 2004;37:549–561. doi: 10.1016/j.clinbiochem.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Talmadge K, Vamvakopoulos NC, Fiddes JC. Evolution of the genes for the beta subunits of human chorionic gonadotropin and luteinizing hormone. Nature. 1984;307:37–40. doi: 10.1038/307037a0. [DOI] [PubMed] [Google Scholar]
- Toth P, Lukacs H, Gimes G, Sebestyen A, Pasztor N, Paulin F, Rao CV. Clinical importance of vascular LH/hCG receptors - a review. Reprod Biol. 2001;1:5–11. [PubMed] [Google Scholar]
- Willey KP. An elusive role for glycosylation in the structure and function of reproductive hormones. Hum Reprod Update. 1999;5:330–355. doi: 10.1093/humupd/5.4.330. [DOI] [PubMed] [Google Scholar]
- Zygmunt M, Herr F, Keller-Schoenwetter S, Kunzi-Rapp K, Munstedt K, Rao CV, Lang U, Preissner KT. Characterization of human chorionic gonadotropin as a novel angiogenic factor. J Clin Endocrinol Metab. 2002;87:5290–5296. doi: 10.1210/jc.2002-020642. [DOI] [PubMed] [Google Scholar]


