Abstract
Molecular characterization of Candida albicans isolates is essential for understanding the epidemiology of nosocomial infections caused by this yeast. Here, we investigated the potential value of multilocus sequence typing (MLST) for characterizing epidemiologically related or unrelated C. albicans strains of various clinical origins. Accordingly, we sequenced the internal regions (loci) of six selected housekeeping genes of 40 C. albicans clinical isolates and 2 reference strains. In all, 68 polymorphic nucleotide sites were identified, of which 65 were found to be heterozygous in at least one isolate. Ten to 24 different genotypes were observed at the different loci, resulting, when combined, in 39 unique genotype combinations or diploid sequence types (DSTs). When MLST was applied to 26 epidemiologically unrelated isolates and the 2 reference strains, it allowed the identification of 27 independent DSTs, thus demonstrating a discriminatory power of 99.7. Using multidimensional scaling together with the minimum spanning tree method to analyze interstrain relationships, we identified six groups of genetically related isolates on the basis of bootstrap values of greater than 900. Application of MLST to 14 epidemiologically related isolates showed that those recovered from patients in the same hospital ward during the same 3 months had specific DSTs, although 73% of these isolates were genetically very close. This suggests that MLST can trace minute variations in the sequences of related isolates. Overall, MLST proved to be a highly discriminatory and stable method for unambiguous characterization of C. albicans.
Candida albicans is a commensal yeast which is also a leading cause of nosocomial infections. The number of patients infected by C. albicans has risen sharply over the last two decades, and these infections are associated with high mortality rates despite of the introduction of novel antifungal agents. Prevention of nosocomial infections caused by C. albicans and related species is difficult, because little is yet known about either the dynamics of transmission among hospitalized patients or the characteristics of the infecting strains. During the last 10 years, the sources of the C. albicans isolates responsible for infecting hospital patients have been identified in epidemiological studies by using molecular typing techniques. It is now well accepted that strains which colonize patients prior to the occurrence of a systemic infection are usually those that are at the origin of the infection (17, 20, 29). Colonized patients are the main reservoir of C. albicans in hospitals, and the cross-contamination that occurs between patients (21, 22, 24, 28) suggests that the source of colonization can be either a commensal strain belonging to the patient's microflora prior to hospitalization or a strain acquired through cross-contamination within the hospital. Longitudinal surveillance studies have shown that several endemic strains were sometimes carried by patients from different wards (17, 21).
However, the typing methods used to characterize C. albicans isolates in epidemiological studies are very diverse. They include restriction fragment length polymorphism analysis (20, 28), Southern blot hybridization with discriminating probes (17, 21, 24), electrophoretic karyotyping (29), and randomly amplified polymorphic DNA analysis (22). Results are laboratory dependent, which precludes interlaboratory comparison of infecting isolates. As no consensual typing method is presently available to characterize C. albicans isolates circulating within or between hospitals, it is not clear whether some strains have a particular ability to cause nosocomial infections or have a local or international distribution. Thus, no global view of the population dynamics and epidemiology of the C. albicans strains that cause nosocomial infections has yet emerged.
These questions are common to the study of nosocomial infections caused by other microorganisms, including bacteria. For bacteria, the multilocus sequence typing (MLST) method has been recently proposed to overcome these flaws (16). MLST is a highly resolutive method based on the analysis of nucleotide polymorphisms of the sequences of approximately 450- to 500-bp internal fragments (loci) of housekeeping genes. For each housekeeping locus, the different sequences present within a bacterial species are assigned as distinct alleles, and for each isolate, the alleles at each of the sequenced loci define an allelic profile or sequence type. Each isolate of a species is therefore unambiguously characterized by a series of integers which correspond to the alleles at the housekeeping loci studied. A major advantage of MLST over other typing methods is that sequence data can be easily compared between laboratories, thus permitting the exchange of molecular typing data via the internet for global epidemiology (16). MLST has been used to characterize many bacterial pathogens, including Neisseria meningitidis, Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Campylobacter jejuni (4, 6-8).
Although MLST was originally developed for haploid organisms, a similar sequence typing methodology has been described for C. albicans, which is an asexual diploid organism. It is based on the sequencing of 12 anonymous loci and has been successfully used for genetic population analysis (9). However, this method has not been evaluated for analysis of isolates from the standpoint of understanding nosocomial infections.
In the present investigation, we developed an MLST scheme that appeared to be highly resolutive, in order to clarify the relationships between C. albicans isolates with well-defined clinical and epidemiological links.
MATERIALS AND METHODS
Strains.
Two groups of C. albicans strains were selected for MLST analysis. The first group included 26 epidemiologically unrelated clinical isolates recovered between 1993 and 2000 from 26 patients in 10 different hospitals located in four French regions and also the two reference strains C. albicans ATCC 36232 and SC5314. The genome sequence of strain SC5314 is currently being determined at the Stanford Genome Technology Center (27). The second group of strains studied comprised 14 epidemiologically related clinical isolates, including 11 which came from 11 patients hospitalized in the same intensive care unit and were isolated during the same 3-month period and 3 isolated from a pregnant woman and her fetus during an episode of systemic candidiasis which ended in abortion. Clinical isolates of Candida glabrata, Candida tropicalis, Candida dubliniensis, Candida krusei, Candida parapsilosis, Candida lusitaniae, and Saccharomyces cerevisiae were also tested. Strains were stored at −80°C in Cryo-bille tubes (Laboratoire AES, Combourg, France).
DNA extraction.
After subculture on Sabouraud agar, two colonies of each isolate were inoculated into 10 ml of YPD broth (2% glucose, 2% Bacto Peptone, and 1% yeast extract) in a sterile tube and grown overnight at 37°C with shaking. Two milliliters of the culture was centrifuged, the pellet was resuspended in 200 μl of buffer (50 mM Tris-HCl [pH 8], 25 mM EDTA [pH 8], and 1% [vol/vol] β-mercaptoethanol) containing 0.5 mg of Zymolyase (25,000 U; ICN Pharmaceuticals, Costa Mesa, Calif.), and the mixture was incubated for 1 h at 37°C. After centrifugation, the pellet was resuspended in 200 μl of lysis buffer (200 mM diethanolamine, 80 mM EDTA [pH 9], and 1% [wt/vol] sodium dodecyl sulfate) and incubated for 30 min at 65°C. Following addition of 100 μl of 5 M potassium acetate, incubation was continued for 45 min on ice. DNA was precipitated with 3.5 volumes of ethanol-7.5 M ammonium acetate (6:1), rinsed with 70% ethanol, dried, and dissolved in 50 μl of sterile water.
Choice of loci.
Murad et al. (18) reported the identification of 3,313 putative open reading frames (ORFs) from assembly 3 of the C. albicans genome sequence carried at the Stanford DNA Sequencing and Genome Technology Center (27). Comparison of these ORFs to the S. cerevisiae proteome revealed a set of C. albicans ORFs with very high similarity to S. cerevisiae ORFs, all of them encoding housekeeping functions. The amino acid sequences corresponding to these C. albicans ORFs and their homologues in S. cerevisiae sequences were aligned, and the most variable regions were identified. Among these regions, those of roughly 150 to 200 amino acids in length and flanked by highly conserved regions were selected. Primers were designed by using the nucleotide sequences of the conserved regions in the C. albicans ORF. Each pair of primers was designed to amplify a 500- to 700-bp fragment.
Amplification and nucleotide sequence determination.
PCRs were carried out in 100-μl reaction volumes containing about 0.1 μg of extracted DNA, 100 pmol of each primer, 2.5 U of Taq DNA polymerase (Amersham Pharmacia Biotech, Orsay, France), 10 μl of 10× buffer (supplied with the Taq polymerase), and a 200 μM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim, Meylan, France). PCRs were performed with an initial 5-min denaturation step at 93°C, followed by 30 cycles of 93°C for 30 s, 55°C for 1 min, and 72°C for 1 min, with a final extension step of 4 min at 72°C. The amplified fragments were purified using a PCR purification kit (Qiagen, Courtaboeuf, France). Purified fragments were sequenced on both strands by using the same primers as those used in the initial amplification. Sequencing reactions were prepared using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) according to the manufacturer's recommendations for cycle sequencing on the GeneAmp PCR 9700 system. The reactions were analyzed on an ABI PRISM 3700 Genetic Analyzer (PE Applied Biosystems).
Sequences and computations.
Sequence analysis was performed using programs of the Staden package (26). Defined regions in all sequences obtained from a single locus were aligned using Gap4. This program automatically highlights disagreements between sequences that exhibit potential polymorphisms. All such sites were visually inspected, and the polymorphic nucleotide sites within these sequences were identified. Each polymorphic site could be either homozygous or heterozygous. For each site, the nature of the two bases resulting from the coamplification and simultaneous sequencing of the two complementary loci was recorded by inspection of the sequence chromatogram, peak by peak. Heterozygosity was identified by the presence of two peaks showing incorporation of the two bases resulting from their coamplification. The one-letter code for nucleotides from the International Union of Pure and Applied Chemistry nomenclature was used to designate homozygous and heterozygous polymorphic sites.
A matrix of distances between pairwise sequences was constructed using the following distances: 0 for identical homozygous or heterozygous sites, 0.5 for homozygous or heterozygous sites sharing one nucleotide, and 1 otherwise. Relationships among isolates were then assessed by cluster analysis, using the unweighted pair-group method with arithmetic averages and the percent disagreement distance measure (Statistica version 4.1; StatSoft).
To obtain a two-dimensional representation of the links between isolates, the matrix of distances was also computed by using the multidimensional scaling (MDS) method (14). Relationships between isolates were then calculated by the minimum spanning tree (MST) method (14, 19), which also determines the lengths of MST connections. The statistical robustness of MST connections was evaluated by bootstrap resamplings (1,000 iterations).
Reproducibility and stability of the MLST method.
To evaluate the stability of the MLST method, the reference strain ATCC 36232 was subcultured daily for 30 days (approximately 400 generations) in YPD broth. The last subculture was plated on Sabouraud agar, and six subclones were subjected to MLST. To test for reproducibility, the DNAs from nine different isolates were extracted from two independent cultures and used for MLST.
RESULTS
Sequence diversity in C. albicans MLST.
Using the genome sequence of the C. albicans reference strain SC5314 available at the Stanford DNA Sequencing and Genome Technology Center (27), we identified regions in six C. albicans housekeeping genes showing relatively high divergence from the corresponding regions in the S. cerevisiae ortholog and flanked by regions that are highly conserved between C. albicans and S. cerevisiae (Table 1). Table 2 lists the oligonucleotides used to amplify these regions (loci) from all 42 C. albicans isolates tested. Analysis of the sequences obtained from all strains and loci showed that all of the sequences obtained from a single locus could be aligned without gaps or insertions. In all, 2,354 nucleotides from six loci were studied for each isolate. Sixty-eight polymorphic nucleotide sites (2.9%) were detected among the 42 C. albicans isolates. The number of polymorphic sites for each locus ranged from 6 in locus CaACC1 to 16 in locus CaVPS13 (Table 3). The number of nucleotide substitutions from the C. albicans SC5314 genome sequence that were not conservative at the amino acid level ranged from one at locus CaRPN2 to nine at locus CaVPS13 (Table 3).
TABLE 1.
Housekeeping loci used
| Locus | C. albicans ORF no.a | Size of C. albicans ORF (bp) | Locus position | S. cerevisiae ORF ortholog no.b | Putative function of gene product |
|---|---|---|---|---|---|
| CaACC1 | 6-8718 | 6,816 | 3251-3769 | YNR016c | Acetyl-coenzyme A carboxylase |
| CaVPS13 | 6-4223 | 9,252 | 5151-5891 | YLL040c | Vacuolar protein sorting protein |
| CaGLN4 | 6-8489 | 2,400 | 109-591 | YOR168w | Glutaminyl-tRNA synthetase |
| CaADP1 | 6-2855 | 3,117 | 918-454 | YCR011c | ATP-dependent permease |
| CaRPN2 | 6-7682 | 2,859 | 1083-1529 | YIL075c | 26S proteasome regulatory subunit |
| CaSYA1c | 6-2925 | 2,910 | 2400-2942 | YOR335c | Alanyl-RNA synthetase |
ORF numbers refer to the genome sequence of C. albicans SC5314 published by Stanford University (http://sequence-www.stanford.edu/group/candida).
ORF numbers refer to the genome sequence of S. cerevisiae (http://genome- www.stanford.edu/Saccharomyces).
CaSYA1 is the ortholog of the ScALA1 gene encoding alanyl-tRNA synthetase; however, the designation CaALA1 could not be used because it corresponds to a gene encoding an agglutinin-like adhesin (11).
TABLE 2.
Oligonucleotide primers used for C. albicans MLST
| Locus | Primer | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|
| CaACC1 | 466 forward | GCAAGAGAAATTTTAATTCAATG | |
| 466 reverse | TTCATCAACATCATCCAAGTG | 519 | |
| CaVPS13 | 552 forward | TCGTTGAGAGATATTCGACTT | 741 |
| 552 reverse | ACGGATGGATCTCCAGTCC | ||
| CaGLN4 | 598 forward | GAGATAGTCAAGAATAAAAAAGT | 483 |
| 598 reverse | ATCTCTTTCATCTTTTGGACC | ||
| CaADP1 | 904 forward | GAGCCAAGTATGAATGATTTG | 537 |
| 904 reverse | TTGATCAACAAACCCGATAAT | ||
| CaRPN2 | 1041 forward | TTCATGCATGCTGGTACTAC | 447 |
| 1041 reverse | TAATCCCATACCCAAAGCAG | ||
| CaSYA1 | 1369 forward | AGAAGAATTGTTGCTGTTACTG | 543 |
| 1369 reverse | GTTACCTTTACCACCAGCTTT |
TABLE 3.
Characteristics of the six housekeeping loci studied
| Locus | Size of sequenced fragment (bp) | No. of genotypes identified | No (%). of polymorphic nucleotide sites | No. of poly- morphic amino acid sites |
|---|---|---|---|---|
| CaACC1 | 407 | 10 | 6 (1.5) | 2 |
| CaVPS13 | 403 | 24 | 16 (4) | 9 |
| CaGLN4 | 404 | 15 | 11 (2.7) | 7 |
| CaADP1 | 443 | 16 | 13 (2.9) | 6 |
| CaRPN2 | 306 | 16 | 11 (3.6) | 1 |
| CaSYA1 | 391 | 13 | 11 (2.8) | 4 |
C. albicans is almost always diploid as isolated, and evidence has been obtained for heterozygosity of complementary chromosomes (23). Therefore, amplification of genomic DNA is expected to result in PCR products that arise from both chromosomes and that upon direct sequencing yield profiles that correspond to the superimposition of the two heterozygous alleles. In the six loci investigated here, polymorphic nucleotide sites were indeed either homozygous or heterozygous, confirming that the oligonucleotides used in this study could amplify both chromosomal alleles. None of the isolates was 100% homozygous. The presently available genome sequence for C. albicans strain SC5314 is composed mostly of homozygous nucleotide sequences derived from the two complementary chromosomes. However, in the present study, we found one heterozygosity at position 35 in locus CaADP1. Thus, altogether, and including reference strain SC5314, at least one heterozygosity was observed among the 68 polymorphic sites in the 42 isolates investigated. These results underline the fact that local heterozygosity is frequent in the sequences of the six housekeeping loci studied.
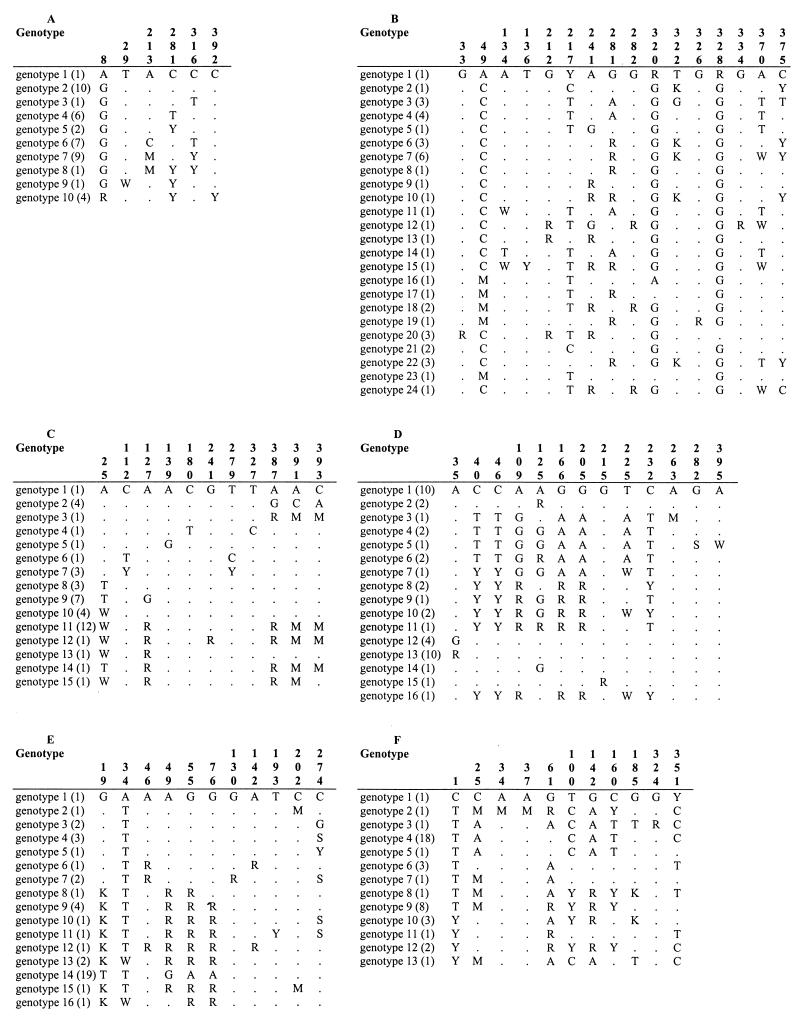
Each of the different sequences in a locus defined a distinct genotype, even if it differed from the others by only a single nucleotide. The different genotypes were numbered in the order of their identification. The number of genotypes identified for each of the six loci investigated ranged from 10 for CaACC1 to 24 for CaVPS13 (Table 3). The positions of the polymorphic nucleotide sites, the different genotypes identified at each of the six loci, and their frequency of detection among the 42 C. albicans isolates are shown in Fig. 1. We noted with interest that at each locus except CaVPS13 some genotypes were more frequent than others and that at least one was found in 20% or more of the isolates.
FIG. 1.
Positions of the polymorphic nucleotide sites and genotypes identified at loci CaACC1 (A), CaVPS13 (B), CaGLN4 (C), CaADP1 (D), CaRPN2 (E), and CaSYA1 (F). The nucleotides present at each variable site among the 42 C . albicans strains tested are shown for genotype 1. For the other genotypes, only sites that differ from those in genotype 1 are shown; sites that are the same as those in genotype 1 are shown by dots. The numbers of isolates with the same genotype are indicated in parentheses. The position of each polymorphic site relative to the fragment sequenced is shown at the top in vertical format. Y, C or T; R, A or G; K, G or T; M, A or C; S, G or C; W, A or T.
For each C. albicans isolate, we defined a diploid sequence type (DST) as the unique combination of the genotypes of the six loci studied. Thirty-nine DSTs were found among the 42 C. albicans isolates (Table 4). One was shared by three isolates (DST 29; isolates CP7, CP8, and CP9), and one was shared by two isolates (DST 14; isolates 17 and 21). The remaining 37 DSTs (95%) were each found in a single isolate only. Among these 37 DSTs, 7 pairs differed at only one locus (DSTs 18 and 20, 31 and 33, 32 and 33, 31 and 38, 33 and 34, 14 and 36, and 16 and 30). The other DSTs differed at two to six loci. When MLST analysis of our 42 C. albicans isolates was performed using the data from only the three loci CaVPS13, CaGLN4, and CaADP1, 38 unique combinations of genotypes could be identified instead of the 39 DSTs identified using the data from the six loci.
TABLE 4.
Characteristics and genotypes of the 42 C. albicans isolates tested
| Isolate | Source
|
Epidemiological linkc | Genotype
|
DST | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinicala | Geographicb | CaACC1 | CaVPS13 | CaGLN4 | CaADP1 | CaRPN2 | CaSYA1 | |||
| 1 | Skin | Paris area, H1 | U | 8 | 5 | 8 | 2 | 9 | 8 | 21 |
| 2 | Blood | Paris area, H2 | U | 7 | 7 | 9 | 13 | 14 | 4 | 19 |
| 3 | Vagina | Paris area, H3 | U | 6 | 2 | 11 | 12 | 14 | 4 | 16 |
| 4 | Blood | Paris area, H4 | U | 7 | 6 | 2 | 13 | 14 | 4 | 17 |
| 5 | Blood | Paris area, H7 | U | 2 | 7 | 11 | 13 | 14 | 4 | 9 |
| 6 | Blood | Paris area, H1 | U | 2 | 4 | 10 | 1 | 2 | 9 | 5 |
| 7 | Blood | Lyon | U | 10 | 18 | 12 | 10 | 13 | 9 | 4 |
| 8 | Blood | Lyon | U | 4 | 12 | 2 | 1 | 10 | 7 | 12 |
| 9 | Blood | Paris area, H4 | U | 3 | 14 | 10 | 1 | 3 | 6 | 11 |
| 10 | Blood | Paris area, H5 | U | 10 | 16 | 2 | 9 | 13 | 9 | 23 |
| 11 | Blood | Strasbourg | U | 2 | 4 | 10 | 1 | 4 | 6 | 6 |
| 12 | Blood | Nantes | U | 2 | 17 | 8 | 6 | 7 | 12 | 10 |
| 13 | Blood | Paris area, H5 | U | 4 | 13 | 6 | 3 | 3 | 13 | 13 |
| 14 | Blood | Nantes | U | 2 | 4 | 8 | 1 | 4 | 9 | 3 |
| 15 | Blood | Lyon | U | 10 | 18 | 11 | 4 | 1 | 9 | 24 |
| 16 | Blood | Paris area, H1 | U | 9 | 1 | 3 | 11 | 12 | 5 | 22 |
| 17 | Blood | Paris area, H5 | U | 4 | 20 | 7 | 8 | 9 | 10 | 14 |
| 18 | Blood | Paris area, H1 | U | 2 | 9 | 5 | 7 | 8 | 11 | 8 |
| 19 | Sputum | Paris area, H4 | U | 5 | 19 | 1 | 5 | 11 | 1 | 15 |
| 20 | Blood | Strasbourg | U | 2 | 8 | 9 | 1 | 6 | 2 | 7 |
| 21 | Vagina | Paris area, H8 | U | 4 | 20 | 7 | 8 | 9 | 10 | 14 |
| 22 | ATCC 36232d | U | 1 | 15 | 4 | 4 | 5 | 3 | 1 | |
| 23 | Gland | Paris area, H1 | U | 7 | 10 | 11 | 12 | 14 | 4 | 20 |
| 24 | Urine | Paris area, H1 | U | 7 | 6 | 11 | 12 | 14 | 4 | 18 |
| 25 | SC5314d | U | 2 | 3 | 9 | 13 | 14 | 4 | 2 | |
| 26 | Blood | Strasbourg | U | 5 | 11 | 10 | 15 | 4 | 9 | 35 |
| 27 | Blood | Nantes | U | 4 | 20 | 7 | 16 | 9 | 10 | 36 |
| 28 | Blood | Lyon | U | 10 | 24 | 11 | 10 | 16 | 9 | 37 |
| CP1 | Feces | Paris area, H1 | R | 7 | 22 | 11 | 13 | 14 | 4 | 33 |
| CP2 | Feces | Paris area, H1 | R | 6 | 7 | 11 | 12 | 14 | 4 | 30 |
| CP3 | Urine | Paris area, H1 | R | 6 | 6 | 11 | 13 | 14 | 4 | 28 |
| CP4 | Feces | Paris area, H1 | R | 2 | 21 | 13 | 2 | 15 | 6 | 26 |
| CP5 | Bronchoalveolar fluid | Paris area, H1 | R | 7 | 3 | 11 | 13 | 14 | 4 | 31 |
| CP6 | Feces | Paris area, H1 | R | 2 | 4 | 9 | 6 | 7 | 12 | 25 |
| CP7 | Placenta | Paris area, H6 | R | 6 | 7 | 9 | 1 | 14 | 4 | 29 |
| CP8 | Fetal lung | Paris area, H6 | R | 6 | 7 | 9 | 1 | 14 | 4 | 29 |
| CP9 | Catheter | Paris area, H6 | R | 6 | 7 | 9 | 1 | 14 | 4 | 29 |
| CP10 | Bronchoalveolar fluid | Paris area, H1 | R | 7 | 22 | 11 | 1 | 14 | 4 | 32 |
| CP11 | Wound | Paris area, H1 | R | 4 | 23 | 2 | 14 | 14 | 9 | 27 |
| CP12 | Urine | Paris area, H1 | R | 7 | 22 | 15 | 13 | 14 | 4 | 34 |
| CP13 | Feces | Paris area, H1 | R | 7 | 3 | 14 | 13 | 14 | 4 | 38 |
| CP14 | Feces | Paris area, H1 | R | 6 | 21 | 11 | 13 | 14 | 4 | 39 |
Primary clinical sample from which the isolate was recovered.
All geographic sources were located in France. H1, Ambroise-Paré Hospital; H2, Tenon Hospital; H3, Saint Louis Hospital; H4, Broussais Hospital; H5, Saint-Antoine Hospital; H6, Necker Hospital; H7, Versailles Hospital; H8, Villejuif Hospital.
U, epidemiologically unrelated; R, epidemiologically related.
Reference strain.
Species specificity of primers of loci.
No amplification was obtained with the CaACC1 and CaGLN4 oligonucleotides when the DNAs of the six Candida non-C. albicans species, or of S. cerevisiae, were used as templates. When oligonucleotides for loci CaVPS13, CaADP1, and CaRPN2 were used, an amplicon was generated from C. dubliniensis DNA but not from the DNA of any of the six other species tested. When the oligonucleotides for CaSYA1 were used, an amplicon was generated from all of the species tested except C. krusei. These results demonstrated that the oligonucleotides used for CaACC1 and CaGLN4 were specific for C. albicans; that those used for CaVPS13, CaADP1, and CaRPN2 were specific for both C. albicans and C. dubliniensis; but that those used for CaSYA1 were not species specific. The sequences of the PCR products obtained from the C. dubliniensis CdVPS13, CdADP1, and CdSYA1 loci were compared to those of the corresponding loci in C. albicans. CdSAY1 displayed polymorphic nucleotide sites similar to those observed in C. albicans. In contrast, comparison of the CdVPS13 and CdADP1 sequences with the corresponding ones in C. albicans showed 41 and 35 divergent nucleotides, respectively. These divergent nucleotides were not at sites associated with polymorphism in C. albicans (data not shown). These results are in agreement with the close relationship between C. albicans and C. dubliniensis, which were nevertheless recently demonstrated to be separate species (5).
Stability and reproducibility of C. albicans MLST.
The sequence stability of the six loci was evaluated by sequencing the PCR products obtained using DNAs from six subclones of C. albicans strain ATCC 36232 that were obtained after 30 successive subcultures (about 400 generations). No difference was observed between the subclones and the original isolate. Reproducibility was tested with nine separate isolates by using DNAs from two independent extractions. Again, the sequences obtained from the two extractions were strictly identical, thus confirming the high reproducibility of the method.
MLST analysis of related and unrelated C. albicans isolates.
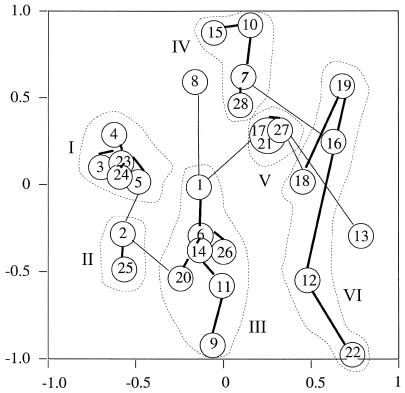
In all, 27 DSTs were found among the 28 epidemiologically unrelated isolates investigated (26 clinical isolates and 2 reference strains). Only two isolates (isolates 17 and 21) had identical DSTs. All of the other 26 had a unique DST and could be separated from each other (Table 4). The DSTs of 16 of these 26 isolates (61%) differed from each other at 3 loci or more. For the present sample of 28 epidemiologically unrelated isolates, the discriminatory power of the MLST scheme for typing C. albicans, calculated using a numerical index as previously described (13), was 99.7. The results of MDS and MST analyses showed that six groups of genetically related isolates were identifiable on the basis of bootstrap values of greater than 900 (Fig. 2). Isolates 8 and 13 were separated, respectively, from groups III and V on the basis of low bootstrap values, which ranged from 554 to 896. Within groups, the lengths of the branches linking the strains were variable, suggesting that some isolates were genetically very close whereas others were more distant. Contrasting patterns were observed. In groups I, II, and V, the isolates were connected by short branches, whereas in group VI, the branches were longer. These results provided a measure of the genetic diversity at multiple loci within a set of epidemiologically unrelated clinical isolates.
FIG. 2.
MDS of 28 epidemiologically unrelated isolates of C. albicans. Each isolate is represented by a circle. Connections between isolates and the lengths of the branches linking them were obtained by the MST method as described in Materials and Methods. The groups of genetically related isolates are numbered I to VI. Bootstrap values of greater than 900 are indicated by thick lines.
We observed that among the 14 epidemiologically related clinical isolates, isolates CP7, CP8, and CP9, which were recovered from a single episode of infection (mother and fetus) shared the same DST (Table 4). The 11 other isolates, which were isolated during the same 3-month period from patients hospitalized in the same intensive care unit, had different DSTs. In 8 of these 11 isolates, the DSTs differed by only one or two of the six genotypes.
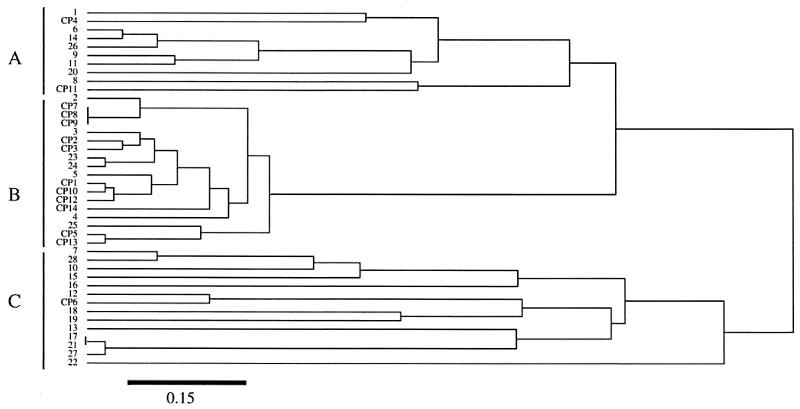
A dendrogram depicting the relationships between the 42 isolates studied showed that 8 of the 11 isolates from the patients hospitalized in the same unit (CP1, CP2, CP3, CP5, CP10, CP12, CP13, and CP14) were grouped in a cluster, whereas the 28 unrelated isolates were not distributed in a specific cluster (Fig. 3).
FIG. 3.
Dendrogram of the genetic relationships between 42 isolates of C. albicans, based on the six housekeeping loci investigated. The dendrogram was constructed by using the unweighted pair-group method with arithmetic averages and the matrix of distances, as described in Materials and Methods. The linkage distance is indicated at the bottom.
DISCUSSION
In this study we showed that from an epidemiological standpoint, MLST provides valuable information for the characterization of C. albicans isolates. MLST is a highly resolutive method based on analysis of the nucleotide polymorphism of internal fragments from several housekeeping genes present in all isolates within a given species (16). It has been used to characterize several bacterial pathogens which are strictly haploid microorganisms (4, 6-8). By contrast, the possible occurrence of heterozygosity in the sequences of diploid organisms makes MLST analysis more difficult. However, sequence typing of randomly amplified DNA fragments from C. albicans anonymous DNA regions has been used to investigate the genetic structures of typical and atypical populations of C. albicans from different geographic origins (9). This analysis has demonstrated that the structure of these populations was predominantly clonal but that there was also evidence for recombination and very low levels of gene flow between them (9, 12).
In the present study, we characterized six loci, each corresponding to an internal fragment of a distinct C. albicans housekeeping gene. At each locus, we observed a wide variety of genotypes among the isolates studied, although the number of polymorphic nucleotide sites was relatively limited. This was the consequence, at least in part, of a high frequency of heterozygosity, which increased sequence diversity at each locus, and thus allowed the identification of a greater number of genotypes. Indeed, heterozygosity was detected at all polymorphic nucleotide sites except three (positions 139, 180, and 327 at locus CaGLN4), and each C. albicans isolate had at least one heterozygous site within the 68 polymorphic sites studied. The presence of heterozygosity at the DNA level has been previously reported for C. albicans (2, 3, 12). When using 13 anonymous DNA regions to analyze the genetic structure of populations of typical C. albicans isolates, Forche et al. found that the frequency of polymorphic nucleotide sites in these regions was 1.1% and that there was evidence for heterozygosity at 35 of the 56 polymorphic nucleotide sites that they identified (9). The presence of heterozygosity has also been reported in the sequence of the CaERG11 gene, which encodes lanosterol 14-α-demethylase (10). However, the overall sequence diversity and levels of heterozygosity previously reported for these DNA regions were lower than those we observed at the six loci which we investigated and which included 68 polymorphic sites (2.9%). Of these, 65 were heterozygous.
In this study, we confirmed the reproducibility of the MLST method and the genomic stability of the loci studied. The fact that neither a loss nor a gain in the heterozygosity was detected at any polymorphic nucleotide site when the subclones obtained after successive subcultures were compared also confirmed that heterozygosity was stable in our isolates. These results agree with previous observations, based on an experimental C. albicans population, that over 330 generations, no change occurred in five DNA regions known to be heterozygous in the progenitor genotype (2).
The MLST method proved to be highly resolutive for C. albicans strain differentiation. The number of theoretically different DSTs resulting from the combination of the 10 to 24 genotypes present at each of the six loci is in the range of 107, and this should increase as new genotypes are described after the use of MLST with additional C. albicans isolates. The discriminatory power of the method, measured with a sample of 28 epidemiologically unrelated strains, was 99.7. Two isolates (isolates 17 and 21) had the same DST, which was unexpected because they were epidemiologically unrelated. However, due to the diploid nature of C. albicans, strains with the same DST may be different if they are heterozygous at several sites, and these two isolates were indeed heterozygous at 21 of the 68 polymorphic sites. Sequencing did not allow us to determine which allele is associated with which nucleotide at heterozygous positions, a limit common to all methods using nucleotide polymorphism analysis at several heterozygous loci in diploid organisms. Despite this limitation, the discriminatory power of our MLST method was greater than that of the other methods previously proposed for typing of C. albicans by using DNA sequence data. The discriminatory power of a method using analysis of the polymorphism of one microsatellite region was only 88 (1). A multilocus genotyping system using oligonucleotide probes for identification of the nucleotide state at only one of the polymorphic nucleotide sites of each of 16 distinct loci distinguished only 64 genotypes in a sample of 84 C. albicans isolates (3). Recently, it was shown by this method that the ability to invade the bloodstream is widespread among C. albicans isolates (15). The discriminatory power of our MLST method was similar to that of fingerprinting with the moderately repetitive sequence Ca3, which provided the highest resolution reported so far for typing of C. albicans isolates (25). However, MLST has several advantages over fingerprinting. First, the technology used, based on PCR amplification followed by the sequencing of six well-characterized loci, is easy to perform and can be done quickly as automatic sequencing becomes more widely available. The PCR conditions used in this study were very robust, as judged by the 100% success rate obtained in more than 700 assays. Second, the results of MLST are unambiguous, and sequence data can be shared and compared between different laboratories.
The C. albicans isolates that we studied in this work were specifically chosen to constitute a sampling of strains, with some having specific epidemiological links and some being unrelated. We have described the genetic relationships within a set of epidemiologically unrelated isolates of C. albicans by using a two-dimensional representation (Fig. 2). This representation allowed to identify groups of strains which are genetically close. We have observed that within the 28 isolates investigated, 26 were associated within six different groups. This method appears to be suitable for identification of the presence or absence of specific groups of strains among C. albicans isolates and could be useful for understanding the transmission and population dynamics in hospitalized patients or in healthy carriers. We showed that MLST can distinguish not only between epidemiologically unrelated strains but also between epidemiologically related strains and can identify a unique strain represented by different isolates. The dendrogram depicting the relationships between all of the isolates studied (Fig. 3) showed that as high a proportion as 73% of the related isolates were grouped in a cluster, whereas the unrelated isolates were not. This suggests that the epidemiologically related isolates, which had been recovered within a short period of time from patients hospitalized in the same intensive care unit, were genetically very close, although they were not strictly identical. These results agreed with those reported by others, who found that the isolates from patients in the same ward were genetically closer than other isolates (17, 24). By contrast, the three isolates recovered from a mother and her fetus were strictly identical. Thus, together our results emphasize that MLST can detect minute variations in the genomes of related isolates of C. albicans and can be used by epidemiologists to trace strain transmission. During epidemiological investigations, a large number of isolates often have to be typed. The fact that MLST was highly discriminatory even when only three loci were studied should make the method highly suitable for such settings.
Taken together, our results showed that MLST is highly reproducible and discriminatory with C. albicans. The sequence data obtained with this method can be used to construct an online global database which should allow laboratories to compare their local isolates and should prove useful both for global epidemiology and for investigating the transmission of C. albicans infections.
Acknowledgments
We are grateful to C. Bouchiez and S. Sibuet, Genopole, Institut Pasteur, Paris, France, for providing sequencing facilities and to C. Hennequin for helpful discussions. We are also indebted to S. Bretagne for constructive comments on the manuscript. We thank S. Challier, O. Eloy, G. Kac, O. Morin, H. Kening, M. Feuilhade de Chauvin, V. Lavarde, J.-L. Poirot, M.-A. Piens, and F. Robert for providing isolates.
This work was supported by a grant from the Ministère de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires-Réseau Infections Fongiques).
REFERENCES
- 1.Bretagne, S., J. M. Costa, C. Besmond, R. Carsique, and R. Calderone. 1997. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J. Clin. Microbiol. 35:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowen, L. E., C. Sirjusingh, R. C. Summerbell, S. Walmsley, S. Richardson, L. M. Kohn, and J. B. Anderson. 1999. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 43:2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly, S. M., D. J. Sullivan, D. B. Shanley, and D. C. Coleman. 1999. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology 145:1871-1882. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Feil, E. J., M. C. Enright, and B. G. Spratt. 2000. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 151:465-469. [DOI] [PubMed] [Google Scholar]
- 9.Forche, A., G. Schonian, Y. Graser, R. Vilgalys, and T. G. Mitchell. 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28:107-125. [DOI] [PubMed] [Google Scholar]
- 10.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaur, N. K., and S. A. Klotz. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gräser, Y., M. Volovsek, J. Arrington, G. Schönian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and C. A. Fraser. 1989. Application of a numerical index of discriminatory power to a comparison of four physiochemical typing methods for Candida albicans. J. Clin. Microbiol. 27:2156-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd ed., p. 381-385. Elsevier, New York, N.Y.
- 15.Luu, L. N., L. E. Cowen, C. Sirjusingh, L. M. Kohn, and J. B. Anderson. 2001. Multilocus genotyping indicates that the ability to invade the bloodstream is widespread among Candida albicans isolates. J. Clin. Microbiol. 39:1657-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marco, F., S. R. Lockhart, M. A. Pfaller, C. Pujol, M. S. Rangel-Frausto, T. Wiblin, H. M. Blumberg, J. E. Edwards, W. Jarvis, L. Saiman, J. E. Patterson, M. G. Rinaldi, R. P. Wenzel, and D. R. Soll. 1999. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J. Clin. Microbiol. 37:2817-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prim, R. C. 1957. Shortest connection networks and some generalizations. Bell System Tech. J. 36:1389-1401. [Google Scholar]
- 20.Reagan, D. R., M. A. Pfaller, R. J. Hollis, and R. P. Wenzel. 1990. Characterization of the sequence of colonization and nosocomial candidemia using DNA fingerprinting and a DNA probe. J. Clin. Microbiol. 28:2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reagan, D. R., M. A. Pfaller, R. J. Hollis, and R. P. Wenzel. 1995. Evidence of nosocomial spread of Candida albicans causing bloodstream infection in a neonatal intensive care unit. Diagn. Microbiol. Infect. Dis. 21:191-194. [DOI] [PubMed] [Google Scholar]
- 22.Robert, F., F. Lebreton, M. E. Bougnoux, A. Paugam, D. Wassermann, M. Schlotterer, C. Tourte-Schaefer, and J. Dupouy-Camet. 1995. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J. Clin. Microbiol. 33:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherer, S., and P. T. Magee. 1990. Genetics of Candida albicans. Microbiol. Rev. 54:226-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid, J., Y. P. Tay, L. Wan, M. Carr, D. Parr, and W. McKinney. 1995. Evidence for nosocomial transmission of Candida albicans obtained by Ca3 fingerprinting. J. Clin. Microbiol. 33:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid, J., E. Voss, and D. R. Soll. 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28:1236-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 27.Tzung, K. W., R. M. Williams, S. Scherer, N. Federspiel, T. Jones, N. Hansen, V. Bivolarevic, L. Huizar, C. Komp, R. Surzycki, R. Tamse, R. W. Davis, and N. Agabian. 2001. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl. Acad. Sci. USA 98:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez, J. A., V. Sanchez, C. Dmuchowski, L. M. Dembry, J. D. Sobel, and M. J. Zervos. 1993. Nosocomial acquisition of Candida albicans: an epidemiologic study. J. Infect. Dis. 168:195-201. [DOI] [PubMed] [Google Scholar]
- 29.Voss, A., R. J. Hollis, M. A. Pfaller, R. P. Wenzel, and B. N. Doebbeling. 1994. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J. Clin. Microbiol. 32:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]