Abstract
A variety of genes have been used to type Pneumocystis carinii. In the present study, nucleotide sequence variations in the ITS1 and ITS2 internal transcribed spacer (ITS) regions of the rRNA genes were used to type Pneumocystis carinii f. sp. hominis DNA obtained from the lungs of 60 human immunodeficiency virus-infected individuals. These regions were amplified by PCR, cloned, and sequenced. Multibase polymorphisms were identified among samples. Several new genotypes are reported on the basis of the nucleotide sequence variations at previously unreported positions of both the ITS1 and the ITS2 regions. Twelve new ITS1 sequences were observed, in addition to the nine sequence types reported previously. The most common was type E, which was observed in 60.5% of the samples. The sequence variations in the ITS1 region were mainly located at positions 5, 12, 23, 24, 45, 53, and 54. Sixteen new ITS2 types were also identified, in addition to the 13 types reported previously. The most common was type g (26.6%). The sequences of the ITS2 regions in most specimens were different from the previously published sequence at bases 120 and 166 through 183. The most common variations observed were deletions at positions 177 through 183. The presence of more than one sequence type in some patients (60%) suggested the occurrence of coinfection with multiple P. carinii strains. The genetic polymorphism observed demonstrates the degree of diversity of Pneumocystis strains that infect humans. Furthermore, the high degree of polymorphism suggests that these genes are evolving faster than other genes. Consequently, the sequence information derived is useful for purposes such as examination of the potential of person-to-person transmission and recurrent infections but perhaps not for other genotyping applications that rely on more stable genetic loci.
Pneumocystis carinii is an opportunistic pathogen that causes potentially fatal pneumonia in immunocompromised or immunosuppressed patients. Despite the decrease in the incidence of the disease in human immunodeficiency virus HIV-infected patients (3), P. carinii pneumonia (PCP) continues to be a common AIDS-defining illness associated with significant mortality in both adults (8%) (3) and children (30%) (19).
Although recent research has clarified our understanding of certain biological aspects of the organism, no standard method for the typing of P. carinii has emerged. Pure isolates of the organism are difficult to obtain in large amounts due to the lack of a standardized method of in vitro cultivation of human-derived P. carinii. Standard methods of typing have similarly been slow to develop (24).
Despite evidence of genetic variations among human P. carinii isolates (12, 20), the degree of diversity among strains that infect humans still lacks clarification. The existence of multiple strains of P. carinii that infect humans was first demonstrated by examination of the restriction fragment length polymorphisms (RFLPs) of P. carinii genomes from three patients (21). However, RFLP analyses are not widely applicable to human clinical specimens because of the difficulties associated with obtaining large numbers of organisms.
The development of molecular biology-based techniques has enabled typing of human-derived P. carinii (P. carinii f. sp. hominis) on the basis of the DNA sequence variations at a number of different genetic loci (2, 26) including the internal transcribed spacers (ITSs) of the nuclear rRNA genes (7, 13, 15). Some of the more recent epidemiologic and clinical applications of molecular biology-based typing techniques have included studies on geographic variations and transmission modes (2), analysis of the potential emergence of antimicrobial resistance (8, 16, 18), and analysis of a potential linkage of genotype with disease severity (17).
The nucleotide sequences of the ITS1 and ITS2 regions of the nuclear rRNA operon have been shown to vary among different P. carinii strains (7, 13, 15, 23). ITS1 is located between the coding regions of the 18S rRNA and the 5.8S rRNA genes, and ITS2 is located between the coding regions of the 5.8S rRNA and the 26S rRNA genes. Fifteen types of ITS1 sequences (designated types A to O) and 14 types of ITS2 sequences (designated types a to n) have been reported (13). The ITS1 and ITS2 regions evolve more rapidly than the functional domains that they flank and are therefore potentially more useful at demonstrating differences among closely related populations of organisms (4). The ITS region has also been used to examine differences in P. carinii isolates from different parts of the world (13) and from patients with recurrent episodes of PCP (10, 23). The high level of variation in the ITS genes compared to those in other genes used for typing (e.g., the mitochondrial large-subunit [mt LSU] rRNA gene) demonstrates the utility of this genetic locus in epidemiologic studies and in the detection of genetic diversity.
The aim of the present study was to characterize the genetic polymorphism observed at the ITS locus in P. carinii isolates from HIV-infected individuals in the cities of Atlanta, Ga.; San Francisco, Calif.; and Seattle, Wash. Several new P. carinii genotypes are reported on the basis of an analysis of the nucleotide sequence variations of the ITS gene regions that have been amplified by PCR from clinical specimens and cloned in a vector.
MATERIALS AND METHODS
Samples.
Sixty induced sputum or bronchoalveolar lavage (BAL) specimens were collected from 60 HIV-infected patients as a part of routine diagnostic procedures in three different U.S. cities (Atlanta, Ga.; San Francisco, Calif.; and Seattle, Wash.), as described previously (2). The clinical diagnosis of PCP was microscopically confirmed in all cases. The specimens used in this study were collected between 1996 and 1999 from patients who gave informed consent to be involved in the study, which was approved through the institutional review board of each organization involved in the study. A portion of the specimen was immediately preserved with an equal volume of absolute ethanol and was stored at −4°C for DNA extraction.
DNA extraction.
Specimens were divided into approximately 1-ml aliquots and were centrifuged at 14,000 × g for 5 to 7 min. The resulting pellet was resuspended in 1 ml of phosphate-buffered saline (PBS; 0.01 M, pH 7.2) containing 1 mM EDTA (PBS-EDTA), washed twice in PBS-EDTA, centrifuged as described above, and stored at −80°C for later DNA extraction. DNA extraction was performed with a commercial purification system (Wizard Genomic DNA Purification kit; Promega, Madison, Wis.), according to the instructions of the manufacturer, for the purification of DNA from blood. Final pellets were resuspended in 50 μl of TE (10 mM Tris, 1 mM EDTA [pH 7.2]).
DNA amplification.
Nested PCR was performed on all specimens to amplify sufficient amounts of DNA fragments from the two ITS regions for analysis, as described previously (13). A 500-bp fragment was amplified from the nuclear rRNA ITS regions. The first PCR was performed with primers 1724F and ITS2R. The cycling conditions were 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 1 min at 72°C, with extension at 72°C for 10 min.
Three microliters of the first-round product was used as a template for the second round in a total volume of 50 μl under the same conditions as those used for the first round, except that primers ITS1F and ITS2R1 were used and annealing was done at 65°C for 30 s, followed by 90 s at 72°C and extension at 72°C for 10 min. DNA extracted from the lungs of rats infected with P. carinii was used as a positive control, and DNase-free water was used as a negative control to monitor for cross contamination.
Sample cross-contamination problems were avoided by use of a number of precautions, including the use of aerosol-guarded tips; performance of DNA extraction in a laminar-flow hood with subsequent irradiation with UV light; and the use of three separate areas for DNA extraction, preparation of the PCR mixture, and PCR amplification and running of the gels.
Five microliters of the PCR product was run on a 1.2% agarose gel stained with ethidium bromide.
Cloning and sequencing of the purified PCR product.
The amplified products were purified with a Wizard PCR Preps DNA Purification system (Promega) and cloned into a plasmid vector, the pGEM-T Easy Vector system (Promega). Recombinant plasmids were transformed in JM109 cells (108 CFU/μg of DNA), which were plated in triplicate for each specimen on Luria-Bertani agar containing ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and isopropyl-β-d-thiogalactopyranoside. For each specimen, five separate white colonies that contained inserts were picked from the transformant plates and were inoculated into Luria-Bertani broth with ampicillin. The specimen number and a letter from A to E identified these colonies. After overnight incubation at 37°C, plasmid DNA was purified with a Wizard Plus Minipreps DNA Purification system (Promega).
Two microliters of the purified product from each sample was digested with restriction endonuclease EcoRI, and the digest was run on a 1.2% ethidium bromide-stained agarose gel to confirm the presence of cloned inserts. The DNA fragments were sequenced by the use of dye terminator chemistry (BigDye Terminator Ready Reaction Mix (Perkin-Elmer Applied Biosystems, Foster City, Calif.) on an ABI Prism 377 automated DNA sequencer (PE Applied Biosystems), according to the manufacturer's protocol. The DNAs of all clones were sequenced in both directions. The prototype sequences of Lee et al. (13) were used as consensus sequences; variations at bases 62 to 71 of ITS1 were not included in the analysis. Discrepancies between the two strands as well as differences among clones were clarified by careful examination of the positive- and negative-sequence strands by use of Sequence Navigator (version 1.0.1; PE Applied Biosystems) to identify specific differences among clones. This program was also used to construct multiple-sequence alignments, which were further compared by use of the Wisconsin computer program package (version 9.1, 1997; Genetics Computer Group, Madison, Wis.)
Sequences with variations from the prototype sequence at five or more positions were considered to represent new ITS types (12, 13).
Nucleotide sequence accession numbers.
The 12 newly reported ITS1 sequences have been submitted to the GenBank database under accession numbers AF374238, AF374239, AF374240, AF374241, AF374242, AF374243, AF374244, AF374245, AF374246, AF374247, AF374248, and AF374249, respectively. The 16 newly reported ITS2 sequences have been submitted to the GenBank database under accession numbers AF374250, AF374251, AF374252, AF374253, AF374254, AF374255, AF374256, AF374257, AF374258, AF374259, AF374260, AF374261, AF374262, AF374263, AF374264, and AF374265, respectively.
RESULTS
Five separate colonies were selected from the transformant plates and examined for each of the 60 BAL and induced sputum specimens obtained from HIV-infected individuals. The DNAs of three of these colonies were sequenced for each specimen, resulting in a total of 180 sequences. A total of 9 ITS1 sequence types and 13 ITS2 sequence types reported previously were identified (Table 1). Twelve new ITS1 types and 16 new ITS2 types were also observed, and more than one ITS1-ITS2 combination types were observed in 36 (60%) specimens.
TABLE 1.
P. carinii sp. f. hominis ITS1 and ITS2 sequence types found in 180 sequences cloned from 60 specimens
| ITS1 type | No. (%) of ITS1 sequences | ITS2 type | No. (%) of ITS2 sequences |
|---|---|---|---|
| A | 2 (1.1) | a | 5 (2.8) |
| B | 7 (3.9) | b | 6 (3.3) |
| C | 1 (0.6) | c | 7 (3.9) |
| D | 2 (1.1) | e | 42 (23.3) |
| E | 109 (60.5) | f | 1 (0.6) |
| F | 2 (1.1) | g | 48 (26.7) |
| I | 1 (0.6) | h | 15 (8.3) |
| K | 5 (2.8) | i | 1 (0.6) |
| N | 39 (21.7) | FR004 | 3 (1.7) |
| Newa | 12 (6.7) | IT007 | 3 (1.7) |
| DK029 | 5 (2.8) | ||
| DK323 | 1 (0.6) | ||
| DK594 | 3 (1.7) | ||
| Newa | 16 (8.9) | ||
| Subtypesb | 24 (13.3) |
Sequences with variations from the prototype sequence at five or more positions.
Sequences with variations from the prototype sequence at less than five positions.
ITS1 sequences.
The ITS1 sequences from these samples were aligned. The consensus sequence of Lee et al. (13) was selected as the prototype, and all sequences were compared with this sequence. This comparison revealed nine ITS1 genotypes reported previously (13). The most common of these were type E, which was found in 109 individual clones (60.5%), and type N, which was noted in 39 clones (21.7%) (Table 1). Sequences similar but not identical to the prototype sequence were frequently identified, and the differences often involved variations at positions 62 to 71, which comprised a run of 10 T residues. One or more T residues were missing from this region in 84 of 180 (46.7%) ITS clones analyzed. Variations in sequences at these positions were not included in the analyses to define new ITS1 types. No subtypes were observed.
The 12 newly reported ITS1 types varied from the consensus sequence of Lee et al. (13) by from 5 to 11 nucleotides. These newly reported sequences are shown in Fig. 1.
FIG. 1.
Nucleotide sequences of 12 new types of ITS1 sequences of P. carinii f. sp. hominis. Bases that are identical to those of the prototype sequence are indicated by periods, missing bases are indicated by hyphens, and bases that are different from those of the prototype sequence are shown. The sequences after position 130 are not shown because no sequence variations were found in these areas. The inserted base A observed at position 50 in clone 10B is shown.
In the ITS1 region, sequence variations were mainly found at nucleotide positions 5, 12, 23, 24, 45, 53, and 54, with unique changes occurring at positions 5 and 45. Variations were also found at positions 21, 50, 79, 80, 82 to 84, 105, and 115 to 121 (Fig. 1). Substitution, deletion, or insertion of a nucleotide at one or more positions was observed. These variations, in addition to the missing bases in the same sequences, might represent new types of ITS1 sequences. Several sequences had one or more deleted bases. All but 5 of the 180 sequences had a missing T residue at position 12; there were two missing T residues at positions 23 and 24 in all but 4 sequences. All sequences except sequence 13A were missing AT residues at positions 53 and 54; sequence 13A was missing one T residue at position 54 and had a C residue instead of an A residue at position 53. Eighty-four (46.7%) sequences showed variations in the number of T residues at positions 62 to 71. Fourteen sequences were missing GA residues at positions 79 and 80 (sequences 15B and 60A are shown in Fig. 1), and six sequences were missing TTTA residues at positions 115 to 118 (for example, sequence 60A in Fig. 1).
Twenty of the 60 (33.3%) specimens contained two or more ITS1 types; 16 specimens simultaneously contained two ITS1 types, and 4 simultaneously contained three ITS1 types. The most frequent ITS1 combination observed was a mixture of types E and N, which was seen in 14 of the 20 specimens. The presence of multiple ITS1 types within these patients suggests that they were simultaneously infected with different strains of P. carinii.
ITS2 sequences.
The ITS2 sequences from the same specimens were also aligned and compared to the prototype sequence. This comparison revealed 13 ITS2 sequences reported previously (13). The most common of these were type g, which was found in 48 individual clones (26.7%), and type e, which was found in 42 clones (23.3%) (Table 1). Lee et al. (13) described sequences FR004, IT007, DK029, DK323, and DK594 as unique and indicated that they may represent new types. As with the ITS1 sequences, sporadic ITS2 sequence variations were also observed in the specimens. Not every sequence had the same numbers of nucleotides in both ITS regions; all sequences were missing bases at certain positions, which made them shorter than the prototype sequence. All sequences except sequence 100B were missing a C residue at position 120; sequence 100B was missing a T residue at position 114 (Fig. 2). Typing of the 180 sequences was made on the basis of variations at other positions, with disregard to the variation at position 120.
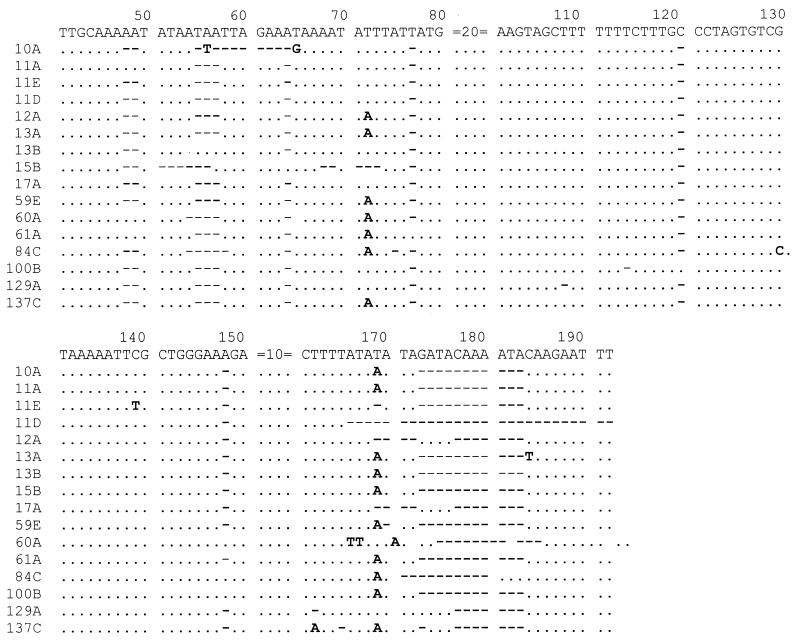
FIG. 2.
Nucleotide sequences of 16 new types of ITS2 sequences of P. carinii f. sp. hominis. Bases that are identical to those of the prototype sequence are indicated by periods, missing bases are indicated by hyphens, and bases that are different from those of the prototype sequence are shown. The sequences before position 41 are not shown because no sequence variations are found in these areas except for sequence 84C, where G was replaced by A at position 19. The inserted bases observed in clones 60A (TT at positions 166 and 167) and 84C (C at position 130) are shown.
There were 16 new ITS2 types (which differed from the consensus sequence by from 5 to 25 nucleotides) and 24 new subtypes (which differed from the consensus sequence by 1 or 2 nucleotides).
The nucleotide sequence variations in ITS2 were much more extensive than those in ITS1 and were not randomly distributed. They were located primarily at positions 48, 49, 55 to 57, 64, 72, 77, 120, 148, and 166 to 183. The most common variations observed were deletions at positions 177 through 183. Variations were also found at positions 51 to 54, 58 to 63, 65, 68 to 69, 71, 73, 75, 108, 114, 130, 139, 162, 165, and 184 to 192 (Fig. 2). Substitution, deletion, or insertion of a nucleotide at one or more positions was observed. All but seven sequences (two clones from specimen 11, three clones from specimen 60, and two clones from specimen 61) were missing AA residues at positions 48 and 49. All but three sequences (clones 13B, 13C, and 73A) were missing TAA residues at positions 55 to 57. All but two sequences were missing the CAAAATA sequence at positions 177 to 183.
The DNA sequences of strains from 27 of the 60 (45%) specimens contained two or more ITS2 types; strains from 18 specimens simultaneously contained two ITS1 types, and strains from 9 specimens simultaneously contained three ITS2 types. The most frequent ITS2 combination observed was a mixture of types e and g, which was seen in 8 of the 27 specimens. The presence of multiple ITS2 types within these patients suggests that they were simultaneously infected with different strains of P. carinii.
P. carinii f. sp. hominis types.
The P. carinii sequence types were designated by a two-letter code, the ITS1 type (A to O) followed by the ITS2 type (a to n). Three new combination types were found. There were a total of 36 ITS1-ITS2 combination types in the 60 specimens (60%) of P. carinii sp. f. hominis DNA that were typed. Type Eg was the most prevalent, represented by 27 (45%) of the 60 specimens; type Ne was the second most common type and was found in 16 (26.7%) of the 60 specimens.
Among the 60 specimens examined, 24 were found to contain strains of a single ITS genotype and 36 contained strains of more than one genotype. These 36 specimens included 22 specimens with strains of two types and 10 specimens with strains of three types. Multiple genotypes were observed more frequently in specimens from Atlanta (75% of the specimens with strains of multiple genotypes) than in specimens from San Francisco or Seattle (16.7 and 8.3% of the specimens with strains of multiple genotypes, respectively).
DISCUSSION
Data from previous studies suggest that genetic diversity exists among isolates of P. carinii infecting certain mammalian hosts (20). DNA sequence analysis has revealed differences among the DNAs of Pneumocystis strains isolated from human lungs. In most cases, the level of genetic diversity appears to be much lower than that among isolates from different host species (26). Typing of P. carinii, based on nucleotide sequence variations, has been conducted by using several genetic loci (1, 11, 23, 25), but for the most part typing studies with these genetic loci all have limitations, depending on the intended application. Sequence variations were not found in the thymidylate synthase gene. The 5S rRNA gene also has limited sequence variation, with polymorphisms reported at five positions. Analysis of the sequence of a portion of the nuclear rRNA operon of the organism from BAL samples from five HIV-infected patients in the United States failed to show any differences (14). It is also possible, however, that the polymorphism could be explained in part by the presence of heterozygosity in diploid-stage organisms in the sample. The sequence of the gene encoding the mt LSU rRNA similarly shows limited sequence variations between these isolates, with polymorphism being detected at only two nucleotide positions (25). However, this locus, along with dihydropteroate synthase, has successfully been used recently to demonstrate the geographical variations of isolates (2). A similar lack of genetic variation was seen at a portion of the arom locus in human-derived P. carinii isolates from different geographical areas (1). This small amount of sequence variation may not have sufficient discriminatory power for typing and may limit the use of these loci, depending on the specific typing objective. It was reported that isolates that have the same mitochondrial rRNA genotype may have different ITS types (9). The 18S, 5.8S, and 26S rRNA gene sequences amplified from specimens from 15 patients were found to be identical, whereas the sequences of the ITS regions were found to be variable (15). Data from other studies suggest that the ITS locus may provide a useful tool for epidemiologic studies.
In this study, we have used the variations in the DNA sequences of the ITS1 and ITS2 regions of the nuclear rRNA genes to type 180 P. carinii clones from 60 specimens. The specimens were collected from HIV-infected patients from three cities in the United States. The relatively large number of P. carinii isolates studied helped identify several new types on the basis of variations at more than five positions, in addition to previously reported types. Our data confirm the genetic heterogeneity of P. carinii f. sp. hominis. The diversity is demonstrated by the nucleotide polymorphism in the ITS1 region at numerous positions, including positions 5, 12, 23, 24, 45, 53, and 54. Variation was also found at positions 21, 50, 79, 80, 82 to 84, 105, and 115 to 121. The ITS2 region primarily had nucleotide polymorphisms at positions 48, 49, 55 to 57, 64, 72, 77, 120, 148, and 166 to 183. The technique used in this study involved cloning of the amplified fragment prior to sequencing. This approach, rather than direct sequencing, has provided data indicating that multiple types of P. carinii f. sp. hominis are present in individual samples. The base changes were not randomly distributed but occurred at certain sites; some positions were previously shown to be variable (13).
Our study identified several new ITS1 and ITS2 types. Definition of a new ITS genotype as a sequence with variations from the prototype sequence at five or more positions identified 12 new ITS1 types, 16 new ITS2 types, and 3 new ITS1-ITS2 types. These new and unique sequences varied from the prototype sequence at 5 to 25 positions by either deletion, substitution, or less frequently, insertion of a base. The observed variations in the ITS1 sequences at positions 21, 45, 82 to 84, and 118 and the variations in the ITS2 sequences at positions 58 to 61, 120, 148, 170 to 176, and 185 to 190 have not been reported previously. We also noted 9 ITS1 types (types A, B, C, D, E, F, I, K, and N) and 13 ITS2 types (types a, b, c, e, f, g, h, i, FR004, IT007, DK029, DK323, and DK594) reported previously. However, several previously reported ITS1 types (types G, H, J, L, M, and O) and ITS2 types (types d, j, k, l, m, and n) were not observed in this study. These ITS1 and ITS2 types were also less frequently reported by others (13), and the variations in these types include either deletion or deletion and substitution of a base at one or more positions. Lastly, our study confirmed the high prevalence of previously reported common type Eg, followed by type Ne (13).
Our study found evidence of infection with mixtures of genotypes in a majority of specimens from patients with PCP. On the basis of the assumption that P. carinii f. sp. hominis has only one copy of the rRNA gene (22), our sequence analysis indicated the presence of two different bases at one position in strains from 36 (60%) specimens and provides evidence in support of infection with more than one type of P. carinii (14). An incidence of 33% mixed infections has previously been reported for AIDS patients (23) and from analysis of the mt LSU rRNA and ITS loci (10, 11, 14). In this study, we found mixed infections in 60% of the specimens that we analyzed. Hauser et al. (5), using single-strand conformation polymorphism analysis, reported a slightly higher level of mixed infection, 69%.
In conclusion, the typing of sequences of Pneumocystis DNA at a locus where multibase polymorphisms occur, such as the ITS regions of the nuclear rRNA operon, can be useful for epidemiologic studies and specifically for investigation of the possibility of person-to-person transmission (6). It has also made it possible to determine whether recurrent infections are due to the relapse of a previous infection or a new infection (10, 23). The study of the genetic diversity of organisms isolated from different geographic areas may help provide an understanding of the differences in strain virulence that may correlate with the severity of the disease (17).
Acknowledgments
The work done at the Centers for Disease Control and Prevention was supported in part by the Emerging Infectious Disease Fellowship from the Association of Public Health Laboratories.
We thank Brian Holloway and the staff of the NCID Biotechnology Core Facility for synthesis of the oligonucleotide primers.
REFERENCES
- 1.Banerji, S., E. B. Lugli, R. F. Miller, and A. E. Wakefield. 1995. Analysis of genetic diversity at the arom locus in isolates of Pneumocystis carinii. J. Eukaryot. Microbiol. 42:675-679. [DOI] [PubMed] [Google Scholar]
- 2.Beard, C. B., J. L. Carter, S. P. Keely, et al. 2000. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg. Infect. Dis. 6:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1999. Surveillance for AIDS-defining opportunistic illnesses, 1992-1997. Morb. Mortal. Wkly. Rep. 48:1-22. [PubMed] [Google Scholar]
- 4.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulin-Barrett, K. Lafe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphism in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser, P. M., D. S. Blanc, J. Bille, and P. Francioli. 1998. Typing methods to approach Pneumocystis carinii genetic heterogeneity. FEMS Immunol. Med. Microbiol. 22:27-35. [DOI] [PubMed] [Google Scholar]
- 6.Helweg-Larsen, J., A. G. Tsolaki, R. F. Miller, B. Lundgren, and A. E. Wakefield. 1998. Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. Q. J. Med. 91:813-820. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, B., J.-J. Lu, B. Li, X. Tang, M. S. Bartlett, J. W. Smith, and C. H. Lee. 1996. Development of type-specific PCR for typing Pneumocystis carinii f. sp. hominis based on nucleotide sequence variations of internal transcribed spacer regions of rRNA genes. J. Clin. Microbiol. 34:3245-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazanjian, P., W. Armstrong, P. A. Hossler, L. Huang, C. B. Beard, J. Carter, L. Crane, J. Duchin, W. Burman, J. Richardson, and S. R. Meshnick. 2001. Pneumocystis carinii cytochrome b mutations are associated with atovaquone exposure in patients with AIDS. J. Infect. Dis. 183:819-822. [DOI] [PubMed] [Google Scholar]
- 9. Keely, S. P., and J. R. Stringer. 1997. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J. Clin. Microbiol. 35:2745-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keely, S. P., J. R. Stringer, R. P. Baughman, M. J. Linke, P. D. Walzer, and A. G. Smulian. 1995. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J. Infect. Dis. 172:595-598. [DOI] [PubMed] [Google Scholar]
- 11.Latouche, S., E. Ortona, E. Mazars, P. Margutti, E. Tamburrini, A. Siracusano, K. Guyot, M. Nigou, and P. Roux. 1997. Biodiversity of Pneumocystis carinii hominis: typing with different DNA regions. J. Clin. Microbiol. 35:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, C. H., J. J. Lu, M. S. Bartlett, M. M. Durkin, T. H. Liu, J. Wang, B. Jiang, and J. W. Smith. 1993. Nucleotide sequence variation in Pneumocystis carinii strains that infect humans. J. Clin. Microbiol. 31:754-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, C. H., J. Helweg-Larsen, X. Tang, S. Jin, B. Li, M. S. Bartlett, J. J. Lu, B. Lundgren, J. D. Lundgren, M. Olsson, S. B. Lucas, P. Roux, A. Cargnel, C. Atzori, O. Matos, and J. W. Smith. 1998. Update on Pneumocystis carinii f. sp. hominis typing based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J. Clin. Microbiol. 36:734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, Y., and M. J. Leibowitz. 1993. Variation and in vitro splicing of group I introns in rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 21:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu, J.-J., M. S. Bartlett, M. M. Shaw, S. F. Queener, J. W. Smith, M. Ortiz-Rivera, M. J. Leibowitz, and C. H. Lee. 1994. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations in internal transcribed spacer regions of rRNA genes. J. Clin. Microbiol. 32:2904-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, L., L. Borio, H. Masur, and J. A. Kovacs. 1999. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J. Infect. Dis. 180:1969-1978. [DOI] [PubMed] [Google Scholar]
- 17.Miller, R. F., and A. E. Wakefield. 1999. Pneumocystis carinii genotypes and severity of pneumonia. Lancet 353:2039-2040. [DOI] [PubMed] [Google Scholar]
- 18.Navin, T. R., C. B. Beard, L. Huang, C. del Rio, S. Lee, N. J. Pieniazek, J. L. Carter, T. Le, A. Hightower, and D. Rimland. 2001. Effect of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of P. carinii pneumonia in patients with HIV-1: a prospective study. Lancet 358:545-549. [DOI] [PubMed] [Google Scholar]
- 19.Simonds, R. J., M. L. Lindegren, P. Thamas, D. Hanson, B. Caldwell, G. Scott, M. Rogers, and the Pneumocystis carinii Pneumonia Prophylaxis Evaluation Working Group. 1995. Prophylaxis against Pneumocystis carinii pneumonia among children with perinatally acquired human immunodeficiency virus infection in the United States. N. Engl. J. Med. 332:786-790. [DOI] [PubMed] [Google Scholar]
- 20.Stringer, J. R. 1993. The identity of Pneumocystis carinii: not a single protozoan, but a diverse group of exotic fungi. Infect. Agents Dis. 2:109-117. [PubMed] [Google Scholar]
- 21.Tanabe, K., M. Fuchimoto, K. Egawa, and Y. Nakamura. 1988. Use of Pneumocystis carinii genomic DNA clones for DNA hybridization analysis of infected human lungs. J. Infect. Dis. 157:593-596. [DOI] [PubMed] [Google Scholar]
- 22.Tang, X., M. S. Bartlett, J. W. Smith, J.-J. Lu, and C.-H. Lee. 1998. Determination of copy number of rRNA genes in Pneumocystis carinii f. sp. hominis. J. Clin. Microbiol. 36:2491-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsolaki, A. G., R. F. Miller, A. P. Underwood, S. Banerji, and A. E. Wakefield. 1996. Genetic diversity at the internal transcribed spacer regions of rRNA operon among isolates of Pneumocystis carinii from AIDS patients with recurrent pneumonia. J. Infect. Dis. 174:141-156. [DOI] [PubMed] [Google Scholar]
- 24.Tsolaki, A. G., P. Beckers, and A. E. Wakefield. 1998. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotypic similarity with contemporary isolates. J. Clin. Microbiol. 36:90-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakefield, A. E., C. C. Fritscher, A. S. Malin, L. Gwanzura, W. T. Hughes, and R. F. Miller. 1994. Genetic diversity in human-derived Pneumocystis carinii isolated from four geographical locations shown by analysis of mitochondrial ribosomal RNA gene sequences. J. Clin. Microbiol. 32:2959-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakefield, A. E. 1995. Re-examination of epidemiologic concepts. Bailliere's Clin. Infect. Dis. 2:431-448. [Google Scholar]