Abstract
The COBAS AMPLIPREP instrument for automated sample preparation has recently been introduced. In this study, the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test, which includes this new molecular device, was evaluated and compared to the COBAS AMPLICOR HCV MONITOR test, which includes a manual extraction protocol. Interassay and intra-assay variation, precision, and linearity were determined, and a total of 130 clinical specimens were investigated. For determination of interassay variation, coefficients of variation were found to be between 9 and 59% for the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and between 13 and 69% for the COBAS AMPLICOR HCV MONITOR test. For determination of intra-assay variation, coefficients of variation were found to be between 7 and 13% for the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and between 8 and 16% for the COBAS AMPLICOR HCV MONITOR test. When precision of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was tested, all results were found to be within ±0.5 log of the expected results. Determination of linearity resulted in a quasilinear curve over 3 logs. When clinical samples were tested with the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and compared with the COBAS AMPLICOR HCV MONITOR test, all results were found within ±0.5 log. In conclusion, the assay, which included the new molecular device, proved to be suitable for the routine molecular laboratory. It was found to be laborsaving and easy to handle.
Molecular techniques have been shown to be effective tools for direct detection of hepatitis C virus (HCV). Such assays for detection of pathogens basically consist of several steps: extraction of HCV RNA (also called sample preparation), reverse transcription (RT), amplification of cDNA, hybridization of amplified products, and detection of nucleic acid hybrids. Molecular techniques can be labor intensive and time consuming with the manual home-brew methods (8). To meet the needs of the routine diagnostic laboratory, PCR amplification and detection of amplified products have recently been automated with the COBAS AMPLICOR (Roche Molecular Systems, Inc., Branchburg, N.J.) analyzer (1, 3, 4, 6, 11). For detection of serum or plasma HCV RNA, both qualitative and quantitative tests are now available.
Sample preparation is currently considered the major weakness in molecular detection of HCV RNA. Conventional sample preparation protocols are usually time consuming, labor intensive, and susceptible to contamination. It has been demonstrated that the probability of false-positive results because of contamination increases in relation to the number of manipulations involved in sample processing (2, 10). To save time and labor, more rapid nucleic acid extraction protocols with fewer manipulation steps have largely replaced conventional protocols. Several ready-to-use sample preparation kits, available either separately or as part of entire molecular kits, were brought on the market and found to be suitable for inclusion in molecular assays for detection of RNA viruses (7, 9, 12). Recently a new automated specimen preparation instrument, the COBAS AMPLIPREP, was developed to automate sample preparation (5).
The aim of this study was to evaluate performance of the COBAS AMPLIPREP analyzer for sample preparation followed by quantitative detection of HCV RNA with the COBAS AMPLICOR HCV MONITOR test. Both interassay and intra-assay variations were determined and compared with those of the manual sample preparation. Precision was tested with a reference material; linearity was tested by a dilution series of high-titer samples. Performance of the new sample preparation instrument in the routine clinical laboratory was evaluated with routine clinical samples.
MATERIALS AND METHODS
Molecular assays.
The COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test, version 2.0 (Roche), which includes automated sample preparation on the COBAS AMPLIPREP analyzer, and the COBAS AMPLICOR HCV MONITOR test, version 2.0 (Roche), which includes a manual sample preparation protocol, were performed according to the manufacturer's package insert instructions.
Automated sample preparation on the COBAS AMPLIPREP instrument.
For the automated sample preparation, 250 μl of the controls and samples were transferred to bar code-labeled tubes. A known number of HCV Quantitation Standard RNA molecules were introduced by the instrument into each sample together with the lysis reagent and the biotinylated capture probe reagent. The system automatically released the target nucleic acid and captured the target with specific oligonucleotide probes, which became attached to magnetic beads via a biotin-streptavidin binding reaction. After attachment to the beads, the target was purified and concentrated automatically by the instrument. Processed samples and controls were manually pipetted into the prealiquoted master mixes.
Manual sample preparation protocol.
HCV RNA was isolated from serum according to the manufacturer's instructions. A known number of HCV Quantitation Standard RNA molecules were introduced into each specimen and carried through the whole procedure.
Reverse transcription, amplification, hybridization, and detection.
Steps after sample preparation and the preparation of reaction mixtures were automatically done on the COBAS AMPLICOR analyzer. HCV RNA was reverse transcribed, followed by PCR amplification of target cDNA using HCV-specific, biotinylated primers. Amplification products were hybridized to capture probes, and hybridization products were detected by colorimetric determination.
Study design.
All experiments were done in the Molecular Diagnostics Laboratory, Institute of Hygiene, by the same technician.
In a first step, interassay variation of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was determined and compared with that of the COBAS AMPLICOR HCV MONITOR test, which included the manual sample preparation protocol. Fourteen dilutions from a high-titer HCV RNA routine clinical sample in pooled normal human plasma were prepared. Samples contained different amounts of HCV RNA ranging from 3.1 × 102 to 1.6 × 106 IU/ml and were tested five times on five different days.
In a second step, intra-assay variation of the assays was compared. Four clinical routine samples were aliquoted and analyzed five times each in a single run.
In a third step, precision of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was determined with the Second European Union Concerted Action Hepatitis C Virus Proficiency Panel (www.qcca.org.uk). Samples of this panel contained different concentrations of HCV genotype 1 (2.0 × 102, 1.9 × 103, 1.9 × 103, and 4.4 × 104 IU/ml), HCV genotype 3 (7.1 × 104 IU/ml), and HCV genotype 4 (3.1 × 103 IU/ml).
In a fourth step, linearity of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was determined. Three routine clinical sera, which exceeded 1.0 × 106 IU of HCV RNA/ml, were taken. A dilution series (alternating 5- and 2-fold) was prepared using HCV-negative human plasma, which is included in the COBAS AMPLICOR kits. Each dilution was analyzed four times, and the mean HCV RNA titer of each sample was determined.
In a fifth step, performance of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test in the routine clinical laboratory was evaluated and compared with that of the COBAS AMPLICOR HCV MONITOR test. A total of 130 clinical sera from patients with chronic hepatitis C with or without anti-HCV therapy were investigated. Another aliquot of each of the samples had been tested earlier with the COBAS AMPLICOR HCV MONITOR test in the routine-diagnostic laboratory.
Statistical analysis.
For computerized statistical analysis, SPSS 10.0 (SPSS Inc., Chicago, Ill.) was employed. A log transformation was used to adjust for inhomogeneity of the variance structure. Linearity was tested by comparison of the results of polynomial regression by Mallows Cp statistic with a quadratic regression.
RESULTS
For determination of interassay variation, mean HCV RNA titers of 14 samples ranged between 2.1 × 102 and 6.8 × 105 IU/ml. Coefficients of variation were found to be between 9 and 59% for the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and between 13 and 69% for the COBAS AMPLICOR HCV MONITOR test (Table 1).
TABLE 1.
Interassay variation of results obtained by the automated versus manual sample preparation protocol
| Sample no. | Mean amt of HCV RNA (IU/ml) detected
|
SD
|
Coefficient of variation (%)
|
|||
|---|---|---|---|---|---|---|
| Automateda | Manualb | Automateda | Manualb | Automateda | Manualb | |
| 1 | 2.1 × 102 | 3.8 × 102 | 1.3 × 102 | 1.1 × 102 | 59 | 29 |
| 2 | 4.2 × 102 | 8.8 × 102 | 2.0 × 102 | 6.1 × 102 | 48 | 69 |
| 3 | 7.2 × 103 | 1.0 × 102 | 2.5 × 102 | 3.7 × 102 | 35 | 38 |
| 4 | 2.3 × 103 | 2.2 × 103 | 6.5 × 102 | 8.7 × 102 | 27 | 39 |
| 5 | 4.5 × 103 | 4.2 × 103 | 1.1 × 103 | 8.6 × 102 | 25 | 21 |
| 6 | 7.3 × 103 | 7.6 × 103 | 1.2 × 103 | 1.5 × 103 | 17 | 20 |
| 7 | 1.6 × 104 | 1.4 × 104 | 3.7 × 103 | 3.0 × 103 | 23 | 21 |
| 8 | 3.2 × 104 | 2.6 × 104 | 4.3 × 103 | 6.1 × 103 | 14 | 23 |
| 9 | 6.7 × 104 | 4.8 × 104 | 9.5 × 103 | 2.1 × 104 | 14 | 44 |
| 10 | 1.1 × 105 | 7.6 × 105 | 1.6 × 104 | 1.1 × 104 | 14 | 15 |
| 11 | 2.1 × 105 | 1.3 × 105 | 2.0 × 104 | 2.5 × 104 | 9 | 20 |
| 12 | 3.2 × 105 | 2.2 × 105 | 5.7 × 104 | 9.3 × 104 | 18 | 42 |
| 13 | 4.2 × 105 | 2.8 × 105 | 5.4 × 104 | 3.7 × 104 | 13 | 13 |
| 14 | 6.8 × 105 | 4.2 × 105 | 1.3 × 105 | 1.3 × 105 | 18 | 32 |
Automated sample preparation on the COBAS AMPLIPREP analyzer.
Manual sample preparation with the COBAS AMPLICOR HCV MONITOR test.
Determination of intra-assay variation was tested with four routine clinical samples by analyzing them five times. Coefficients of variation were found to be between 7 and 13% for the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and between 8 and 16% for the COBAS AMPLICOR HCV MONITOR test (Table 2).
TABLE 2.
Intra-assay variation of results obtained by the automated versus manual sample preparation protocols
| Sample no. | Mean amt of HCV RNA (IU/ml detected)
|
SD
|
Coefficient of variation (%)
|
|||
|---|---|---|---|---|---|---|
| Automateda | Manualb | Automateda | Manualb | Automateda | Manualb | |
| 1 | 9.4 × 105 | 7.0 × 105 | 1.3 × 105 | 9.9 × 104 | 13.3 | 14.2 |
| 2 | 6.3 × 105 | 4.6 × 105 | 6.8 × 104 | 7.2 × 104 | 10.6 | 15.6 |
| 3 | 3.0 × 105 | 2.5 × 105 | 3.6 × 104 | 2.4 × 104 | 11.9 | 9.5 |
| 4 | 1.9 × 104 | 2.0 × 104 | 1.4 × 103 | 1.7 × 103 | 7.3 | 8.3 |
Automated sample preparation on the COBAS AMPLIPREP analyzer.
Manual sample preparation with the COBAS AMPLICOR HCV MONITOR test.
When six samples of the Second European Union Concerted Action Hepatitis C Virus Proficiency Panel were tested with the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test, all results were found to be within ±0.5 log of the expected panel results (Table 3). Sample number 3 containing 200 IU of HCV RNA/ml was found to be below the detection limit.
TABLE 3.
Results of precision testing performed with the Second European Union Concerted Action HCV Proficiency Panel
| Vial no. | Genotype | HCV RNA (IU/ml)
|
|
|---|---|---|---|
| Panel resultsa | Obtained results | ||
| 1 | 4 | 3.1 × 103 | 9.88 × 102 |
| 2 | 1 | 5.4 × 104 | 2.18 × 105 |
| 3 | 1 | 2.0 × 102 | BDLb |
| 4 | 1 | 1.9 × 103 | 5.59 × 103 |
| 5 | 3 | 7.1 × 104 | 4.01 × 104 |
| 6 | 1 | 1.9 × 103 | 4.68 × 103 |
Average of results obtained by the production laboratory (Boston Biomedica Inc., Boston, Mass.) using the AMPLICOR HCV MONITOR test, version 2.0.
BDL, below detection limit.
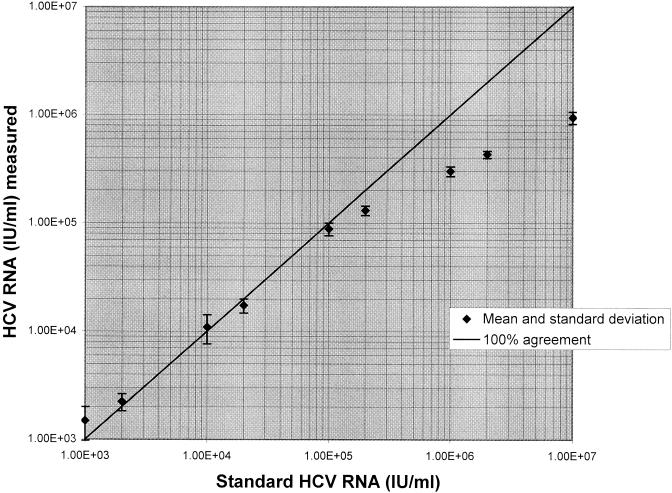
Linearity was tested with a dilution series of three high-titer routine clinical samples. Up to 1.00 × 105 IU/ml, a quasilinear curve was observed. For higher HCV-RNA values, an increasing underestimation of expected values was found (Fig. 1).
FIG. 1.
Linearity results produced by the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test, version 2.0, with dilution of a high-titer patient sample.
When 130 clinical samples were tested with the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test and results were compared with those of the COBAS AMPLICOR HCV MONITOR test, all results were found to be within ±0.5 log. All sera which tested negative with the COBAS AMPLICOR HCV MONITOR test were also found to be negative with the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test.
The automated sample preparation proved to be quick and labor saving. For 24 sera, sample preparation took 2.5 h. The time required for steps of maintenance and loading of reagents and disposables was 20 min, followed by a 10-min pipetting of samples. Extraction of samples took 2 h without any hands-on work. In contrast, the manual sample preparation could be finished within 2 h with continuous user interventions.
For both assays, aliquots of master mixes had to be transferred into PCR tubes and extracted samples had to be added manually.
DISCUSSION
In the present study, the performance of the COBAS AMPLIPREP analyzer for sample preparation was evaluated and compared to that of the COBAS AMPLICOR HCV MONITOR test, which includes a manual extraction protocol.
According to the manufacturer's package insert of the COBAS AMPLICOR HCV MONITOR test, version 2.0, the interassay variation for quantitation of serum HCV RNA ranges between 8 and 91% and the intra-assay variation ranges between 7 and 51%. In this study, the interassay variation of the COBAS AMPLICOR HCV MONITOR test ranged between 13 and 69% and the intra-assay variation between 8 and 16%. With the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test, similar results were found for both interassay (9 to 59%) and intra-assay (7 to 13%) variation. Inter- and intra-assay variations obtained by both assays were found to be at the lower border of the range indicated in the manufacturer's package insert mentioned above. No significant differences in interassay and intra-assay variation were observed between the manual and the automated sample preparation protocol. This may be explained by the skills and experience of the technician involved in this study.
For a routine diagnostic laboratory, it is of major importance to report accurate and reliable results of molecular assays. To meet the requirements of quality assurance and quality control, it is essential to participate in an external quality assessment program. In this study, the Second European Union Concerted Action Hepatitis C Virus Proficiency Panel was used to evaluate precision of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test. All results were found to be correct, i.e., within ±0.5 log of the expected panel results.
The linear range of the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was determined by analysis of a series of dilutions of high-titer HCV-RNA sera. According to the manufacturer's package insert of the COBAS AMPLICOR HCV MONITOR test, this assay is linear between 600 IU/ml and 850,000 IU/ml. In this study, the automated assay test revealed sufficient linearity up to 1.00 × 105 IU/ml. HCV-RNA titers above this value are underestimated. To obtain accurate results for samples with HCV RNA levels above 1.00 × 105 IU/ml, it may be advisable to prepare appropriate dilutions prior to sample preparation.
When the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test was used for routine samples, results were comparable to those obtained by the COBAS AMPLICOR HCV MONITOR test. No discrepant results were observed during the whole study.
The new analyzer for sample preparation showed good overall functionality and user friendliness. Compared with the manual sample preparation, a 10% increase of total time was observed with the automated sample preparation protocol. In contrast, hands-on time was reduced by approximately 70%. Pipetting of the extracted samples into the tubes containing the master mixes, however, is still not automated and thus has to be done manually. Nevertheless, because of the significantly lower number of manipulations required, there may be less probability of getting false-positive results because of contamination.
In conclusion, the COBAS AMPLIPREP/COBAS AMPLICOR HCV MONITOR test proved to be suitable for the routine-diagnostic laboratory. Compared to the manual assay, it saved hands-on work and was easy to use. Due to automation more accurate results may be obtained, especially for quantitative tests.
REFERENCES
- 1.Albadalejo, J., R. Alonso, R. Antinozzi, M. Bogard, A. M. Bourgault, G. Colucci, T. Fenner, H. Petersen, E. Sala, J. Vincelette, and C. Young. 1998. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J. Clin. Microbiol. 36:862-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clewley, J. P. 1989. The polymerase chain reaction, a review of the practical limitations for human immunodeficiency virus diagnosis. J. Virol. Methods 25:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Doglio, A., C. Laffont, F. X. Caroli-Bosc, P. Rochet, and J. C. Lefebvre. 1999. Second generation of the automated COBAS AMPLICOR HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J. Clin. Microbiol. 37:1567-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jungkind, D., S. DiRenzo, K. G. Beavis, and N. S. Silverman. 1996. Evaluation of an automated COBAS AMPLICOR PCR system for detection of several infectious agents and the impact on laboratory management. J. Clin. Microbiol. 34:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction--our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Kessler, H. H., E. A. Dragon, K. Pierer, B. I. Santner, Y. Liao, D. Stünzner, E. Stelzl, and E. Marth. 1997. Performance of the automated COBAS AMPLICOR system for the detection of hepatitis C virus RNA. Clin. Diagn. Virol. 7:139-145. [DOI] [PubMed] [Google Scholar]
- 7.Kessler, H. H., K. Pierer, B. I. Santner, S. K. Vellimedu, E. Stelzl, E. Marth. P. Fickert, and R. E. Stauber. 1999. Evaluation of molecular parameters for routine assessment of viremia in patients with chronic hepatitis C who are undergoing antiviral therapy. J. Hum. Virol. 1:314-319. [PubMed] [Google Scholar]
- 8.Klapper, P., D. Jungkind, T. Fenner, R. Antinozzi, J. Schirm, and C. Blanckmeister. 1998. Multicenter international workflow study of an automated polymerase chain reaction instrument. Clin. Chem. 44:1737-1739. [PubMed] [Google Scholar]
- 9.Kramis, A., S. Bukofzer, and M. C. Kew. 1996. Comparison of hepatitis B virus DNA extractions from serum by the QIAamp blood kit, GeneReleaser, and the phenol-chloroform method. J. Clin. Microbiol. 34:2731-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature (London) 339:237-238. [DOI] [PubMed] [Google Scholar]
- 11.Poljak, M., K. Seme, and S. Koren. 1997. Evaluation of the automated COBAS AMPLICOR hepatitis C virus PCR system. J. Clin. Microbiol. 35:2983-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafer, R. W., D. J. Levee, M. A. Winters, K. L. Richmond, D. Huang, and T. C. Merigan. 1997. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1-RNA from plasma. J. Clin. Microbiol. 35:520-522. [DOI] [PMC free article] [PubMed] [Google Scholar]