Abstract
Rationale: ABCA3 is a member of the ATP-binding cassette family of proteins that mediate the translocation of a wide variety of substrates, including lipids, across cellular membranes. Mutations in the gene encoding ABCA3 were recently identified in full-term neonates with fatal surfactant deficiency.
Objective: To test the hypothesis that ABCA3 mutations are not always associated with fatal neonatal lung disease but are a cause of pediatric interstitial lung disease.
Methods: DNA samples were obtained from 195 children with chronic lung disease of unknown etiology. The 30 coding exons of the ABCA3 gene were sequenced in four unrelated children with a referring diagnosis of desquamative interstitial pneumonitis and who were older than 10 years at the time of enrollment.
Results: Three of four patients (ages 16, 23, and 11 years) with desquamative interstitial pneumonitis had ABCA3 mutations identified on both alleles. All three had the same missense mutation (E292V) and a second unique mutation. The E292V mutation was not found on 200 control alleles from adults without lung disease, but seven additional patients of the remaining study patients had the E292V mutation on one allele. Immunohistochemical analysis of surfactant protein expression in three patients revealed a specific staining pattern for surfactant protein-B, which was the same pattern observed in several infants with fatal lung disease due to ABCA3 mutations.
Conclusion: ABCA3 mutations cause some types of interstitial lung disease in pediatric patients.
Keywords: desquamative interstitial pneumonitis, pulmonary alveolar proteinosis, surfactant
Pulmonary surfactant is a mixture of lipids and specific proteins needed to reduce alveolar surface tension. Surfactant is produced and stored by alveolar type II cells, is secreted by exocytosis, and forms a lipid-rich monolayer at the air–liquid interphase, which lowers surface tension and prevents end-expiratory atelectasis. Deficiency of surfactant due to prematurity is the primary cause of respiratory distress syndrome in some infants (1). Lung disease in some full-term infants and older children with interstitial lung disease (ILD) has a genetic basis related to surfactant metabolism. Single-gene defects that encode the hydrophobic surfactant proteins, SP-B and SP-C, result in severe neonatal and chronic ILD, respectively (2–4). Recently, mutations in the gene encoding the ATP-binding cassette protein A3 (ABCA3) were identified in full-term neonates with fatal surfactant deficiency (5). The ABCA3 gene encodes a 1,704–amino acid protein that is highly expressed in the lung and has been localized to the limiting membrane of lamellar bodies in alveolar type II cells (6–9). Its location within the alveolar type II cell, the role of other ABCA subfamily proteins in lipid transport, and the induction of ATPase activity by lipids (10) suggested ABCA3 as a candidate gene for neonatal surfactant deficiency. The finding of ABCA3 mutations as an autosomal recessive cause of fatal surfactant deficiency supported this hypothesis, indicated an essential role for ABCA3 in surfactant metabolism, and expanded the previously recognized genetic etiologies of neonatal lung disease.
The lung histopathology in six of the infants with ABCA3 mutations in the initial study (5) was interpreted as desquamative interstitial pneumonitis (DIP), a form of pediatric ILD (pILD). pILD is a heterogeneous group of rare disorders of largely unknown etiology associated with diffuse infiltrates and disordered gas exchange (11, 12). The one infant with prolonged survival in that initial report had a missense mutation identified on one allele, but no second mutation identified on the other allele (5). These observations are consistent with the hypotheses that reduced, rather than absent, ABCA3 function caused a milder phenotype in this child and that ABCA3 is a candidate gene for pILD. However, although the inability to identify a mutation on the other allele in this child may have been due to its location in a region of the gene that was not examined, other explanations are possible. For example, a mutation on one allele in combination with some other genetic or environmental factor or factors may have resulted in lung disease. Alternatively, the finding of a missense mutation in this infant may have been coincidental and not causal for his lung disease. The present study was designed to test the hypotheses that some ABCA3 mutations allow for prolonged survival and that such mutations are the etiology for some forms of pILD. Preliminary results of this study were presented at the 2005 annual meeting of the Pediatric Academic Societies and reported in abstract form (13).
METHODS
Patients
From July 1995 through December 2003, DNA samples were obtained from 195 children with chronic lung disease of unknown etiology as part of a study to identify genetic defects in surfactant metabolism. The children were referred by their primary providers and classified according to their referring information. The entry criteria included the following: (1) gestational age of 36 weeks or older and age 30 days or older or discharged from the neonatal intensive care unit, or gestational age younger than 36 weeks and age older than 3 months or discharged from the neonatal intensive care unit; and (2) indications of parenchymal lung disease as manifested by two of three of the following factors: need for supplemental oxygen; clinical signs of lung disease, including tachypnea, cough, retractions, or rales; and an abnormal chest radiograph. Children with lung biopsy findings interpreted as a form of ILD qualified for enrollment, and infants younger than 30 days were eligible for enrollment if there was a family history of ILD. A history of neonatal lung disease was not a requirement for inclusion. The institutional review boards of the participating institutions approved the protocols for these evaluations, and written consent for genetic testing was obtained from the families. The patients have not been excluded based on sex or race/ethnicity. Of the patients, 55% were male, 41% female, and 4% unknown. The distribution of samples by racial/ethnic background was 131 (67%) white (non-Hispanic), 10 (5%) African American, 28 (14.5%) Hispanic, 5 (2.5%) Asian, less than 1% each American Indian/Alaskan Native or Native Hawaiian/other Pacific Islander, and 17 (8.5%) unknown. Age distribution at the time of enrollment was as follows: 47 (23.5%) aged 1 to 6 months, 38 (18%) aged 6 months to 1 year, 29 (15%) aged 1 to 2 years, 34 (17%) aged 2 to 5 years, 35 (18%) aged 6 to 17 years, 12 (6%) aged 18 years and older. Control DNA samples were obtained from adults of northern European descent who did not have a history of known lung disease and were analyzed anonymously. Genomic DNA was prepared from blood leukocytes with use of a commercially available kit (Gentra Systems, Minneapolis, MN).
Mutational Analysis
Primers were designed to amplify the 30 coding exons of the ABCA3 gene and their respective splice junctions. Closely spaced exons were grouped within the same amplicon to reduce the total number of reactions. Primers and Taq polymerase were purchased from Invitrogen (Carlsbad, CA). The buffer and MgCl2, were included with the polymerase used in the polymerase chain reactions. Deoxyribonucleotide triphosphates were purchased from Applied Biosystems (Foster City, CA). Polymerase chain reaction conditions contained 1.5 mM MgCl2, 0.5 μM of each specific primer, and 125 μM of each deoxyribonucleotide triphosphate, with annealing temperatures and conditions, as outlined in Table E1 in the online supplement, using a PX2 thermal cycler (Thermohybaid, Franklin, MA). Amplicons were analyzed by agarose gel electrophoresis, purified with spin columns (Qiagen, Valencia, CA) and quantitated by comparison with standards of known size and concentration. For amplicons greater than 700 bp, additional primers corresponding to internal sequences were used for DNA sequencing. Automated DNA sequencing was performed through the Johns Hopkins University School of Medicine's genetic resources core DNA sequencing facility using Applied Biosystems 3730 DNA Analyzer (Applied Biosystems). DNA sequencing chromatograms were analyzed with the aid of Sequencher 4.1.4 software (Gene Codes Corporation, Ann Arbor, MI) and compared with the reference ABCA3 sequence. Sequence analysis of the coding exons of the SP-C gene was performed on all patients as previously described (4).
Restriction Digest Analysis
Restriction endonucleases BsrG1 and BanII were purchased from the New England Biolabs (Beverly, MA) and used with supplied reagents according to the manufacturer's instructions.
Immunohistochemistry
Immunohistochemical staining for the surfactant proteins SP-A, mature B, proSP-B, and proSP-C was performed using antisera and methods as previously described (14).
RESULTS
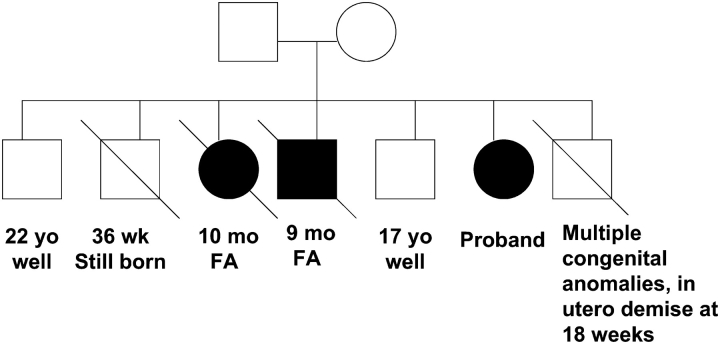
Of the 195 children enrolled, 28 (14.4%) were found to have an SP-C mutation and 2 (1%) had SP-B mutations, which were identified as the cause of their lung disease. Of the remaining 165 children in whom the cause of lung disease remained unknown, 4 were initially selected for further study on the basis of the following criteria: age older than 10 years at the time of enrollment, full-term gestation, development of respiratory symptoms at less than 1 year of age, and lung biopsy findings that had been interpreted as consistent with DIP from the referring physicians. The characteristics of these four patients are summarized in Table 1. The clinical course of Patient 1 has been previously reported in detail as illustrative of pediatric DIP (15). Briefly, she had respiratory distress syndrome in the neonatal period from which she recovered, but developed increasing respiratory symptoms and hypoxemia during childhood and subsequently underwent a single-lung transplant at age 21. Patient 2 had a family history of lung disease, with two siblings who died within the first year of life; and she was included in a survey of pILD (16). The pedigree is shown in Figure 1. One male sibling also had lung histopathology interpreted as DIP; histopathology was not available from the affected female sibling. Patients 1, 3, and 4 did not have a family history of lung disease.
TABLE 1.
Characteristics of older children with chronic lung disease
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Race | White | White | White | White |
| Age of symptoms | Birth | < 1 yr | < 1 yr | Birth |
| Sex | Female | Female | Male | Male |
| Lung biopsy | DIP | DIP | DIP | DIP |
| Age at enrollment | 23 yr | 14 yr | 10 yr | 11 yr |
| Current age | 32 yr | 16 yr | 12 yr | 11 yr |
| Family history | No | Yes | No | No |
| Outcome | Lung transplant | Alive | Alive | Alive |
| ABCA3 mutations | E292V/N1076K | E292V/c3704-1 G>T | E292V/c1742-9 G>A | None |
| Maternal allele | NA | c3704-1 G>T | E292V | NA |
| Paternal allele | NA | NA | c1742-9 G>A | NA |
Definition of abbreviations: DIP = desquamative interstitial pneumonitis; NA = not available.
Figure 1.
Pedigree for Patient 2. Clinical information for those with lung disease is shown below their symbol. Circle = female, square = male, open symbol = unaffected by known lung disease, closed symbol = affected by lung disease, slashed symbol = death. FA = fibrosing alveolitis.
ABCA3 mutations were identified in Patients 1, 2, and 3, but not in Patient 4. The results are summarized in Table 1. DNA was available from one parent of Patient 2 and both parents of Patient 3. Analysis of parental DNA in both families was consistent with the ABCA3 mutations in the children being on separate alleles.
The same ABCA3 mutation was identified on one allele in all three patients, consisting of a thymine for adenine substitution at cDNA position 875 and located in the first codon of exon 9. This mutation results in the substitution of valine for glutamic acid in codon 292 (E292V) and introduces a new recognition site for the restriction endonuclease BsrG1. The E292V mutation was not found on any of the 200 alleles examined from 100 adults without lung disease and who were matched for ethnic background by restriction analysis of polymerase chain reaction products spanning exon 9.
DNA samples from 153 of the remaining study patients, for whom the cause of lung disease remained undetermined, were then examined for the E292V mutations by restriction analysis. In eight patients, amplification for exon 9 was unsuccessful using the DNA despite repeated attempts varying polymerase chain reaction conditions and DNA concentrations. Seven additional patients with the E292V mutation on one allele were identified by restriction analysis and confirmed by DNA sequence analysis, including one pair of siblings (Table 2). Sequence analysis of the remaining 29 coding exons of ABCA3 identified mutations in five of the seven patients, with one patient having two potential mutations. A second mutation was not identified in two patients. Analysis of parental DNA of Patient 7 indicated mutations on separate alleles in the child.
TABLE 2.
Characteristics of children with chronic lung disease who screened positive for E292v mutation
| Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | Patient 11 | |
|---|---|---|---|---|---|---|---|
| Race | White | White | White | White | White | Hispanic | White |
| Age of symptoms | Neonate | Neonate | Neonate | 7 yr | 5 yr | Neonate | Neonate |
| Sex | Female | Female | Male | Male | Male | Male | Male |
| Lung biopsy | Not done | Not done | Not done | IP/PF | Not done | BO/fibrosis | Normal |
| Current age | NA | 11 yr | 11 yr | 11 yr | 9 yr | NA | 6 yr |
| Family history | Yes | No | Yes | Yes | Yes | No | No |
| Outcome | Died at 11 yr | Alive | Alive | Alive | Alive | Died at 6 mo | Alive |
| ABCA3 mutations | E292V/c1612-2 A>G/P1301L |
E292V/G1302E | E292V/T1114M | E292V/E690K | E292V/E690K | E292V/none identified |
E292V/none identified |
Definition of abbreviations: BO = bronchiolitis obliterans; IP = interstitial pneumonitis; NA = not available; PF = pulmonary fibrosis.
Lung tissue blocks from Patients 1, 3, and 8 were available for immunohistochemical analysis of surfactant protein expression. A similar staining pattern was observed in all three patients. Staining for SP-A (not shown) and proSP-B was robust in both epithelial cells and the proteinaceous material and cells in the airspaces. ProSP-C staining was also robust, although restricted to alveolar epithelial cells. In contrast, staining for mature SP-B was very weak, but could be detected after antigen retrieval (Figure 2). A similar pattern of robust staining for proSP-B with reduced staining for mature SP-B, which was recovered after antigen retrieval, was observed in Patients 3 and 8 (not shown), as well as in lung tissue from a full-term neonate who was a compound heterozygote for two ABCA3 mutations (W12X/3997delAG; Figure 2). In comparison to these findings in patients with ABCA3 mutations, robust staining for both proSP-B and mature SP-B and markedly reduced staining for proSP-C was observed in lung tissue from a patient with an SFTPC mutation (17).
Figure 2.
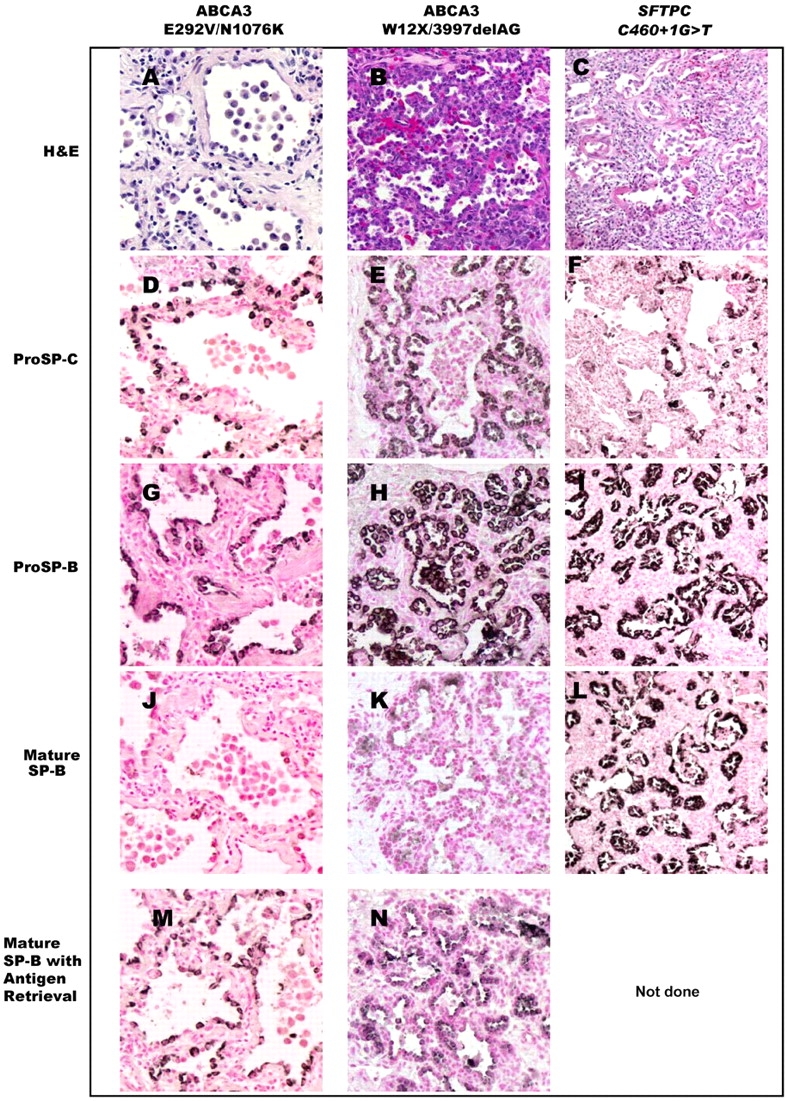
Immunohistochemical analysis for surfactant proteins. Lung tissue from Patient 1, with ABCA3 mutations (E292V/N1076) and prolonged survival (left column: A, D, G, J, M; original magnification, ×20). Robust staining for proSP-C and proSP-B is observed (D, G), in contrast to weak staining for mature SP-B (J), which was enhanced with antigen retrieval (M). A similar pattern is seen in lung tissue from a full-term neonate with fatal lung disease who was a compound heterozygote for ABCA3 null mutations (W12X/3997delAG; middle column: B, E, H, K, N; original magnification, ×20). In contrast, robust staining for mature SP-B was observed in a patient with an SFTPC mutation (c460+1G>T, resulting in skipping of exon 4) and similar histopathology (right column: C, F, I, L; original magnification, ×10; antigen retrieval not done for this patient). H&E = hematoxylin–eosin.
DISCUSSION
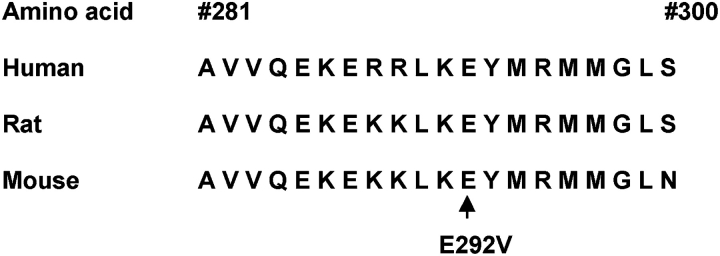
We have demonstrated that the ABCA3 gene is mutated in some patients with pILD. In contrast to the initial description of ABCA3 gene mutations in newborns with severe surfactant deficiency (5), these children survived well beyond infancy and in one case to early adulthood. All 10 patients had the identical ABCA3 mutation on one allele, E292V, a nonconservative amino acid substitution in a region of the protein that has been highly conserved during evolution (Figure 3). The E292V mutation was not found on 200 control alleles, indicating that it is not a common polymorphism. Five of the 10 patients had abnormal lung histopathology interpreted as DIP, chronic pneumonitis of infancy, or pulmonary fibrosis. The early onset of disease, similar lung histopathology, and finding of a common mutation all support the idea that ABCA3 mutations in general, and the E292V mutation in particular, are responsible for the genetic etiology of pILD related to abnormal surfactant function.
Figure 3.
ABCA3 amino acid sequence is highly conserved in region of E292V mutation. Twenty amino acids in the region of codon 292 in the human, rat, and mouse ABCA3 protein sequences are shown, with conservation of protein sequence observed in this region.
To test the hypothesis that some ABCA3 mutations are associated with prolonged survival, we focused our analyses on patients who were older than 10 years at the time of enrollment and who had lung histopathology consistent with those observed in neonates with ABCA3 deficiency. Some of those patients were reported as having had DIP, a form of pILD also associated with mutations in SFTPB and SFTPC (12). The histopathology in many of these patients might be more appropriately described as consistent with either chronic pneumonitis of infancy or nonspecific interstitial pneumonitis; however, the biopsies of the older patients in our study predated the published description of chronic pneumonitis of infancy and classification of nonspecific interstitial pneumonitis (18, 19). DIP is a term adapted from adult ILD, but in pediatric patients, the disease usually has its onset in early infancy, is often familial, and has a worse prognosis than DIP in adults, suggesting that it reflects a disease process with different underlying etiologies (11). The specific histopathology associated with ABCA3 mutations will require additional study for more accurate classification. The criteria for the classification of pILD are currently under revision (20). However, our findings further support a genetic basis for some forms of pILD, which are related to abnormalities in surfactant metabolism.
Mutations were identified on both ABCA3 alleles in a majority of patients (8 of 10). The finding that patients with a similar phenotype had the same variant supports the hypothesis that E292V is deleterious and not a neutral variant, although we cannot exclude the possibility that E292V is linked functionally to a more significant mutation in a region of the ABCA3 gene that we have not analyzed. In addition to the E292V mutation, five additional missense variants (N1076K, G1302E, P1301L, T1114M, E690K) and three splice junction site mutations (c3704-1 G>T, c1742-9 G>A, c1612–2 A>G) that would likely alter RNA splicing were identified. The missense mutations result in nonconservative amino acid substitutions in highly conserved regions of the ABCA3 protein. We did not find these variants in the public polymorphism databases or in 100 ethnically matched control subjects. However, without a functional test or available tissue for RNA analysis, we cannot exclude the possibility that these are neutral variants. Two children did not have a mutation identified on the second allele. A second mutation affecting ABCA3 expression or mRNA splicing may have been present in a region of the gene we did not examine, or other genetic or environmental factors in combination with the E292V mutation on one allele may have accounted for their lung disease. The outcomes of these two infants were very different. One was mildly premature and died of his lung disease at 6 months of age, whereas the other had minimal changes on lung biopsy and is without symptoms at age 5 years.
The finding of the identical missense mutation in multiple patients with a milder phenotype than previously appreciated suggests the possibility of a genotype–phenotype correlation. Little specific information is known concerning the domains important for ABCA3 routing and function. By homology with other ABC proteins, the location of codon 292 corresponds to an intracellular loop connecting two membrane-spanning domains and could potentially be important in substrate binding. Such a mutation could also result in abnormal routing of ABCA3 to lamellar bodies. Additional studies will be needed to address these questions. Moreover, although the phenotype of the patients in this report was milder than those in the previous study, there was still considerable variability in the severity of the lung disease among the 10 patients and no clear correlation with the nature of the second mutation. One patient, who was 16 years old at the time of study, had two siblings with presumably the same genotype who died within the first year of life, and another had a twin who also died in early infancy, suggesting that other genetic and environmental variables may be important in modifying the course of the disease. Variability was also observed in the descriptions of the lung histopathology, which included interstitial pneumonitis, pulmonary fibrosis, bronchiolitis obliterans, and in one lung biopsy that showed minimal abnormalities. Some of this variability may reflect different stages of the disease and/or patchy distribution of disease within the lung, as well as interobserver variability. Most of the patients had some lung disease in the immediate neonatal period. However, both siblings with the E292V/E690K mutations reportedly did not develop symptoms of lung disease until 5 to 7 years of age. Although it is possible that these children had symptoms of lung disease earlier in life that were very mild or not appreciated, they did not have symptoms of surfactant deficiency at birth.
The precedent that some genotypes are associated with milder disease has been recognized in other genetic diseases, including SP-B deficiency and cystic fibrosis (CF) (21, 22). CFTR is an ABC transporter and may thus provide a useful paradigm for genotype–phenotype correlations. The relationship between CFTR genotype and CF disease phenotype has been studied extensively. In general, CFTR genotype correlates well with pancreatic disease but is highly variable with pulmonary disease (23). In addition, studies involving chronic rhinosinusitis have demonstrated that many carriers of known CF mutations had a second variant mutation on a separate allele that was not associated with typical CF phenotype but was found in individuals with chronic rhinosinusitis (24). It has been speculated that the variable CF pulmonary and chronic rhinosinusitis phenotypes result from reduced CFTR function and/or CFTR dysfunction that under stress and/or in combination with other genetic factors or specific environmental exposures alter the course of disease. In the case of SP-B deficiency, which was originally recognized as a lethal disease within the first year of life, some children with partial deficiency and prolonged survival have been described (14). Similar to these other genetic diseases, variability in ABCA3 expression might be based on genotype, whereas phenotype may be modulated by other alleles or environmental factors that further downregulate its expression.
The immunohistochemical staining pattern for the surfactant proteins in three of our E292V patients was abnormal, with robust staining for proSP-B but markedly reduced staining for mature SP-B, which was recovered with antigen retrieval. The mechanisms underlying this decreased staining for mature SP-B are unknown and require further investigation. Because proSP-B expression was robust, this may indicate that processing of proSP-B to mature SP-B is impaired in ABCA3 deficiency. ABCA3 deficiency is associated with abnormal lamellar body formation (5), and the final steps for proSP-B processing occur in multivesicular or lamellar bodies (25, 26). Reduced mature SP-B staining may therefore reflect an impairment of processing of proSP-B to mature SP-B due to abnormal lamellar body formation. Alternatively, because the lamellar bodies in ABCA3 deficiency are very small, with tightly packed membranes that may hinder staining, the reduced staining may reflect decreased availability of the SP-B epitopes for the specific antibodies used. Recovery of staining with antigen retrieval could reflect unmasking of epitopes rather than enhanced sensitivity for a reduced amount of mature SP-B. Tissue for ultrastructural studies was not available to examine lamellar body formation in these patients. Reduced staining for mature SP-B could also be nonspecific and secondary to lung injury. However, the identical immunohistochemical staining pattern was observed in a neonate who was a compound heterozygote for two clear loss-of-function ABCA3 mutations, supporting the hypothesis of a common mechanism. This pattern has also been observed in other patients with ABCA3 mutations and is rarely found in association with SFTPC mutation (S.E.W., unpublished observations). With SFTPB mutations, the alveolar material stains intensely for proSP-C, distinguishing it from the pattern of staining observed in ABCA3 deficiency (14). Further studies are needed to determine the specificity and sensitivity of this staining pattern for ABCA3 deficiency, and the interaction of ABCA3 with SP-B metabolism.
Of the 195 patients enrolled in our study, 28 had SFTPC mutations as the basis for their disease, and 10 have now been identified with ABCA3 mutations. The relative contribution of ABCA3 mutations to lung disease may be even greater than we currently appreciate, because the majority of patients in our study group were screened only for the E292V mutation and have not yet had their ABCA3 gene sequenced or screened for other mutations. In addition, the finding of patients with disease who have an ABCA3 mutation identified on only one allele suggests that there are ABCA3 mutations that may have escaped our current detection methods, some of which could be associated with an even milder phenotype. Our study was not population based. The patients were identified by their primary physicians on the basis of the unusual nature of their lung disease, and the majority were younger than 1 year at the time of enrollment. Thus, there is significant ascertainment bias that limits interpretation of the relative frequency of these genetic causes of lung disease, and the unusual nature of the lung disease in these patients may exaggerate the apparent contribution of E292V and other ABCA3 mutations to pILD. Additional studies will be needed to fully define the roles of ABCA3 mutations and other gene mutations in pILD. The finding of a common mutation may prove useful in population-based studies to determine the incidence of lung disease due to ABCA3 mutations (27).
In summary, we identified 10 children with pILD that was associated with a common ABCA3 mutation and whose lung disease was considerably less severe than previously described. The mechanism for decreased severity of lung disease in these children remains unclear, but we speculate that it may be related to reduced function rather than complete absence of the ABCA3 protein. Children and young adults with forms of pILD, particularly DIP, chronic pneumonitis of infancy, and nonspecific interstitial pneumonitis, or who have a family history of neonatal lung disease, should be evaluated for ABCA3 mutation.
Supplementary Material
Acknowledgments
The authors thank Kristen Cowperthwaite, Zachary Nevin, and Paula Blair for technical assistance, and the patients and their families for their willingness to participate in the study, and the physicians and nurses who cared for them.
Supported by grants from the National Institutes of Health (HL-54703 [L.M.N.], HL-56387 [S.E.W., L.M.N., J.A.W.]) and the Eudowood Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Conflict of Interest Statement: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Johansson J, Curstedt T. Molecular structures and interactions of pulmonary surfactant components. Eur J Biochem 1997;244:675–693. [DOI] [PubMed] [Google Scholar]
- 2.Cole FS, Hamvas A, Nogee LM. Genetic disorders of neonatal respiratory function. Pediatr Res 2001;50:157–162. [DOI] [PubMed] [Google Scholar]
- 3.Weaver TE, Conkright JJ. Function of surfactant proteins B and C. Annu Rev Physiol 2001;63:555–578. [DOI] [PubMed] [Google Scholar]
- 4.Nogee LM, Dunbar AE, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med 2001;344:573–579. [DOI] [PubMed] [Google Scholar]
- 5.Shulenin S, Nogee LM, Annilo T, Wert SE, Whitsett JA, Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med 2004;350:1296–1303. [DOI] [PubMed] [Google Scholar]
- 6.Connors TD, Van Raay TJ, Petry LR, Klinger KW, Landes GM, Burn TC. The cloning of a human ABC gene (ABC3) mapping to chromosome 16p13.3. Genomics 1997;39:231–234. [DOI] [PubMed] [Google Scholar]
- 7.Yamano G, Funahashi H, Kawanami O, Zhao LX, Ban N, Uchida Y, Morohoshi T, Ogawa J, Shioda S, Inagaki N. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett 2001;508:221–225. [DOI] [PubMed] [Google Scholar]
- 8.Mulugeta S, Gray JM, Notarfrancesco KL, Gonzales LW, Koval M, Feinstein SI. Ballard PL, Fisher AB, Shuman H. Identification of LBM180, a lamellar body limiting membrane protein of alveolar type II cells, as the ABC transporter protein ABCA3. J Biol Chem 2002;277:22147–22155. [DOI] [PubMed] [Google Scholar]
- 9.Zen K, Notarfrancesco K, Oorschot V, Slot JW, Fisher AB, Shuman H. Generation and characterization of monoclonal antibodies to alveolar type II cell lamellar body membrane. Am J Physiol 1998;275:L172–L183. [DOI] [PubMed] [Google Scholar]
- 10.Nagata K, Yamamoto A, Ban N, Tanaka AR, Matsuo M, Kioka N, Inagaki N, Ueda K. Human ABCA3, a product of a responsible gene for abca3 for fatal surfactant deficiency in newborns, exhibits unique ATP hydrolysis activity and generates intracellular multilamellar vesicles. Biochem Biophys Res Commun 2004;324:262–268. [DOI] [PubMed] [Google Scholar]
- 11.Fan LL, Langston C. Chronic interstitial lung disease in children. Pediatr Pulmonol 1993;16:184–196. [DOI] [PubMed] [Google Scholar]
- 12.Fan LL, Deterding RR, Langston C. Pediatric interstitial lung disease revisited. Pediatr Pulmonol 2004;38:369–378. [DOI] [PubMed] [Google Scholar]
- 13.Bullard JE, Wert SE, Whitsett JA, Dean M, Nogee LM. An ABCA3 mutation associated with pediatric interstitial lung disease [abstract]. Presented at: PAS annual meeting; May 14–17, 2005; Washington, DC. [DOI] [PMC free article] [PubMed]
- 14.Nogee LM, Wert SE, Proffit SA, Hull WM, Whitsett JA. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am J Respir Crit Care Med 2000;161:973–981. [DOI] [PubMed] [Google Scholar]
- 15.Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 49-1993. A 21-year-old woman with lifelong progressive interstitial lung disease. N Engl J Med 1993;329:1797–1805. [DOI] [PubMed] [Google Scholar]
- 16.Sharief N, Crawford OF, Dinwiddie R. Fibrosing alveolitis and desquamative interstitial pneumonitis. Pediatr Pulmonol 1994;17:359–365. [DOI] [PubMed] [Google Scholar]
- 17.Nogee LM, Dunbar AE, Wert S, Askin F, Hamvas A, Whitsett JA. Mutations in the surfactant protein C gene associated with interstitial lung disease. Chest 2002;121:20S–21S. [DOI] [PubMed] [Google Scholar]
- 18.Katzenstein AL. Chronic pneumonitis of infancy: a unique form of interstitial lung disease occurring in early childhood. Am J Surg Pathol 1995;19:439–447. [PubMed] [Google Scholar]
- 19.Katzenstein AL, Fiorelli RF. Non-specific interstitial pneumonia/fibrosis: histologic patterns and clinical significance. Am J Surg Pathol 1994;18:136–137. [PubMed] [Google Scholar]
- 20.Deutsch GH, Dishop MK, Deterding R, Cutz E, Langston C, Albright E, Chou PM, Cool CD, Coventry S, Davis MM, et al. Defining the spectrum of diffuse lung disease in infancy: a working classification of the pediatric interstitial lung disease (pILD) multidisciplinary cooperative group [abstract]. Presented at: Society of Pediatric Pathology; February 25–27, 2005; San Antonio, TX. Mod Pathol 2005;18:301–313.15915554 [Google Scholar]
- 21.Nogee LM. Genetic mechanisms of surfactant deficiency. Biol Neonate 2004;85:314–318. [DOI] [PubMed] [Google Scholar]
- 22.Nogee LM. Alterations in SP-B and SP-C expression in neonatal lung disease. Annu Rev Physiol 2004;66:601–623. [DOI] [PubMed] [Google Scholar]
- 23.Rowntree RK, Harris A. The phenotypic consequences of CFTR mutations. Ann Hum Genet 2003;67:471–485. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Moylan B, Leopold DA, Kim J, Rubenstein RC, Togias A, Proud D. Zeitlin PL, Cutting GR. Mutation in the gene responsible for cystic fibrosis and predisposition to chronic rhinosinusitis in the general population. JAMA 2000;284:1814–1819. [DOI] [PubMed] [Google Scholar]
- 25.Weaver TE, Na C-L, Stahlman M. Biogenesis of lamellar bodies, lysosome-related organelles involved in storage and secretion of pulmonary surfactant. Semin Cel Dev Biol 2002;13:263–270. [DOI] [PubMed] [Google Scholar]
- 26.Weaver TE, Conkright JJ. Functions of surfactant B and C. Annu Rev Physiol 2001;63:555–578. [DOI] [PubMed] [Google Scholar]
- 27.Hamvas A. Trusgnich M, Brice H, Baumgartner J, Hong Y, Nogee LM, Cole FS. Population-based screening for rare mutations: high-throughput DNA extraction and molecular amplification from Guthrie cards. Pediatr Res 2001;50:666–668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.