Abstract
Between 9 October and 12 November 1996, an outbreak of bloody diarrhea occurred in the neonatal nursery ward of the Policlínico Neuquén, in Neuquén, a city in the southwestern region of Argentina. Seven patients of the intermediate care unit were affected. Isolates of Escherichia coli O18ac:H31 that were non-lactose and -sorbitol fermenting were recovered from outbreak cases. Although the strains were negative for a number of virulence factors typically found in diarrheagenic groups of E. coli, all isolates from the present neonatal outbreak possessed the enterohemolysin gene, ehl1. All isolates showed resistance to the antibiotics ampicillin and chloramphenicol. These isolates showed a low adherence property in HeLa cells without any recognizable pattern. In order to evaluate the outbreak dissemination in the neonatology ward, a prevalence study was conducted on 13 November. Stool specimens were obtained from 16 neonates hospitalized in the sector and from 33 medical staff members. E. coli isolates with identical biochemical characteristics of the outbreak strain were recovered from 11 of 16 inpatients and from 4 of 33 staff members during the prevalence study. A total of 15 E. coli strains recovered both from the outbreak and the prevalence study were processed by random amplified polymorphic DNA (RAPD)-PCR and pulsed-field gel electrophoresis (PFGE). By RAPD-PCR 14 of 15 strains showed patterns with 85 to 100% similarity, and by PFGE these strains were identical, each showing a unique pattern with 15 bands between 40 and 400 kb. One strain isolated from a nurse during the prevalence study presented a pattern not related to the others, and this was characterized as E. coli O81:HNM resistant to ampicillin only and negative for all the virulence factors studied. This outbreak occurred despite strict regulations in place to prevent cross-infection in the hospital. Postoutbreak prevalence studies were performed weekly thereafter without detecting new cases.
Infections in the community with respect to summer diarrhea and outbreaks of infantile gastroenteritis, particularly associated with hospitals and day nurseries, are more common in developing countries. Infants who have low birth weight or who are premature are highly susceptible to enteric infections, due in large part to poorly developed immune systems. Moreover, many supportive therapies frequently applied in a neonatal intensive care unit (NICU) may breach natural barriers to infection and may further complicate the ability of newborns to resist infection. Therefore, infants in NICUs frequently acquire nosocomial infections, while NICUs are the site of numerous outbreaks of infectious disease.
There are four distinct categories of Escherichia coli causing diarrheal disease: enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli, and enterohemorrhagic E. coli (EHEC). Epidemiologic studies have also implicated two additional categories, enteroaggregative and diffusely adherent E. coli. However, outbreaks of nosocomial diarrheal disease acquired by infants have traditionally been associated with EPEC, while ETEC predominates in the young in developing countries. Sporadic cases and outbreaks of food-borne disease often associated with hemorrhagic colitis and hemolytic-uremic syndrome are caused by EHEC strains that produce Shiga-like toxins and enterohemolysins (12, 16). Serotyping and molecular markers for E. coli have been used successfully in epidemiologic investigations associated with outbreaks of disease. Pulsed-field gel electrophoresis (PFGE) provides a useful molecular subtyping method which demonstrates that isolates of similar serotype from a common source can be characterized based on identical banding patterns. Epidemiologically, this is highly significant in terms of differentiating between outbreak-related strains and non-outbreak-related strains (2).
Most of the reported E. coli outbreaks in neonates have been associated with diarrhea or pyelonephritis (6). It is known that the prevalence of serogroups O1, O2, O4, O5, O18, O20, O25, and O75 in the human fecal flora is greater than that of other serogroups, and it is thought that this is in some way due to the capacity of some of these serogroups to persist in the bowel (19).
Frequently, cases in the community precede hospital outbreaks, and these usually result from the admission to the hospital of a baby with either established disease or a carrier state in which the pathogenic strain is excreted. An outbreak of bloody diarrhea due to E. coli O18ac:H31 occurred in the neonatal nursery ward of the Policlínico Neuquén. This outbreak was not associated with disease in the community and occurred in the hospital despite a strict program established for the prevention of nosocomial infections.
MATERIALS AND METHODS
Outbreak investigation.
Between 9 October and 12 November 1996, an outbreak of bloody diarrhea occurred in the neonatal nursery ward of the Policlínico Neuquén in Neuquén City, located 750 miles from Buenos Aires in the southwestern region of Argentina. The ward is divided into three sections: intensive care unit, intermediate care unit, and general care unit. Seven patients (cases A to G) of the intermediate care unit were affected. Three cases (cases A to C) occurred during the period of 15 October through 17 October, and four further cases (cases D to G) occurred between 6 November and 12 November.
The index case, a 5-day-old female patient, was admitted with a diagnosis of an abdominal tumor to the intermediate care unit on 7 October in order to undergo scheduled surgery. On 9 October the patient exhibited bloody diarrhea in the absence of other clinical symptoms, including fever, vomiting, abdominal pain, or dehydration. Bacteriologic culture of stool yielded polymicrobial flora with no recognized enteropathogens. The child was discharged on 11 October. Subsequently, five of six neonatal patients presenting with diarrhea showed abundant macroscopic blood in their stools (Table 1). These patients exhibited neither signs of inflammatory bowel disease nor general compromise. Another symptomatic patient (case F) with Down's syndrome died 3 days after birth due to necrotizing enterocolitis (NEC) grade III with intestinal perforation, peritonitis, and septicemia. Three of the six patients (cases A to C) involved in the outbreak were in the hospital at the same time as the index case.
TABLE 1.
Strains recovered during the outbreak and prevalence study periods
| Case identifier | Age (days)a | Weight (g)a | Gestational age (wk) | Duration of symptoms (days) | Diagnosis | Date of sample collection (mo/day/yr) | E. coli serotype |
|---|---|---|---|---|---|---|---|
| A | 33 | 1,920 | 31 | 5 | Bloody diarrhea | 10/15/96 | O18ac:H31 |
| B | 13 | 1,390 | 34 | 10 | Bloody diarrhea | 10/16/96 | O18ac:H31 |
| C | 36 | 1,580 | 31 | 5 | Bloody diarrhea | 10/17/96 | O18ac:H31 |
| D | 20 | 1,200 | 32 | 23 | Bloody diarrhea | 11/06/96 | O18ac:H31 |
| E | 10 | 2,460 | 35 | 1 | Diarrhea | 11/08/96 | O18ac:H31 |
| F | 2 | 2,500 | 38 | 3b | Bloody diarrhea/NEC | 11/11/96 | O18ac:H31 |
| Gc | 24 | 2,040 | 34 | 25 | Bloody diarrhea | 11/12/96 | O18ac:H31 |
| Hc | 25 | 2,020 | 34 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| I | 7 | 2,195 | 35 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| J | 7 | 1,980 | 33 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| K | 6 | 3,130 | 36 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| L | 20 | 2,940 | 36 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| M | 45 | 4,650 | 40 | Asymptomatic | 11/13/96 | O18ac:H31 | |
| N | Adult | Asymptomatic | 11/13/96 | O81:HNM | |||
| O | Adult | Asymptomatic | 11/13/96 | O18ac:H31 |
Age and weight at time of sample collection.
Patient died on day 3 after birth.
Patients G and H were twins.
A prevalence study was conducted on 13 November, in order to evaluate the outbreak dissemination in the neonatology ward. Stool specimens were all obtained on the same day from the remaining five symptomatic patients (cases A, B, C, D, and G). Stools were also received from the 11 neonates hospitalized in the sector for reasons other than enteric illness, such as low birth weight, premature status, and febrile syndrome, as well as from 33 staff members comprising 6 physicians, 21 nurses, and 6 auxiliary personnel.
Microbiological examination.
Stool specimens from all symptomatic and asymptomatic children and from all of the clinical staff were collected and processed at the hospital microbiology laboratory for E. coli, Salmonella, Shigella, Vibrio, Aeromonas, and Campylobacter species as well as for rotavirus using an enzyme-linked immunosorbent assay kit (Abbott Laboratories, Abbott Park, Ill.). For E. coli detection, each stool sample was plated on to MacConkey agar and sorbitol MacConkey agar (Difco Laboratories, Detroit, Mich.). From each case, five colonies, characterized as E. coli by standard biochemical tests (7), were selected, cultured on nutrient agar slants, and sent to the Instituto Nacional de Enfermedades Infecciosas reference laboratory for further investigations.
All isolates (seven from symptomatic patients, six from asymptomatic patients, and two from staff) were tested by agglutination assay using specific O antisera: O1, O6, O8, O15, O18, O20, O25, O26, O27, O28ac, O29, O44, O55, O63, O78, O86a, O111, O112ac, O114, O115, O119, O124, O125, O126, O127a, O128, O136, O142, O143, O144, O146, O148, O151, O152, O153, O157, O158, O159, O164, O166, O167, O168, and O169 (Denka Seiken, Co., Ltd., Tokyo, Japan). Complete serotyping was performed at the National Laboratory for Enteric Pathogens, National Microbiology Laboratory, Winnipeg, Canada. Antimicrobial susceptibility patterns were established according to the Kirby-Bauer method for ampicillin, carbenicillin, cefixime, cefotaxime, cefuroxime, cephalothin, colistin, chloramphenicol, gentamicin, nalidixic acid, norfloxacin, streptomycin, tetracycline, and trimethoprim-sulfamethoxazole, according to NCCLS standards and methods (12a).
Detection of virulence factors.
Isolates from seven neonates associated with the outbreak (cases A to G), together with those recovered during the prevalence study period, including six from asymptomatic infants not related to the outbreak (cases H to M) and two from staff members (cases N and O), were examined at the Instituto Nacional de Enfermedades Infecciosas and National Laboratory for Enteric Pathogens for the presence of established E. coli virulence factors.
Cytotoxic activity was assayed on Vero cells according to the method of Karmali et al. (10). Specific monoclonal antibodies (13C4 and BC5BB12) against Shiga toxins 1 and 2 (verotoxins 1 and 2), respectively, were used in neutralization testing and were kindly provided by Nancy Strockbine, Centers for Disease Control and Prevention, Atlanta, Ga. Diarrheagenic E. coli virulence factors, including bundle-forming pilus (bfp), the attaching-and-effacing gene (eaeA), Stx1 and Stx2, and DA fimbriae (daa) were analyzed by colony blot hybridization assay under stringent washing conditions with specific UTP-digoxigenin-labeled gene probes (kindly provided by J. Kaper and J. Nataro, Center for Vaccine Development, University of Maryland, School of Medicine, Baltimore, Md.) (20). Heat-stable and heat-labile toxin production of ETEC was analyzed by enzyme-linked immunosorbent assay (3).
The outer membrane protein of enteroinvasive E. coli was studied by enzyme-linked immunosorbent assay (4). A modified form of the HeLa cell adherence assay described by Scaletsky et al. (15) was performed to characterize E. coli according to the localized, diffuse, and aggregative patterns published by Cravioto et al. (5).
Enteroaggregative E. coli and EHEC-hlyA detection was performed by PCR as described by Schmidt et al. (17). Furthermore, all 15 isolates were analyzed by PCR for the presence of the enterohemolysin gene ehl1, which is genetically unrelated to EHEC-hlyA (21.8% sequence similarity). ehl1 is frequently expressed by E. coli strains that produce Shiga-like toxins and in toxin-negative EPEC strains (18). The oligonucleotide primers used in this study were forward (5′-CTCCTGATGTGCTTCTCGGTGTTT-3′) and reverse (5′-AGGCCATCAGCATTTCTTCT-3′). These amplified a 195-bp fragment from the ehl1 allele and were designed based on enterohemolysin sequence 1 (GenBank accession no. X70047). The PCR procedure and conditions used in this study were as previously described (16). All isolates were further examined for a variety of additional virulence factors, including pInv; pCDV432; CDT I, II, and III; CNF; ial; IpaH; K1; and adherence patterns.
RAPD-PCR.
Random amplified polymorphic DNA (RAPD)-PCR was carried out as described by Pacheco et al. (14) in 30-μl reaction volumes containing 3 μl of undiluted, boiled DNA containing 20 pmol of each primer (254, 5′-CCGCAGCCAA-3′; 256, 5′-AACGCGCAAC-3′; 257, 5′-GTGGATGCGA-3′; and 258, 5′-AGCCAGTTTC-3′), 2 U of Taq DNA polymerase (Gibco BRL, Life Technologies Ltd., Gaithersburg, Md.), and a 250 μM concentration of each deoxynucleoside triphosphate (Gibco BRL) in 10 mM Tris HCl (pH 8.4), 50 mM KCl, and 3 mM MgCl2. Amplification reactions were performed in a Biometra apparatus (Biometra GmbH, Göttingen, Germany). Reaction products were analyzed by electrophoresis on 1.2% agarose (Bio-Rad Laboratories, Hercules, Calif.).
PFGE.
PFGE was performed as described by Barrett et al. (2), with minor modifications. In brief, each bacterial cell suspension (optical density at 600 nm, 1.35) was embedded in low-melting-temperature agarose (Gibco BRL). The plugs were incubated at 50°C in lysis buffer overnight and sliced into aliquots 2 mm in thickness. The DNA-containing plugs were subjected to restriction endonuclease digestion with 20 U of XbaI (Promega Corp., Madison, Wis.) at 37°C for 18 h.
Restriction fragments were separated by electrophoresis through 1% PFGE agarose (Bio-Rad) in 0.5× Tris-borate-EDTA buffer at 14°C at 200 V in a CHEFF DR III apparatus (Bio-Rad). A linearly ramped switching time from 5 to 40 s was applied for 20 h. The gels were stained with ethidium bromide and photographed in a UV transilluminator. E. coli O18ac:H31 isolates recovered from a chicken and a calf, kindly provided by Flemming Scheutz, Statens Serum Institut, Copenhagen, Denmark, were included as controls for RAPD-PCR and PFGE.
Data analysis.
Following an analysis of the profiles derived from RAPD and PFGE, the relatedness among the patterns was estimated by the proportions of shared bands after applying Jaccard's similarity coefficient. Data recording and calculations were performed using the RAPDistance program, version 1.03 (1). The resulting matrix of pairwise distance was used to generate a phenogram based on the unweighted pair group method with arithmetic mean, included in the MEGA software (11).
RESULTS
All of the symptomatic patients (outbreak related) were premature, except the child with NEC (case F). The gestational age range was 31 to 38 weeks, and the mean weight was 1,870 g (range, 1,200 to 2,500 g). The symptoms of most patients decreased without treatment. The gestational age and mean weight in the asymptomatic children in the prevalence study were 34 to 40 weeks and 2,819 g (range, 1,980 to 4,650 g), respectively (Table 1). Although both values were higher than those observed in the symptomatic children, these differences were not statistically significant (P > 0.5 in the Kruskal-Wallis test).
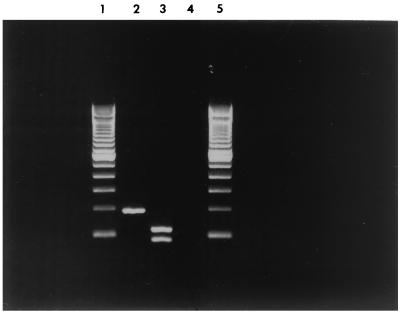
Non-lactose- and -sorbitol-fermenting E. coli O18ac:H31 isolates were recovered from outbreak cases. All the isolates from the outbreak were positive for ehl1. Restriction enzyme digestion using BclI confirmed that the expected restriction PCR fragments of 111 and 84 bp were observed (Fig. 1). In addition, isolates were resistant to ampicillin and cloramphenicol. These strains showed low adherence properties for HeLa cells, without any recognized pattern. During the prevalence study, E. coli isolates that did not ferment lactose or sorbitol were recovered from 11 of 16 (68.7%) inpatients. Five out of 11 isolates were derived from patients associated with the outbreak who were still hospitalized (cases A, B, C, D, and G), while the remaining six isolates were recovered from asymptomatic infants (cases H to M). E. coli isolates with the same biochemical characteristics were also recovered from 4 of 33 staff members examined, and all were asymptomatic. Of these, one was serotyped as an E. coli O18ac:H31 (case N) and a second was E. coli O81:HNM (case O). The remaining two isolates were not serotyped. Eight out of 15 isolates, derived from cases H to O, recovered during the prevalence study (six from inpatients not associated with the outbreak and two from staff) were fully characterized. All of the strains from the inpatients in the prevalence study and one of the two from the staff were identical to those isolated during the outbreak when serotype, antimicrobial susceptibility, and virulence factors were analyzed. The isolate from a nurse was serotyped as E. coli O81:HNM, was resistant to ampicillin, and was negative for all virulence factors studied, including ehl1.
FIG. 1.
PCR detection of E. coli enterohemolysin ehl1. Lanes 1 and 5, 100-bp ladder (Life Technologies, Rockville, Md.); lane 2, O18ac:H31 (case A); lane 3, BclI digestion of O18ac:H31 PCR amplification product; and lane 4, O81:HNM (case N).
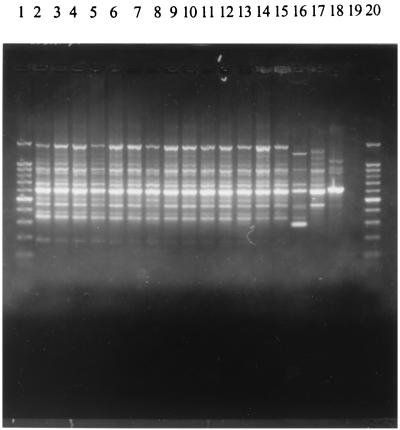
The results obtained from 15 strains (seven from the outbreak and eight from the prevalence study [Table 1]) processed by RAPD-PCR using different primers were analyzed. Patterns with 13 to 15 bands were obtained with 14 E. coli O18ac:H31 strains which had a high clonal relationship and showed 85 to 100% similarity using the primers 254, 256, 257, and 258. In contrast, the E. coli O81:HNM strain recovered from the nurse during the prevalence study was not related to the others and presented a 54 to 77% similarity. This strain showed a total of seven to nine amplified bands. The E. coli O18ac:H31 strains of animal origin used as out-of-group controls gave two RAPD profiles with less than 40% similarity with respect to the human outbreak strains. Patterns obtained with primer 258 are shown in Fig. 2.
FIG. 2.
RAPD profiles of E. coli strains recovered from the diarrhea outbreak and the prevalence study obtained with primer 258. Lanes 1 and 20, 100-bp DNA ladder (Promega); lanes 2 to 15, E. coli O18ac:H31 strains from the outbreak and prevalence study; lane 16, E. coli O81:HNM from the nurse; lane 17, a chicken isolate of E. coli O18ac:H31; lane 18, a calf isolate of E. coli O18ac:H31; and lane 19, no template. Chicken and calf isolates were unrelated to the outbreak.
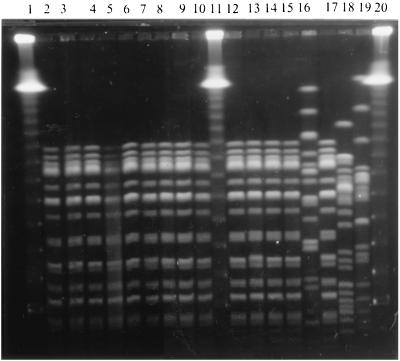
A unique profile distinguished by the presence of a total of 15 bands between 40 and 400 kb (Fig. 3) was obtained when the E. coli O18ac:H31 strains recovered from the outbreak and the prevalence study were analyzed by PFGE. The E. coli O81:HNM strain not associated with the outbreak and the E. coli O18ac:H31 strains used as a control showed unrelated patterns.
FIG. 3.
PFGE patterns of E. coli O18ac:H31 isolates recovered from the diarrhea outbreak and the prevalence study in a neonatal nursery ward. DNA from isolates were digested with the restriction enzyme XbaI. Lanes 1, 11 and 20, lambda molecular size marker ladder (Bio-Rad Laboratories); lanes 2 to 10, 12 to 15, and 17, E. coli O18ac:H31 isolates from the outbreak and prevalence study; lane 16, E. coli O81:HNM isolate from a nurse attending the ward; lane 18, a chicken isolate of E. coli O18ac:H31; and lane 19, a calf isolate of E. coli O18ac:H31. Chicken and calf isolates were unrelated to the outbreak.
DISCUSSION
E. coli has caused several documented outbreaks of gastroenteritis and pyelonephritis in NICUs (6, 9, 13). Up to 38% of newborns examined may carry such organisms in their stool. The origin in the majority of these is the maternal stool. Newborns are particularly vulnerable to acquiring a bacterial infection, and this risk is greater if they are premature. In this study, all 14 E. coli O18ac:H31 isolates were PCR positive for ehl1; however, only the premature infants (7 of 14) showed clinical symptoms. This further confirms the observation made previously (8) that premature infants are antibody deficient, with an immune system not mature enough to avoid infections. Among this group the incidence may be as high as 10 to 15%, depending upon gestational age. The mortality of neonatal bacterial sepsis is also inversely related to gestational age and may be as high as 75 to 90% among infants weighing less than 1,000 g.
In this study, the premature status of the infants may offer explanation for the outbreak of bloody diarrhea associated with an E. coli O18ac:H31 strain producing ehl1; however, it should be noted that there were premature infants in the prevalence study who excreted the organism without symptoms. One hypothesis could be that the association of low birth weight with premature status in infants constitutes a higher risk factor for the development of enteric illness. It has also been postulated that hand carriage by health care workers could be one mode of transmission of the bacteria, given that the patients usually have common caregivers and that their beds were close to each other (6).
Investigations using colony blot DNA hybridization showed that 26 of 357 (7%) of E. coli strains were ehl1 positive and that 50% of these belonged to E. coli serotype O26 (18). The isolates from the present outbreak were characterized as non-lactose and -sorbitol fermenting and were serotyped as E. coli O18ac:H31. RAPD-PCR and PFGE results established the clonal relationship among the isolates and suggested a nosocomial spread within the nursery. This report on a major outbreak of enteric disease in a children's ward in Argentina caused by E. coli O18ac:H31 is the first recorded outbreak associated with this serotype and possessing the present virulence factor.
Serogroup E. coli O18 with a capsular K1 antigen is usually associated with septicemia or meningitis. The strains in this outbreak were K1 negative; however, one case of septicemia was detected. The E. coli O18ac:H31 strains isolated were capable of rapid colonization; indeed, the patient with NEC who died on the 3rd day after birth was fecal culture positive. The organism spread rapidly throughout the hospital ward, and during the prevalence study performed a month after the index case, the same strain confirmed by clonal analysis was recovered from a majority of the inpatients in the ward. Most of the inpatients continued to excrete the organism at the time of discharge, having carried the pathogen for at least 18 days (mean = 28.3 days). This suggested a high capacity for the organism to persist in the bowel. After the infected patients were discharged from the hospital, the strain was not isolated from any newborn patients or staff members. This outbreak occurred in spite of strict rules for preventing nosocomial infections. Following this outbreak, cross-infection prevention protocols were improved and rigorously enforced. Since then, prevalence studies performed weekly, including culture of stools, have yielded no isolates and no new cases of disease due to this pathogen.
REFERENCES
- 1.Armstrong, J., A. Gibbs, R. Peakall, and G. Weiller. 1994. RAPDistance programs, version 1.04 for the analysis of patterns of RAPD fragments. Australian National University, Canberra, Australia.
- 2.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binsztein, N., M. J. Jouve, G. I. Viboud, L. L. Moral, M. Rivas, I. Ørskov, C. Åhrén, and A.-M. Svennerholm. 1991. Colonization factors of enterotoxigenic Escherichia coli isolated from children with diarrhea in Argentina. J. Clin. Microbiol. 29:1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinen, I., M. Rivas, M. I. Caffer, R. O. Cinto, and N. Binsztein. 1993. Diagnóstico de Escherichia coli enteroinvasiva asociada a diarrhea. Rev. Argent. Microbiol. 25:27-35. [PubMed] [Google Scholar]
- 5.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 6.Diekema, D. J., J. Barr, I. D. Boyken, B. J. Buschelman, R. N. Jones, M. A. Pfaller, and L. A. Herwaldt. 1997. A cluster of serious Escherichia coli infections in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 18:774-776. [DOI] [PubMed] [Google Scholar]
- 7.Farmer, J. J., and M. T. Kelly. Enterobacteriaceae, p. 360-383. In A. Balows, W. J. Hausler, Jr., K. L. Hermann, H. D. Isenberg, and H. J. Shadomy (ed.) Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 8.Ferrieri, P. 1991. Neonatal susceptibility and immunity to major bacterial pathogens. Rev. Infect. Dis. 12:S394-S400. [DOI] [PubMed] [Google Scholar]
- 9.Gransden, W. R., S. J. Eykyn, I. Phillips, and B. Rowe. 1990. Bacteremia due to Escherichia coli: a study of 861 episodes. Rev. Infect. Dis. 12:1008-1018. [DOI] [PubMed] [Google Scholar]
- 10.Karmali, M., M. Petric, C. Lim, R. Cheung, and G. S. Arbus. 1985. Sensitive method for detecting low numbers of verotoxin-producing Escherichia coli in mixed cultures by use of colony sweeps and polymyxin extraction of verotoxin. J. Clin. Microbiol. 22:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetics analysis, version 1.02. The Pennsylvania State University, University Park.
- 12.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.National Committee for Clinical Laboratory Standards. 1998. Performance standards for antimicrobial susceptibility testing; 8th informational supplement (1998). Document M100-S8, vol. 18, no. 1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Olesen, B., H. J. Kolmos, F. Ørskov, and I. Ørskov. 1995. A comparative study of nosocomial and community-acquired strains of Escherichia coli causing bacteremia in a Danish university hospital. J. Hosp. Infect. 31:295-304. [DOI] [PubMed] [Google Scholar]
- 14.Pacheco, A. B. F., B. E. C. Guth, D. F. de Almeida, and L. C. F. Ferreira. 1996. Characterization of enterotoxigenic Escherichia coli by random amplification of polymorphic DNA. Res. Microbiol. 147:175-182. [DOI] [PubMed] [Google Scholar]
- 15.Scaletsky, I. C. A., M. L. M. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H., C. Knop, S. Franke, S. Aleksic, J. Heesemann, and H. Karch. 1995. Development of PCR for screening of enteroaggregative Escherichia coli. J. Clin. Microbiol. 33:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroeher, U. H., L. Bode, L. Beutin, and P. A. Manning. 1993. Characterization and sequence of a 33-kDa enterohemolysin (Ehly 1)-associated protein in Escherichia coli. Gene 132:89-94. [DOI] [PubMed] [Google Scholar]
- 19.Sussman, M. 1985. Escherichia coli in human and animal disease, p. 7-45. In M. Sussman (ed.), The virulence of Escherichia coli: reviews and methods. Society for General Microbiology, Washington, D.C.
- 20.Thomas, A., H. R. Smith, G. A. Willshaw, and B. Rowe. 1991. Non-radioactively labelled polynucleotide and oligonucleotide DNA probes, for selectively detecting Escherichia coli strains producing Vero cytotoxins VT1, VT2 and VT2 variant. Mol. Cell. Probes 5:129-135. [DOI] [PubMed] [Google Scholar]