Abstract
By using sequence analysis of Shiga toxin 1 (Stx 1) genes from human and ovine Stx-producing Escherichia coli (STEC) strains, we identified an Stx1 variant in STEC of human origin that was identical to the Stx1 variant from ovine STEC, but demonstrated only 97.1 and 96.6% amino acid sequence identity in its A and B subunits, respectively, to the Stx1 encoded by bacteriophage 933J. We designated this variant “Stx1c” and developed stxB1 restriction fragment length polymorphism and stx1c-specific PCR strategies to determine the frequency and distribution of stx1c among 212 STEC strains isolated from humans. stx1c was identified in 36 (17.0%) of 212 STEC strains, 19 of which originated from asymptomatic subjects and 16 of which were from patients with uncomplicated diarrhea. stx1c was most frequently (in 23 STEC strains [63.9%]) associated with stx2d, but 12 (33.3%) of the 36 STEC strains possessed stx1c only. A single STEC strain possessed stx1c together with stx2 and was isolated from a patient with hemolytic-uremic syndrome. All 36 stx1c-positive STEC strains were eae negative and belonged to 10 different serogroups, none of which was O157, O26, O103, O111, or O145. Stx1c was produced by all stx1c-containing STEC strains, but reacted weakly with a commercial immunoassay. We conclude that STEC strains harboring the stx1c variant account for a significant proportion of human STEC isolates. The procedures developed in this study now allow the determination of the frequency of STEC strains harboring stx1c among clinical STEC isolates and their association with human disease in prospective studies.
During the past 20 years, Shiga toxin (Stx)-producing Escherichia coli (STEC) have emerged as important causes of diarrhea and the hemolytic-uremic syndrome (HUS) throughout the world (2, 9, 14, 32, 33). Stx are believed to be the cardinal virulence factors of STEC (20). Based on toxin neutralization assays (30) and sequence analysis of stx genes (11), two major toxin types, Stx1 and Stx2, have been assigned (11, 20, 30). Stx1 and Stx2 are not cross-neutralized by heterologous antisera in cell culture assays (30), and the structural genes encoding these toxins demonstrate approximately 55% overall nucleotide sequence identity (11). The Stx2 group has been shown to be highly heterogeneous, comprising, in addition to Stx2, several Stx2 variants that have been classified as Stx2c (29), Stx2d (24), Stx2e (34), and Stx2f (28). The substantial sequence heterogeneity observed between members of the Stx2 family (24, 28, 29, 34) enabled the development of PCR techniques that differentiate stx2 from its variants and identify the respective stx2 alleles (7, 24, 28). This is of particular clinical importance, because STEC strains possessing different stx2 variants appear to differ in their capacity to cause HUS (8). Information about the stx2 allele of an infecting STEC strain has, therefore, considerable potential predictive value for the treating physician to assess the risk of HUS development in a patient that presents with STEC infection (8).
In contrast to the Stx2 family, the Stx1 group appears to be more homogeneous. stx1 genes carried in the genomes of bacteriophages H19B (6), H30 (18), and 933J (10) have identical nucleotide sequences (6, 10, 18) and differ by only three nucleotides in their A subunits, resulting in only one amino acid difference from the sequence of stx from Shigella dysenteriae type 1 (31). Paton et al. (21, 22) reported three human STEC strains that possess slight variations in their stx1 genes. Each of these stx1 variants shared more than 99% nucleotide sequence identity with stx1 from phage 933J and with stx from S. dysenteriae type 1 (21, 22). Stx1 encoded by these stx1 variants differed by one and two amino acid residues in their A subunits from Stx of S. dysenteriae type 1 and from Stx1 encoded by phage 933, respectively, whereas their B subunits were identical to those of the latter two toxins (21, 22). A considerably greater degree of sequence heterogeneity was observed in stx1 genes of STEC isolated from sheep (1, 22). Specifically, the stx1 gene from a sheep STEC strain differed from stx1 of phage 933J by 43 nucleotides, resulting in nine and three amino acid substitutions in the A and B subunits, respectively (22). Moreover, phylogenetic analysis based on sequencing of stx genes of STEC strains from various origins places Stx1 of ovine STEC isolates into a different group from the group that contains Stx1 from other STEC (1). Thus, the Stx1 variant of ovine STEC can be considered to be encoded by an allele distinct from previously reported stx1 (6, 10, 18, 21, 22).
In this study, we used nucleotide sequence analysis of stx1 genes to characterize allelic variants from STEC strains isolated from humans. We identified an stx1 variant in STEC strains of human origin, which is identical to the stx1 variant from ovine STEC strains, but which differs markedly from the prototype phage-encoded stx1. We designated this stx1 variant “stx1c,” developed stxB1 restriction fragment length polymorphism (RFLP) and PCR strategies to determine the frequency and distribution of stx1c among human STEC isolates, and characterized STEC strains harboring stx1c.
MATERIALS AND METHODS
Bacterial strains.
Two hundred fourteen STEC strains were investigated in this study. Two hundred twelve STEC strains were isolated from patients with HUS (n = 48) or watery diarrhea (n = 105) or from asymptomatic individuals (n = 59), at the Institute for Hygiene and Microbiology, University of Würzburg, Würzburg, Germany, during microbiological evaluation between 1996 and 2000. STEC strains were isolated from stools by protocols described previously (8, 12, 13, 27). All 212 STEC strains originated from apparently sporadic cases of infection without obvious geographic or temporal linkage. In addition, two STEC strains isolated from healthy sheep in the Czech Republic were included in this study for comparison with human STEC isolates. Ninety-seven of the STEC strains investigated in this study were analyzed for their stx2 gene variants in a previous study (8).
Case definition.
Patients with diarrhea had three or more watery stools without visible blood per day. HUS was defined as a case of microangiopathic hemolytic anemia (hematocrit less than 30% with peripheral evidence of intravascular hemolysis), thrombocytopenia (platelet count less than 150,000/mm3), and renal insufficiency (serum creatinine concentration greater than the upper limit of the normal range for age) (35). Asymptomatic carriers were apparently healthy individuals without diarrhea.
Phenotypic methods.
STEC isolates were serotyped according to Bockemühl et al. (3) with antisera against E. coli O antigens 1 to 173 and H antigens 1 to 56. Stx production was tested by using the Vero cell cytotoxicity assay and a commercial latex agglutination assay (VTEC-RPLA [verotoxin-producing E. coli reverse passive latex agglutination]; Denka Seiken Co., Ltd., Tokyo, Japan), both performed as described by Karmali et al. (15, 16), with slight modifications. Briefly, the strains were grown overnight in Trypticase soy broth (Difco Laboratories, Detroit, Mich.) at 37°C with shaking (180 rpm), supernatants were filtered through 0.22-μm-pore-diameter filters (Schleicher & Schuell, GmbH, Dassel, Germany), and serial twofold dilutions of the culture filtrates were used for the assays. The toxin titers were expressed as the reciprocals of the highest dilutions that caused cytotoxicity in 50% of Vero cells after 2 days of incubation and a clear agglutination of the Stx1 and Stx2 latex reagents after overnight incubation. The culture filtrate of E. coli O157:H7 strain EDL 933 (stx1+, stx2+) (19) was used as a positive control in both toxin assays.
PCR.
PCRs to detect STEC-specific sequences were performed in the GeneAmp PCR System 9600 (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) in a volume of 50 μl containing 5 μl of bacterial suspension (ca. 104 bacteria), 200 μM each deoxynucleoside triphosphate, 30 pmol of each primer, 5 μl of 10-fold-concentrated polymerase synthesis buffer, 1.5 mM MgCl2, and 2.0 U of AmpliTaq DNA polymerase (Perkin-Elmer Applied Biosystems). The PCR primers and conditions are listed in Table 1. To detect stx genes, primer pairs KS7-KS8, GK3-GK4, and VT2-cm-VT2-f (Table 1) were used as described previously (13, 24, 26). stx2 and stx2c were differentiated by restriction analysis of GK3-GK4 amplification products by using HaeIII and FokI as described by Rüssmann et al. (25). Primers Stx1c-1 and Stx1c-2 (Table 1) were designed in this study to specifically amplify a 498-bp fragment of the A subunit of an stx1 variant that we propose below to be termed “stx1c”. The eae gene was detected with primers SK1 and SK2 (Table 1) as described earlier (26). E. coli O157:H7 strain EDL 933 was used as a positive control in PCRs for the detection of stx1, stx2, and eae genes. E. coli strain EH250 (ONT:H12; stx2d) (24) served as a positive control in PCR for the detection of stx2d. In the stx1c-specific PCR, E. coli strain 3115/97 (O128:H2; stx1c+stx2d) (this study) was used as a positive control.
TABLE 1.
PCR primers and conditions used in this study
| Primer pair | Sequence | Target | PCR conditionsa
|
Length of PCR product (bp) | Reference | ||
|---|---|---|---|---|---|---|---|
| Denaturing | Annealing | Extension | |||||
| KS7 | 5′-CCC GGA TCC ATG AAA AAA ACA TTA TTA ATA GC-3′ | stxB1 | 94°C, 30 s | 52°C, 60 s | 72°C, 40 s | 282 | 26 |
| KS8 | 5′-CCC GAA TTC AGC TAT TCT GAG TCA ACG-3′ | stxB1c | |||||
| GK3 | 5′-ATG AAG AAG ATG TTT ATG-3′ | stxB2 | 94°C, 30 s | 52°C, 60 s | 72°C, 40 s | 260 | 13 |
| GK4 | 5′-TCA GTC ATT ATT AAA CTG-3′ | stxB2c | |||||
| VT2-cm | 5′-AAG AAG ATA TTT GTA GCG G-3′ | stxB2d | 94°C, 30 s | 55°C, 60 s | 72°C, 60 s | 256 | 24 |
| VT2-f | 5′-TAA ACT GCA CTT CAG CAA AT-3′ | ||||||
| SK1 | 5′-CCC GAA TTC GGC ACA AGC ATA AGC-3′ | eae | 94°C, 30 s | 52°C, 60 s | 72°C, 60 s | 863 | 26 |
| SK2 | 5′-CCC GGA TCC GTC TCG CCA GTA TTC G-3′ | ||||||
| Stx1c-1b | 5′-TTT TCA CAT GTT ACC TTT CCT-3′ | stxA1c | 94°C, 30 s | 51°C, 60 s | 72°C, 60 s | 498 | This study |
| Stx1c-2 | 5′-CAT AGA AGG AAA CTC ATT AGG-3′ | ||||||
| Oligo 1 | 5′-TCG CAT GAG ATC TGA CC-3′ | stx1, stx1c | 94°C, 30 sc | 55°C, 60 s | 72°C, 90 s | 1,469 | 22 |
| Oligo 2 | 5′-AAC TGA CTG AAT TGA GAT G-3′ | Whole | |||||
All PCRs included 30 cycles followed by a final extension step of 5 min at 72°C.
Primers Stx1c-1 and Stx1c-2 were derived from the positions 511 to 531 and 988 to 1008, respectively, of the stx1c sequence from strain 3115/97 determined in this study (accession no. AJ312232).
PCR conditions were established in this study.
stxB1 PCR-RFLP.
The stxB1 gene was amplified with primers KS7-KS8 (Table 1), and 12 μl of each PCR product was digested with restriction endonuclease FspI or HhaI (New England BioLabs GmbH, Frankfurt, Germany), as recommended by the manufacturer. Restriction fragments were separated on a 2% (wt/vol) agarose gel and visualized by staining with ethidium bromide. The restriction enzymes for the differentiation of classical stx1 and the stx1c variant were selected with the DNASIS program, version 2.0, from Hitachi Software (San Bruno, Calif.) based on the published sequence of stx1 from phage 933J (accession no. M19473) and the sequence of stx1c from E. coli strain 3115/97 determined in this study.
stx1 sequence analysis.
For nucleotide sequencing, the whole stx1 genes from 14 STEC strains were amplified with the PCR primers described by Paton et al. (22) (Table 1). The amplification products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). The sequencing was performed with an automated 377 DNA sequencer (Perkin-Elmer Applied Biosystems) with the PCR primers and customized primers. A fluorescence procedure with the Taq Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems) was applied according to the manufacturer's instructions. Nucleotide sequence analysis was performed with the DNASIS program (Hitachi Software). Homology searches were performed with the EMBL GenBank database.
Nucleotide sequence accession number.
The nucleotide sequences for the stx1c genes from E. coli strains 3115/97 (O128:H2; human), 4756/98 (O70:H−; human), and 295/00 (O128:H−; ovine) have been entered into the EMBL database under accession no. AJ312232, AJ314838, and AJ314839, respectively.
RESULTS
stx genotypes and serotypes of STEC.
As determined by PCR with primers KS7-KS8, GK3-GK4, and VT2-cm-VT2-f, all 214 STEC strains investigated contained stx1, either as the sole stx gene (117 isolates) or in combination with stx2, stx2c, or stx2d (97 isolates) (Table 2). Serotype O157:H7/H− isolates mostly contained stx2 or stx2c (Table 2). In contrast, almost half (49 of 117) of STEC strains harboring stx1 as the only stx gene, and all 29 STEC strains harboring stx1 together with stx2d, belonged to a diversity of serogroups that were different from O157 and from the major non-O157 STEC serogroups (i.e., O26, O103, O111, and O145) (Table 2).
TABLE 2.
stx genotypes, serotypes, and Stx production by 214 STEC strainsa investigated in this study
| stx genotype | No. of isolates | Serotype (no. of isolates)
|
Vero cell cyto- toxicity titer
|
VTEC-RPLA titer with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stx1 reagent
|
Stx2 reagent
|
||||||||||||
| O157:H7/H− | O26:H11/H− | O103:H2/H− | O111:H8/H− | O145:H− | Others | Range | Median | Range | Median | Range | Median | ||
| stx1b | 105 | 1 | 21 | 25 | 10 | 11 | 37c | 16-512 | 64 | 16-256 | 64 | <2 | <2 |
| stx1cb | 12 | 0 | 0 | 0 | 0 | 0 | 12d | 32-256 | 64 | 2-8 | 4 | <2 | <2 |
| stx1+stx2 | 49 | 25 | 12 | 1 | 7 | 1 | 3e | 32-2,048 | 256 | 16-256 | 64 | 32-256 | 128 |
| stx1c+stx2 | 1 | 0 | 0 | 0 | 0 | 0 | 1f | 256 | 256 | 8 | 8 | 128 | 128 |
| stx1+stx2c | 14 | 10 | 0 | 0 | 0 | 1 | 3g | 16-128 | 48 | 8-128 | 32 | 4-32 | 16 |
| stx1+stx2+stx2c | 4 | 2 | 0 | 0 | 0 | 0 | 2h | 64-256 | 128 | 8-64 | 24 | 32-128 | 64 |
| stx1+stx2d | 4 | 0 | 0 | 0 | 0 | 0 | 4i | 16-128 | 64 | 16-32 | 24 | 4 | 4 |
| stx1c+stx2d | 25j | 0 | 0 | 0 | 0 | 0 | 25k | 16-128 | 64 | 2-8 | 4 | 4-8 | 6 |
All but two STEC isolates were of human origin; two strains were isolated from sheep.
Both stx1 and stx1c genes were originally detected as stx1 by PCR with primer pair KS7-KS8, without restriction.
O3:H2, O3:H10, O8:H− (nonmotile) (three strains), O25:H− (two strains), O31:H−, O62:H−, O84:H4, O84:H− (three strains), O91:H14 (two strains), O92:H33, O112:H−, O118:H− (two strains), O119:H2, O128:H−, O129:H−, O146:H20, O152:H4, O156:H−, ONT (nontypeable):H14 (two strains), ONT:H− (four strains), Orough:H− (five strains), Orough:HNT.
O8:H19, O8:H−, O74:H−, O75:H33, O78:H− (three strains), O112:H−, O128:H−, ONT:H− (two strains), Orough:HNT.
O4:H−, O68:H4, O118:H−.
ONT:H−.
O104:H16, O113:H−, O120:H−.
O75:H−, Orough:H25.
O62:H− (two strains), O91:H−, O128:H−.
Twenty-three of 25 STEC strains of the stx1c+stx2d genotype were isolated from humans; two strains (both of serotype O128:H−) originated from sheep.
O22:H8, O70:H−, O75:H8, O75:H21, O96:H−, O113:H− (two strains), O128:H2 (five strains), O128:H− (three strains), O128:HNT, ONT:H8 (three strains), ONT:H− (two strains), Orough:H19, Orough:HNT (three strains).
Stx production.
All 214 STEC strains were cytotoxic for Vero cells in titers ranging from 16 to 2,048 in culture filtrates (Table 2) and reacted positively with the Stx2 and/or Stx1 latex reagent in the VTEC-RPLA assay, in accordance with their stx genotypes (Table 2). However, the Stx1 latex agglutination titers of culture filtrates of 12 of the 117 STEC strains that harbored stx1, but not stx2, were significantly lower than those detected in the remaining 105 isolates (Table 2), although both these groups displayed comparable Vero cell cytotoxicity (Table 2). Similarly, the supernatants of 25 of 29 STEC strains that harbored stx1 in combination with stx2d produced significantly lower Stx1 latex agglutination titers than the other four strains (Table 2), although ranges and medians of the Vero cell cytotoxicity titers of these 29 STEC strains were identical (Table 2).
stx1 sequence analysis.
To investigate possible reasons for the unusually low Stx1 latex agglutination titers observed in the 37 STEC strains described above, the whole stx1 genes from 14 of them, including 12 human and 2 ovine isolates identified by PCR to harbor stx1+stx2d, were amplified and the resulting 1,469-bp products were sequenced. The nucleotide and deduced amino acid sequences of the A and B subunits of the toxin genes from these 14 STEC strains were compared to published sequences for the corresponding subunits of stx1 from phage 933J (accession no. M19473), stx1 variants from human STEC strains including strains PH (O111:H−) (accession no. L04539), 94C (O48:H21) (accession no. Z36899), and CB168 (O111:H−) (accession no. Z36900) (21, 22), and the stx1 variant from the ovine STEC isolate 131/3 (OX3:H8) (accession no. Z36901) (22).
The nucleotide sequences of stx1 amplicons from each of the 14 STEC strains that produced low Stx1 latex agglutination titers were identical, and their A subunits differed from the stxA1 from phage 933J by 30 nucleotides, corresponding to 97% nucleotide sequence identity. Their B subunits differed from stxB1 from phage 933J by 13 nucleotides, resulting in 95% nucleotide sequence identity. Moreover, the sequences of the A and B subunits of the toxin genes from these 14 STEC strains demonstrated 97 and 95% respective identities to the corresponding subunits of the three stx1 variants identified in human isolates PH, 94C, and CB168 by Paton et al. (21, 22). However, the nucleotide sequences of both the A and the B toxin subunits from each of these 14 STEC strains were 100% identical to the sequences of the A and the B subunits, respectively, of the stx1 variant from sheep isolate 131/3 described by that group (22). Stx1 from each of these 14 isolates differed from Stx1 encoded by phage 933J and from Stx1 of human isolates PH, 94C, and CB168 by nine and three amino acid residues in the A and B subunits, respectively, corresponding to 97.1 and 96.6% amino acid sequence identities of the respective subunits to those of the latter four Stx1 proteins. Based on these significant nucleotide and amino acid differences, we propose to term the Stx1 variant identified in this study “Stx1c.”
stxB1 PCR-RFLP and stx1c-specific PCR to differentiate the stx1c variant from stx1.
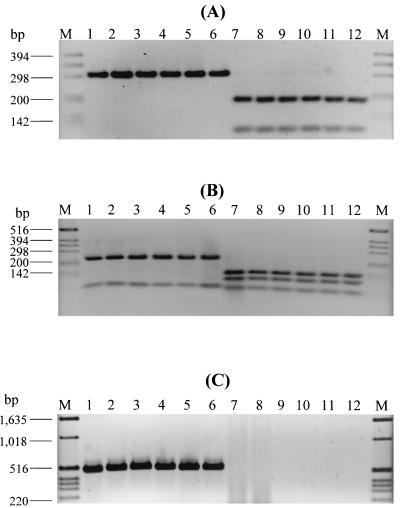
The sequence differences between stx1c and stx1 from phage 933J enabled us to develop PCR strategies to differentiate these alleles. In the stxB1 PCR-RFLP procedure, the B subunit genes were amplified with primers KS7-KS8, and the resulting 282-bp PCR products were digested with FspI or HhaI. Based on the restriction sites identified for the respective endonucleases in the stx1 sequence from phage 933J (accession no. M19473) and in the stx1c sequence from strain 3115/97 (this study) using the DNASIS program, FspI produces two fragments of 189 and 93 bp from the KS7-KS8 PCR product of stxB1, whereas the stxB1c PCR product remains undigested. Digestion with HhaI would yield three fragments 135, 92, and 55 bp long from the stxB1 PCR product, but only two fragments (218 and 64 bp) from the stxB1c KS7-KS8 amplification product. To evaluate this approach, 6 of the 14 STEC strains that were determined by sequencing to contain stx1c (4 human isolates of different serotypes and 2 ovine isolates) and 6 STEC strains harboring stx1 were first investigated (Fig. 1). The latter six STEC strains included strain EDL 933 and five STEC strains from our collection that were assumed to harbor classical stx1 (either alone or in combination with stx2 and/or stx2c) based on their high Stx1 latex agglutination titers (128 to 256) that were comparable with that of strain EDL 933 (titer of 256). As demonstrated in Fig. 1, and in accordance with the computer prediction, digestion with FspI left the KS7-KS8 PCR products from all the six STEC strains harboring stx1c (Fig. 1A, lanes 1 to 6) intact, but produced two fragments of 189 and 93 bp from the KS7-KS8 PCR products of all six strains harboring stx1 (Fig. 1A, lanes 7 to 12). Digestion of the KS7-KS8 PCR products with HhaI yielded two restriction fragments of 218 and 64 bp from each of the six STEC strains harboring stx1c (Fig. 1B, lanes 1 to 6), but three restriction fragments of 135, 92, and 55 bp from each of the six strains harboring stx1 (Fig. 1B, lanes 7 to 12).
FIG. 1.
Agarose gel electrophoresis of KS7-KS8 PCR products digested with FspI (A) or HhaI (B) and of PCR amplification products with primers Stx1c-1 and Stx1c-2 (C). M, molecular weight marker (1-kb DNA ladder; Gibco BRL, Eggenstein, Germany). In lanes 1 to 12, the following STEC strains (genotypes, serotypes, and origins, if not human, in parentheses) are shown: 1, 808/97 (stx1c+stx2d; ONT:H8); 2, 3115/97 (stx1c+stx2d; O128:H2); 3, 521/99 (stx1c+stx2d; Orough:H19); 4, 4756/98 (stx1c+stx2d; O70:H−); 5, 273/00 (stx1c+stx2d; O128:H−; sheep); 6, 295/00 (stx1c+stx2d; O128:H−; sheep); 7, EDL 933 (stx1+stx2; O157:H7); 8, 2544/00 (stx1; O145:H−); 9, 3385/00 (stx1+stx2; O111:H−); 10, 4424/99 (stx1+stx2; O157:H7); 11, 2049/98 (stx1+stx2c; O157:H−); 12, 2050/98 (stx1+stx2+stx2c; O157:H−).
Each of the 12 STEC strains analyzed by the stxB1 PCR-RFLP approach was further subjected to PCR with primers Stx1c-1 and Stx1c-2 (Table 1), which were derived from the sequence of the stxA1c subunit from strain 3115/97. As demonstrated in Fig. 1C, all six STEC strains harboring stx1c (lanes 1 to 6) produced a PCR product of the expected size of 498 bp, whereas all six STEC strains containing the classical stx1 gene (lanes 7 to 12) were negative in this stx1c-specific PCR.
Identification of the stx1c variant in STEC strains isolated from humans.
To determine the frequency and the distribution of stx1c in human STEC strains, stxB1 PCR-RFLP, and stx1c-specific PCR were applied to the 212 STEC strains of human origin, described above. stx1c was identified in 36 (17.0%) of the 212 STEC strains. All of these 36 STEC strains demonstrated the FspI and HhaI restriction patterns, which were identical to those shown for the six representative STEC strains containing stx1c in lanes 1 to 6 of Fig. 1A and B, respectively. Specifically, the KS7-KS8 PCR product from each of these 36 STEC strains remained undigested with FspI, but was digested into two fragments of 218 and 64 bp with HhaI. Moreover, each of these 36 STEC strains produced an amplification product 498 bp long in the stx1c-specific PCR. In contrast, the remaining 176 STEC strains yielded FspI and HhaI stxB1 PCR-RFLP patterns, which indicated the presence of classical stx1 rather than stx1c and were all negative in the stx1c-specific PCR. This demonstrates 100% concordance between the RFLP and PCR assays for the detection of stx1 and stx1c in human STEC strains.
The highest frequency of stx1c was detected among 27 human STEC strains that were initially identified by PCR with primers KS7-KS8, without restriction, to contain stx1 together with stx2d. Based on their stxB1 RFLP patterns after digestion with FspI and HhaI and their positive results in the stx1c-specific PCR, 23 (85.2%) of these 27 STEC strains belonged to genotype stx1c+ stx2d (Table 2). Moreover, positive results in the stx1c-specific PCR and the FspI and HhaI RFLP patterns indicated the presence of stx1c rather than stx1 in 12 (10.3%) of the 117 STEC strains that were initially identified by PCR with primers KS7-KS8 (without stxB1-RFLP) to contain stx1 only (Table 2). In contrast, only 1 of 68 STEC strains originally identified to contain stx1 in combination with stx2 and/or stx2c was found to harbor stx1c (Table 2).
Characteristics of STEC strains harboring stx1c.
To further characterize the 36 human STEC strains that possessed the stx1c variant, the serotypes of these isolates and the presence of eae were determined. Also, production of Stx1c by these isolates was tested with the latex agglutination assay, and their association with clinical symptoms was evaluated. The characteristics of these 36 human STEC strains were compared with those of 2 STEC strains isolated in this study from healthy sheep that were determined to contain stx1c by the sequence analysis, by the stxB1 PCR-RFLP after restriction with FspI (Fig. 1A, lanes 5 and 6) and HhaI (Fig. 1B, lanes 5 and 6), and by the stx1c-specific PCR (Fig. 1C, lanes 5 and 6).
As demonstrated in Table 2, the 36 human STEC strains harboring stx1c belonged to 15 different serotypes, 8 (22.2%) of these strains clustering in serogroup O128. O antigens of eight isolates were not typeable, and five strains autoagglutinated. None of these 36 STEC strains belonged to the major STEC serogroups (i.e., O157, O26, O103, O111, and O145) (Table 2), and none contained eae. All 36 STEC strains harboring stx1c reacted in the latex agglutination test, but the titers were low (Table 2). Nineteen of the 36 STEC strains harboring stx1c were isolated from asymptomatic subjects, and 16 were from patients with uncomplicated diarrhea. The single isolate that contained stx1c in combination with stx2 originated from a patient with HUS.
Both stx1c-harboring STEC strains isolated from sheep possessed stx1c together with the stx2d gene and belonged to serogroup O128 (Table 2). Both were eae negative and displayed low toxin titers in the latex agglutination assay (Table 2).
DISCUSSION
Stx1 produced by human STEC has been considered to have minimal sequence variability (1, 21, 22). However, we have demonstrated above that a substantial subset of human STEC isolates harbor a variant stx1 allele that displays significant sequence deviation from the prototype phage-encoded stx1 (6, 10, 18). This variant was initially identified in E. coli strains of ovine origin (22) and was termed stx1OX3 according to the serogroup of the prototype ovine STEC isolate 131/3 (OX3:H8). Recently, and independently of our study, stx1OX3 has been identified in human STEC strains of selected serotypes and has been demonstrated to be phage encoded in an STEC O146:H21 strain, but not in STEC strains of other serotypes (17). The appreciable frequency of this stx1 variant identified in our study among human STEC strains isolated during a 5-year period demonstrates a need for the expansion of Stx1 nomenclature according to the recommendations proposed for the designation of new toxin variants within the Stx family (5), as have been applied to stx2 variants (24, 28, 29, 34). Following these recommendations, we propose to designate this stx1 variant stx1c.
Our data have several important implications. First, STEC strains harboring the stx1c variant appear to be associated with either mild disease or with asymptomatic carriage. Only 1 of the 36 stx1c-harboring STEC strains identified in the present study was isolated from an HUS patient, and this organism also contained stx2, which might contribute to its ability to cause systemic disease (4, 8). The uniform absence of eae from all of the 36 human STEC isolates that harbored stx1c might be one of the reasons why such STEC strains were predominantly associated with asymptomatic infection or uncomplicated diarrhea. Moreover, most isolates containing stx1c also possessed stx2d. We have recently demonstrated (8) that stx2d is an stx2 allele that is not found in STEC strains isolated from HUS patients. However, our data do not permit us to determine if the apparently milder infections caused by STEC strains containing the stx1c allele are attributable to any of these factors. Moreover, it is not known at present if the structural differences between Stx1c and Stx1 might have any impact on the expression of the former toxin and/or on its binding or enzymatic activity, which, in turn, could influence the pathogenic potential of STEC strains producing Stx1c for humans. Further investigation is necessary to address these questions in order to understand if sequence variations in Stx1 molecules might affect the capacity of STEC strains producing such toxins to cause human disease. Indeed, Stx2 sequence variability has been suggested to be the reason underlying different toxicities in a mouse model (23). The different capacities of STEC strains harboring different stx2 variants to cause severe human disease, including HUS, have been demonstrated recently (8).
Second, an improved understanding of the clinical significance of STEC strains harboring stx1c depends on the availability of appropriate and economical procedures to identify such strains. In this respect, the strategies developed in this study to differentiate stx1c from the classical stx1 can be of a considerable utility for clinical microbiological laboratories to identify STEC strains containing stx1c. For laboratories that cannot use PCR to diagnose these organisms, it is particularly significant that Stx1c can be detected by a commercial latex agglutination assay. The low titers of Stx1c as detected by the Stx1 latex reagent appear to be probably a consequence of antigenic differences between Stx1c and Stx1 that are plausibly derived from the substantial sequence differences between these toxin molecules, rather than reduced Stx1c expression, because Vero cell cytotoxicity was not correspondingly diminished. Karmali et al. (16) and our group (8) have noticed analogous discordance between Stx2 structure and Stx2 antigenic detection. From a practical standpoint, a low Stx1 latex agglutination titer in a STEC culture filtrate may suggest production of Stx1c rather than of the classical Stx1 by the isolate and should stimulate efforts to send such isolates for further analysis to a laboratory that can subtype stx1 genes by molecular approaches designed in this study. The possibility of differentiating stx1c from stx1 in clinical microbiological laboratories has several potential benefits. (i) It allows the identification of possibly less pathogenic STEC strains, which has a direct implication for patient management, if the association of stx1c with milder disease is confirmed. (ii) It can prompt the expeditious epidemiological investigation of unique cases. (iii) Our findings can also be used to more precisely genotype and thereby categorize STEC strains.
Specifically, the introduction of the stx1-subtyping procedures developed in this study enabled us to differentiate three additional stx genotypes not identified by conventional PCR, including stx1c, stx1c+stx2, and stx1c+ stx2d, allowing us to classify the 214 STEC strains investigated in this study into the eight different stx genotypes presented in Table 2. This suggests that the application of these stx1-subtyping procedures in epidemiological investigations could substantially contribute to the identification of reservoirs of STEC strains harboring stx1c, sources of infection for humans, and modes of spreading of these organisms. Our hypothesis that sheep could be a natural reservoir of STEC strains harboring stx1c for humans, which is based on our finding that both STEC strains isolated from sheep in this study demonstrated characteristics that were identified in a significant proportion of human stx1c-containing STEC strains, was also recently and independently proposed by Koch et al. (17). These authors identified stx1c, either alone or in combination with stx2d, in 38 (79%) of 48 STEC strains isolated from sheep, but in none of 28 STEC isolates from cattle or goats (17). The majority of the ovine stx1c-harboring STEC strains belonged to serotypes O128:H2 and O146:H21, which are also most frequently found among human STEC strains harboring this stx1 variant, but were not identified in any of 88 stx1c-negative STEC isolates from different origins (17). Further epidemiological studies with the approaches for the detection of stx1c developed in our study are needed to determine the sources of STEC strains harboring stx1c for humans and the ways of transmitting the infection.
In conclusion, a substantial number of human STEC strains harbor the stx1c variant. This variant markedly differs from the classical stx1. stx1c is mostly associated with stx2d and has not yet been found in STEC strains containing eae. Infections by STEC strains harboring stx1c are usually asymptomatic or manifest as mild diarrhea. The diagnostic procedures developed in this study to differentiate stx1c from stx1 should be applied in prospective studies to further determine the frequency of stx1c in clinical STEC isolates and the association of STEC strains harboring stx1c with clinical symptoms and to better understand the epidemiology of infections caused by these organisms.
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 479) and from the Bundesministerium für Bildung und Forschung (BMBF) Verbundprojekt, Forschungsnetzwerk “Emerging foodborne pathogens in Germany” (no. 01KI 9903).
We thank Barbara Plaschke for excellent technical assistance and Phillip I. Tarr (Children's Hospital and Regional Medical Center, Seattle, Wash.) for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Asakura, H., S. I. Makino, H. Kobori, M. Watarai, T. Shirahata, T. Ikeda, and K. Takeshi. 2001. Phylogenetic diversity and similarity of active sites of Shiga toxin (Stx) in Shiga toxin-producing Escherichia coli (STEC) isolates from humans and animals. Epidemiol. Infect. 127:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielaszewska, M., and H. Karch. 2000. Non-O157:H7 Shiga toxin (verocytotoxin)-producing Escherichia coli strains: epidemiological significance and microbiological diagnosis. World J. Microbiol. Biotechnol. 16:711-718. [Google Scholar]
- 3.Bockemühl, J., S. Aleksic, and H. Karch. 1992. Serological and biochemical properties of Shiga-like toxin (verocytotoxin)-producing strains of Escherichia coli, other than O-group 157, from patients in Germany. Int. J. Med. Microbiol. Virol. Parasitol. Infect. Dis. 276:189-195. [DOI] [PubMed] [Google Scholar]
- 4.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Association between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderwood, S. B., D. W. K. Acheson, G. T. Keusch, T. J. Barrett, P. M. Griffin, N. A. Strockbine, B. Swaminathan, J. B. Kaper, M. M. Levine, B. S. Kaplan, H. Karch, A. D. O'Brien, T. G. Obrig, Y. Takeda, P. I. Tarr, and I. K. Wachsmuth. 1996. Proposed new nomenclature for Shiga-like toxin (verotoxin) family. ASM News 62:118-119. [Google Scholar]
- 6.De Grandis, S., J. Ginsberg, M. Toone, S. Climie, J. Friesen, and J. Brunton. 1987. Nucleotide sequence and promoter mapping of the Escherichia coli Shiga-like toxin operon of bacteriophage H-19B. J. Bacteriol. 169:4313-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franke, S., F. Gunzer, L. H. Wieler, G. Baljer, and H. Karch. 1995. Construction of recombinant Shiga-like toxin-IIv (SLT-IIv) and its use in monitoring the SLT-IIv antibody status in pigs. Vet. Microbiol. 43:41-45. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich, A. W., M. Bielaszewska, W.-L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 10.Jackson, M. P., J. W. Newland, R. K. Holmes, and A. D. O'Brien. 1987. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2:147-153. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, M. P., R. J. Neil, A. D. O'Brien, R. K. Holmes, and J. W. Newland. 1987. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 44:109-114. [DOI] [PubMed] [Google Scholar]
- 12.Karch, H., C. Janetzki-Mittmann, S. Aleksic, and M. Datz. 1996. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J. Clin. Microbiol. 34:516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karch, H., H. I. Huppertz, J. Bockemühl, H. Schmidt, A. Schwarzkopf, and R. Lissner. 1997. Shiga toxin-producing Escherichia coli infections in Germany. J. Food Prot. 11:1454-1457. [DOI] [PubMed] [Google Scholar]
- 14.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 15.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 16.Karmali, M. A., M. Petric, and M. Bielaszewska. 1998. Evaluation of a microplate latex agglutination method (Verotox-F assay) for detecting and characterizing verotoxins (Shiga toxins) in Escherichia coli. J. Clin. Microbiol. 37:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin.2001. Isolation of a lysogenic bacteriophage carrying the stx1OX3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlov, Y. V., A. A. Kabishev, E. V. Lukyanov, and A. A. Bayev. 1988. The primary structure of the operons coding for Shigella dysenteriae toxin and temperate phage H30 Shiga-like toxin. Gene 67:213-221. [DOI] [PubMed] [Google Scholar]
- 19.O'Brien, A. D., A. T. Lively, M. E. Chen, S. W. Rothman, and S. B. Formal. 1983. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet i:702. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien, A. D., V. L. Tesh, A. Donohue-Rolfe, M. P. Jackson, S. Olsnes, K. Sandvig, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action, and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 21.Paton, A. W., J. C. Paton, P. N. Goldwater, M. W. Heuzenroeder, and P. A. Manning. 1993. Sequence of a variant Shiga-like toxin type-I operon of Escherichia coli O111:H−. Gene 129:87-92. [DOI] [PubMed] [Google Scholar]
- 22.Paton, A. W., L. Beutin, and J. C. Paton. 1995. Heterogeneity of the amino-acid sequences of Escherichia coli Shiga-like toxin type-I operons. Gene 153:71-74. [DOI] [PubMed] [Google Scholar]
- 23.Paton, A. W., A. J. Bourne, P. A. Manning, and J. C. Paton. 1995. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect. Immun. 63:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piérard, D., G. Muyldermas, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rüssmann, H., H. Schmidt, J. Heesemann, A. Caprioli, and H. Karch.1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338-343. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt, H., H. Rüssmann, A. Schwarzkopf, S. Aleksic, J. Heesemann, and H. Karch.1994. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Int. J. Med. Microbiol. Virol. Parasitol. Infect. Dis. 281:201-213. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, H., C. Geitz, P. I. Tarr, M. Frosch, and H. Karch. 1999. Non-O157 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J. Infect. Dis. 179:115-123. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scotland, S. M., H. R. Smith, and B. Rowe. 1985. Two distinct toxins active on Vero cells from Escherichia coli O157:H7. Lancet ii:885-886. [DOI] [PubMed] [Google Scholar]
- 31.Strockbine, N. A., M. P. Jackson, L. M. Sung, R. K. Holmes, and A. D. O′Brien. 1988. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J. Bacteriol. 170:1116-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Tarr, P. I., and M. A. Neill. 1996. The problem of non-O157 Shiga toxin (verocytotoxin)-producing Escherichia coli. J. Infect. Dis. 174:1136-1139. [DOI] [PubMed] [Google Scholar]
- 34.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]